|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gülay H, Bora S, Kilicturgay S, Hamaloglu
E and Göksel HA: Management of nipple discharge. J Am Coll Surg.
178:471–474. 1994.PubMed/NCBI
|
|
3
|
Vargas HI, Vargas MP, Eldrageely K,
Gonzalez KD and Khalkhali I: Outcomes of clinical and surgical
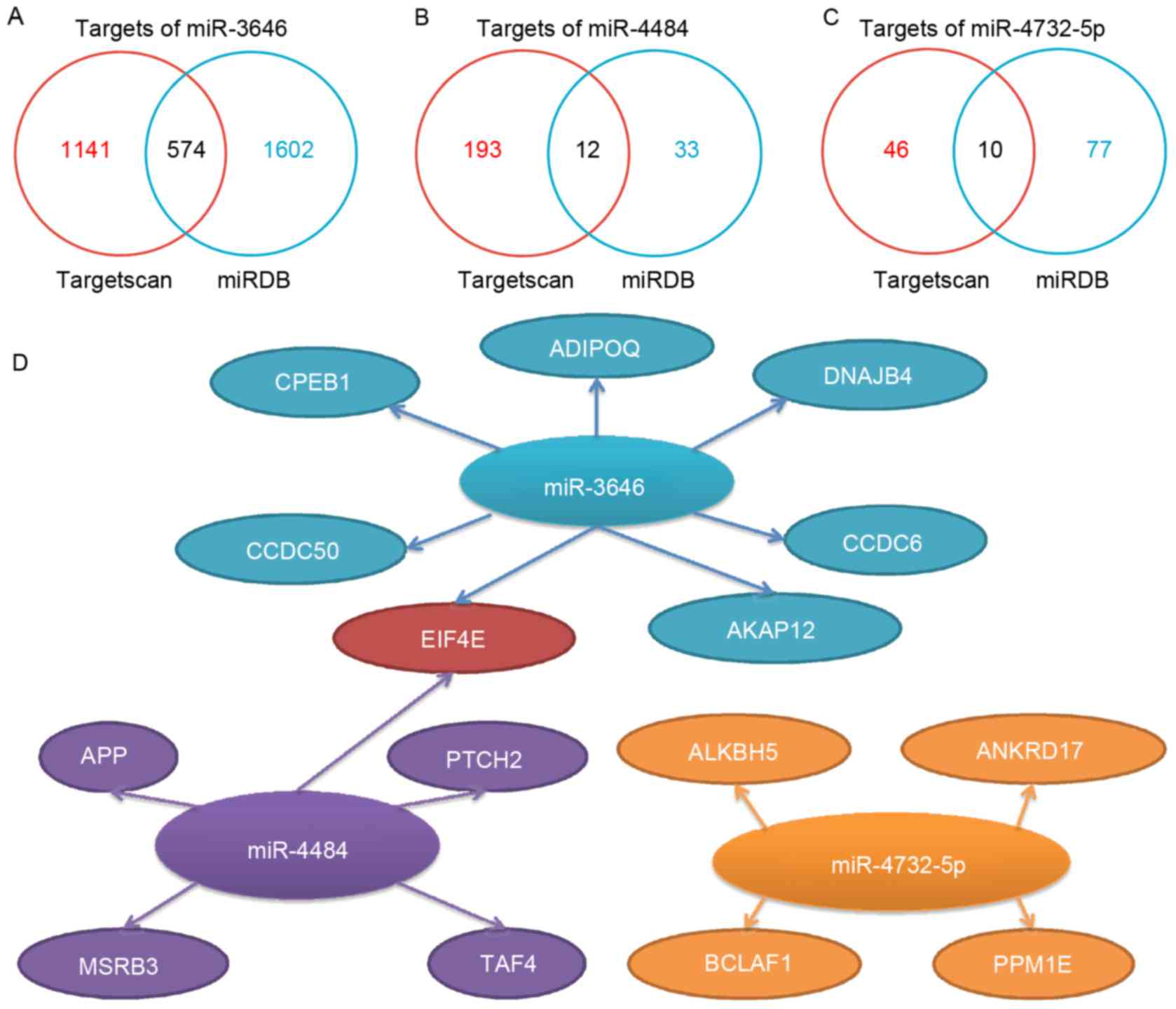
assessment of women with pathological nipple discharge. Am Surg.
72:124–128. 2006.PubMed/NCBI
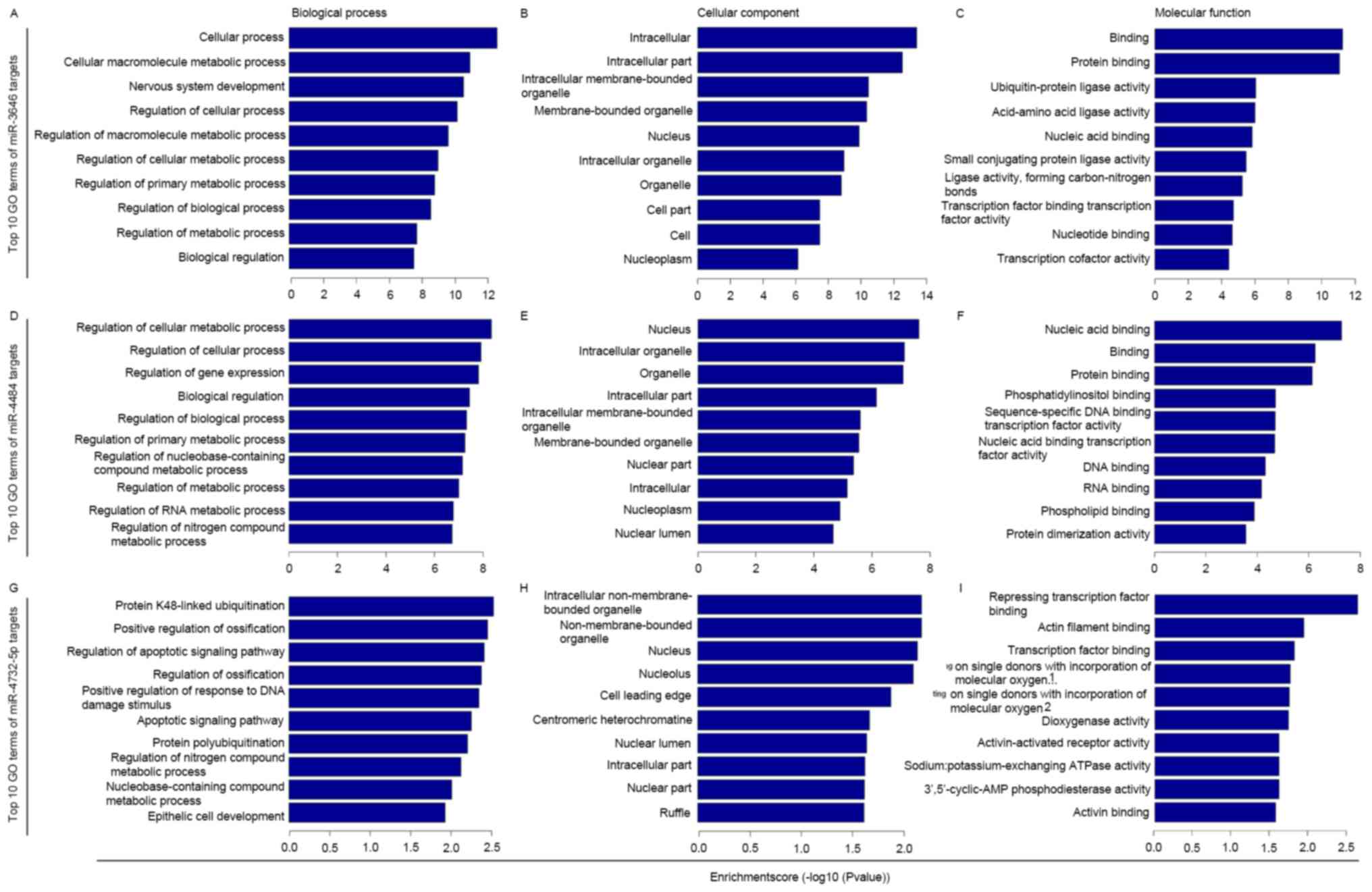
|
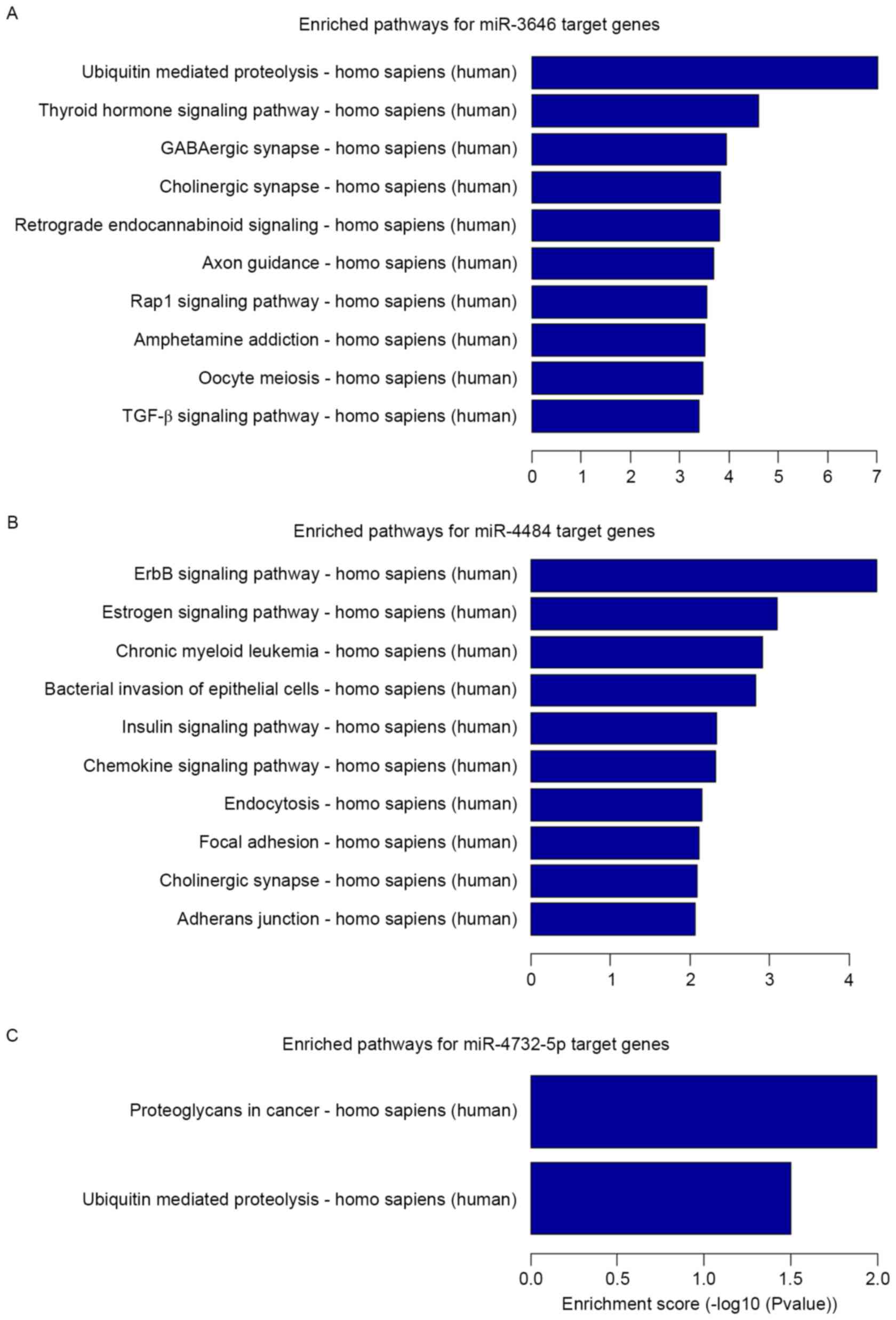
|
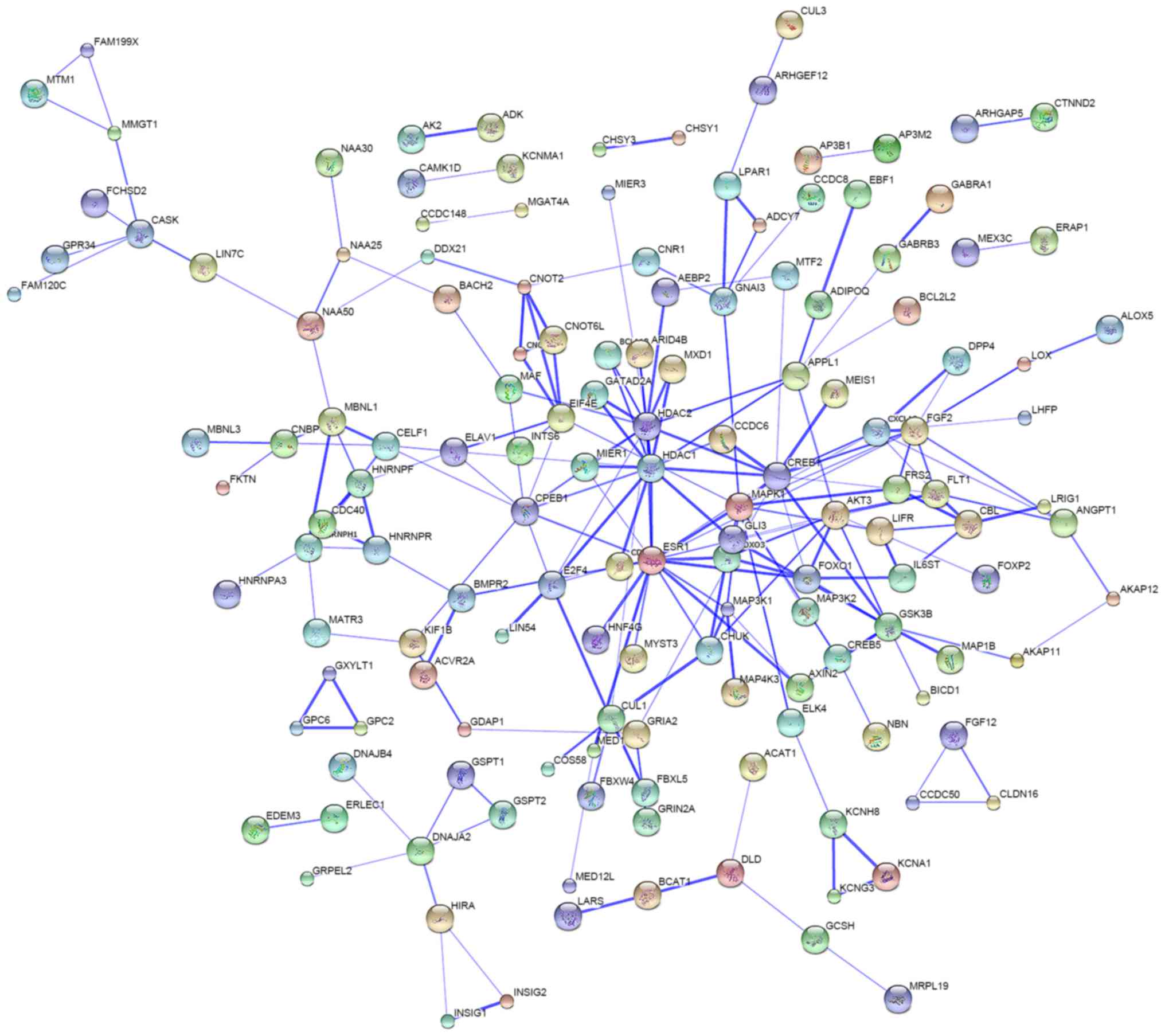
4
|
Jardines L: Management of nipple
discharge. Am Surg. 62:119–122. 1996.PubMed/NCBI
|
|
5
|
Murad TM, Contesso G and Mouriesse H:
Nipple discharge from the breast. Ann Surg. 195:259–264. 1982.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sabel MS, Helvie MA, Breslin T, Curry A,
Diehl KM, Cimmino VM, Chang AE and Newman LA: Is duct excision
still necessary for all cases of suspicious nipple discharge?
Breast J. 18:157–162. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kapenhas-Valdes E, Feldman SM and Boolbol
SK: The role of mammary ductoscopy in breast cancer: A review of
the literature. Ann Surg Oncol. 15:3350–3360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
López-Ríos F, Vargas-Castrillón J,
González-Palacios F and de Agustín PP: Breast carcinoma in situ in
a male. Report of a case diagnosed by nipple discharge cytology.
Acta cytol. 42:742–744. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsukamoto Y, Nakada C, Noguchi T, Tanigawa
M, Nguyen LT, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M and
Moriyama M: MicroRNA-375 is downregulated in gastric carcinomas and
regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer
Res. 70:2339–2349. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang YW, Shi DB, Chen X, Gao C and Gao P:
Clinicopathological significance of microRNA-214 in gastric cancer
and its effect on cell biological behaviour. PloS one.
9:e913072014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fabbri M: miRNAs as molecular biomarkers
of cancer. Expert Rev Mol Diagn. 10:435–444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang K, Zhang Y, Liu C, Xiong Y and Zhang
J: MicroRNAs in the diagnosis and prognosis of breast cancer and
their therapeutic potential (Review). Int J Oncol. 45:950–958.
2014.PubMed/NCBI
|
|
14
|
Zhong Z, Dong Z, Yang L, Chen X and Gong
Z: Inhibition of proliferation of human lung cancer cells by green
tea catechins is mediated by upregulation of let-7. Exp Ther Med.
4:267–272. 2012.PubMed/NCBI
|
|
15
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stahlhut C and Slack FJ: Combinatorial
ction of microRNAs let-7 and miR-34 effectively synergizes with
erlotinib to suppress non-small cell lung cancer cell
proliferation. Cell Cycle. 14:2171–2180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cui L, Zhang X, Ye G, Zheng T, Song H,
Deng H, Xiao B, Xia T, Yu X, Le Y and Guo J: Gastric juice
microRNAs as potential biomarkers for the screening of gastric
cancer. Cancer. 119:1618–1626. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang K, Zhao S, Wang Q, Yang HS, Zhu J
and Ma R: Identification of microRNAs in nipple discharge as
potential diagnostic biomarkers for breast cancer. Ann Surg Oncol.
22:(Suppl 3). S536–S544. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wong N and Wang X: miRDB: An online
resource for microRNA target prediction and functional annotations.
Nucleic Acids Res. 43:D146–D152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gene Ontology Consortium, Blake JA, Dolan
M, Drabkin H, Hill P, Li N, Sitnikov D, Bridges S, Burgess S, Buza
T, et al: Gene ontology annotations and resources. Nucleic Acids
Res. 41:D530–D535. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanehisa M, Goto S, Sato Y, Furumichi M
and Tanabe M: KEGG for integration and interpretation of
large-scale molecular data sets. Nucleic Acids Res. 40:D109–D114.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41:D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vargas HI, Romero L and Chlebowski RT:
Management of bloody nipple discharge. Curr Treat Options Oncol.
3:157–161. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dinkel HP, Trusen A, Gassel AM, Rominger
M, Lourens S, Müller T and Tschammler A: Predictive value of
galactographic patterns for benign and malignant neoplasms of the
breast in patients with nipple discharge. Br J Radiol. 73:706–714.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sauter ER, Ehya H, Babb J, Diamandis E,
Daly M, Klein-Szanto A, Sigurdson E, Hoffman J, Malick J and
Engstrom PF: Biological markers of risk in nipple aspirate fluid
are associated with residual cancer and tumour size. Br J Cancer.
81:1222–1227. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang K, Zhao S, Wang Q, Yang HS, Zhu J
and Ma R: Identification of microRNAs in nipple discharge as
potential diagnostic biomarkers for breast cancer. Ann Surg Oncol.
22:(Suppl 3). S536–S544. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Y, Zhou J, Zhang C, Fu W, Xiao X, Ruan
S, Zhang Y, Luo X and Tang M: HLJ1 is a novel biomarker for
colorectal carcinoma progression and overall patient survival. Int
J Clin Exp Pathol. 7:969–977. 2014.PubMed/NCBI
|
|
28
|
Simões-Correia J, Silva DI, Melo S,
Figueiredo J, Caldeira J, Pinto MT, Girão H, Pereira P and Seruca
R: DNAJB4 molecular chaperone distinguishes WT from mutant
E-cadherin, determining their fate in vitro and in vivo. Hum Mol
Genet. 23:2094–2105. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen HW, Lee JY, Huang JY, Wang CC, Chen
WJ, Su SF, Huang CW, Ho CC, Chen JJ, Tsai MF, et al: Curcumin
inhibits lung cancer cell invasion and metastasis through the tumor
suppressor HLJ1. Cancer Res. 68:7428–7438. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tsai MF, Wang CC, Chang GC, Chen CY, Chen
HY, Cheng CL, Yang YP, Wu CY, Shih FY, Liu CC, et al: A new tumor
suppressor DnaJ-like heat shock protein, HLJ1 and survival of
patients with non-small-cell lung carcinoma. J Natl Cancer Inst.
98:825–838. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mantzoros C, Petridou E, Dessypris N,
Chavelas C, Dalamaga M, Alexe DM, Papadiamantis Y, Markopoulos C,
Spanos E, Chrousos G and Trichopoulos D: Adiponectin and breast
cancer risk. J Clin Endocrinol Metab. 89:1102–1107. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Libby Falk E, Liu J, Li YI, Lewis MJ,
Demark-Wahnefried W and Hurst DR: Globular adiponectin enhances
invasion in human breast cancer cells. Oncol Lett. 11:633–641.
2016.PubMed/NCBI
|
|
33
|
Chen L, Ye C, Huang Z, Li X, Yao G, Liu M,
Hu X, Dong J and Guo Z: Differentially expressed genes and
potential signaling pathway in Asian people with breast cancer by
preliminary analysis of a large sample of the microarray data. Nan
Fang Yi Ke Da Xue Xue Bao. 34:807–812. 2014.(In Chinese).
PubMed/NCBI
|
|
34
|
Nairismägi ML, Vislovukh A, Meng Q,
Kratassiouk G, Beldiman C, Petretich M, Groisman R, Füchtbauer EM,
Harel-Bellan A and Groisman I: Translational control of TWIST1
expression in MCF-10A cell lines recapitulating breast cancer
progression. Oncogene. 31:4960–4966. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hansen CN, Ketabi Z, Rosenstierne MW,
Palle C, Boesen HC and Norrild B: Expression of CPEB, GAPDH and
U6snRNA in cervical and ovarian tissue during cancer development.
APMIS. 117:53–59. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Caldeira J, Simões-Correia J, Paredes J,
Pinto MT, Sousa S, Corso G, Marrelli D, Roviello F, Pereira PS,
Weil D, et al: CPEB1, a novel gene silenced in gastric cancer: A
Drosophila approach. Gut. 61:1115–1123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xiaoping L, Zhibin Y, Wenjuan L, Zeyou W,
Gang X, Zhaohui L, Ying Z, Minghua W and Guiyuan L: CPEB1, a
histone-modified hypomethylated gene, is regulated by miR-101 and
involved in cell senescence in glioma. Cell Death Dis. 4:e6752013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kochanek DM and Wells DG: CPEB1 regulates
the expression of MTDH/AEG-1 and glioblastoma cell migration. Mol
Cancer Res. 11:149–160. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Miyazaki T, Ikeda K, Horie-Inoue K and
Inoue S: Amyloid precursor protein regulates migration and
metalloproteinase gene expression in prostate cancer cells. Biochem
Biophys Res Commun. 452:828–833. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yoshitomi T, Kawakami K, Enokida H,
Chiyomaru T, Kagara I, Tatarano S, Yoshino H, Arimura H, Nishiyama
K, Seki N and Nakagawa M: Restoration of miR-517a expression
induces cell apoptosis in bladder cancer cell lines. Oncol Rep.
25:1661–1668. 2011.PubMed/NCBI
|
|
41
|
Chen Y, Wang Y, Song H, Wang J, Yang H,
Xia Y, Xue J, Li S, Chen M and Lu Y: Expression profile of
apoptosis-related genes potentially explains early recurrence after
definitive chemoradiation in esophageal squamous cell carcinoma.
Tumour Biol. 35:4339–4346. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Carroll M and Borden KL: The oncogene
eIF4E: using biochemical insights to target cancer. J Interferon
Cytokine Res. 33:227–238. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pettersson F, Del Rincon SV, Emond A, Huor
B, Ngan E, Ng J, Dobocan MC, Siegel PM and Miller WH Jr: Genetic
and Pharmacologic inhibition of eIF4E reduces breast cancer cell
migration, invasion and metastasis. Cancer Res. 75:1102–1112. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hu A, Sun M, Yan D and Chen K: Clinical
significance of mTOR and eIF4E expression in invasive ductal
carcinoma. Tumori. 100:541–546. 2014.PubMed/NCBI
|
|
45
|
Yin X, Kim RH, Sun G, Miller JK and Li BD:
Overexpression of eukaryotic initiation factor 4E is correlated
with increased risk for systemic dissemination in node-positive
breast cancer patients. J Am Coll Surg. 218:663–671. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Spanjaard E, Smal I, Angelopoulos N,
Verlaan I, Matov A, Meijering E, Wessels L, Bos H and de Rooij J:
Quantitative imaging of focal adhesion dynamics and their
regulation by HGF and Rap1 signaling. Exp Cell Res. 330:382–397.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Che YL, Luo SJ, Li G, Cheng M, Gao YM, Li
XM, Dai JM, He H, Wang J, Peng HJ, et al: The C3G/Rap1 pathway
promotes secretion of MMP-2 and MMP-9 and is involved in serous
ovarian cancer metastasis. Cancer lett. 359:241–249. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Alemayehu M, Dragan M, Pape C, Siddiqui I,
Sacks DB, Di Guglielmo GM, Babwah AV and Bhattacharya M:
β-Arrestin2 regulates lysophosphatidic acid-induced uman breast
tumor cell migration and invasion via Rap1 and IQGAP1. PloS one.
8:e561742013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ahmed SM, Thériault BL, Uppalapati M, Chiu
CW, Gallie BL, Sidhu SS and Angers S: KIF14 negatively regulates
Rap1a-Radil signaling during breast cancer progression. J Cell
Biol. 199:951–967. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
McSherry EA, Brennan K, Hudson L, Hill AD
and Hopkins AM: Breast cancer cell migration is regulated through
junctional adhesion molecule-A-mediated activation of Rap1 GTPase.
Breast Cancer Res. 13:R312011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Itoh M, Nelson CM, Myers CA and Bissell
MJ: Rap1 integrates tissue polarity, lumen formation, and
tumorigenic potential in human breast epithelial cells. Cancer Res.
67:4759–4766. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhao B and Chen YG: Regulation of TGF-β
Signal Transduction. Scientifica (Cairo).
2014:8740652014.PubMed/NCBI
|
|
53
|
Zarzynska JM: Two faces of TGF-beta1 in
breast cancer. Mediators Inflamm. 2014:1417472014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kotiyal S and Bhattacharya S: Breast
cancer stem cells, EMT and therapeutic targets. Biochem Biophys Res
Commun. 453:112–116. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Imamura T, Hikita A and Inoue Y: The roles
of TGF-β signaling in carcinogenesis and breast cancer metastasis.
Breast Cancer. 19:118–124. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Mehta A and Tripathy D: Co-targeting
estrogen receptor and HER2 pathways in breast cancer. Breast.
23:2–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Rogler A, Hoja S, Socher E, Nolte E, Wach
S, Wieland W, Hofstädter F, Goebell PJ, Wullich B, Hartmann A and
Stoehr R: Role of two single nucleotide polymorphisms in secreted
frizzled related protein 1 and bladder cancer risk. Int J Clin Exp
Pathol. 6:1984–1998. 2013.PubMed/NCBI
|
|
58
|
Chen J, Yao D, Li Y, Chen H, He C, Ding N,
Lu Y, Ou T, Zhao S, Li L and Long F: Serum microRNA expression
levels can predict lymph node metastasis in patients with
early-stage cervical squamous cell carcinoma. Int J Mol Med.
32:557–567. 2013.PubMed/NCBI
|
|
59
|
Pouladi N, Kouhsari SM, Feizi MH, Gavgani
RR and Azarfam P: Overlapping region of p53/wrap53 transcripts:
Mutational analysis and sequence similarity with microRNA-4732-5p.
Asian Pac J Cancer Prev. 14:3503–3507. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Omura T, Shimada Y, Nagata T, Okumura T,
Fukuoka J, Yamagishi F, Tajika S, Nakajima S, Kawabe A and Tsukada
K: Relapse-associated microRNA in gastric cancer patients after S-1
adjuvant chemotherapy. Oncol Rep. 31:613–618. 2014.PubMed/NCBI
|