Introduction
Colorectal cancer (CRC) is one of the most common
malignancies worldwide, accounting for an estimated 1.3 million new
cases and >500,000 mortalities/year (1). CRC is the fourth leading cause of
cancer-associated mortality worldwide, with a rapidly increasing
incidence rate in the worldwide (2,3). In the
Asian population, the annual prevalence rate of CRC has steadily
increased over the past two decades (4). The Ministry of Health and Medical
Education in Iran states that cancer is the third most common cause
of mortality in Iran (5). Currently,
the gold standard method for CRC detection is internal imaging of
the colon with colonoscopy followed by a biopsy examination;
however, it remains an invasive procedure with possible serious
complications and limitations (6).
Cancer is a multistep process resulting from a gradual accumulation
of genetic and epigenetic changes to the genome (7). The methylation of 5′-C-phosphate-G-3′
(CpG) islands has been suggested to be associated with gene
silencing, serving an important role in cancer development
(8,9).
The aberrant promoter methylation of genes is a primary occurrence
in the process of CRC carcinogenesis, which can be considered as a
candidate diagnostic biomarker for CRC (10). A direct association between promoter
methylation and cancer development has caused a growing interest in
the use of plasma methylation as a diagnostic biomarker for
patients with cancer (11). There are
numerous reports of methylation biomarkers tested in fecal and
plasma samples (12–15). Previous epigenomic studies have
revealed that a number of genes, including spastic paraplegia 20
(SPG20), can potentially be silenced in patients with CRC due to
DNA hypermethylation within promoter regions (16–19). This
has prompted the opportunity to implement a reliable, inexpensive
and simple approach for CRC detection (16,17). The
SPG20 gene is located in chromosome band 13q13.3; the SPG20 gene
encodes the spartin protein, which is a multifunctional protein
that has previously been identified to be involved in intracellular
epidermal growth factor receptor trafficking (20), and in lipid droplet turnover (21). It has also been reported to be an
inhibitor of bone morphogenetic protein (BMP) signaling (22) and an adaptor for E3 ubiquitin ligases
(23). The spartin protein contains a
Microtubule-Interacting and Trafficking molecule (MIT) domain in
the N-terminus (24). It has also
been demonstrated that there is a function for the spartin protein
in cytokinesis (18). In a study
performed by Connell et al (25), it was observed that the knockdown of
SPG20 gene expression results in a cytokinesis arrest; however, the
number of cells arrested in cytokinesis was decreased when the
spartin protein was re-expressed. In a gene expression study using
microarray analysis, when colon cancer cell lines were treated with
5-aza-2′deoxycytidine, it was demonstrated that SPG20 could be
considered as a potential epigenetic biomarker for the diagnosis of
CRC (18). As compared with mRNA
expression, methylation studies on a variety of cancer cell lines
have demonstrated that aberrant methylation of the SPG20 promoter
is not only associated with gene silencing but also can
subsequently cause a cytokinesis arrest and aneuploidy, a situation
thought to be correlated with tumorigenesis (26). Early diagnosis of CRC can reduce
mortality and improve the overall survival rate of patients with
CRC. However, currently available diagnostic methods are limited by
their invasiveness, or suboptimal sensitivities and specificities.
Although colonoscopy is currently applied as a routine diagnostic
method for CRC, its application is limited by low acceptability in
a screening setting (6). As an
available non-invasive method, fecal occult blood testing (FOBT)
demonstrates no or low impact on CRC incidence because of its low
sensitivity (27).
Currently, it is generally accepted that methylation
markers may be suggested as reliable markers to effectively
identify patients with CRC at earlier stages. Aberrant promoter
methylation of genes is a common epigenetic alteration found in
CRC, which can be detected in blood or stool samples (28). However, the quantification of SPG20
promoter-methylated DNA in plasma samples has not yet been
reported. In the present study, the presence of SPG20 gene
promoter-methylated DNA was measured in plasma and tissue samples
from patients with CRC. The aim of the current study was to
determine whether the quantity of circulating methylated DNA of the
SPG20 gene could discriminate between patients with CRC and healthy
individuals. In addition, the level of SPG20-methylated DNA in
plasma samples was compared with a routine blood-based tumor
marker, carcinoembryonic antigen (CEA), for the first time.
Materials and methods
Tissue and plasma samples
A total of 37 patients with CRC and 37 healthy
individuals were included in the present study. The age range and
gender distribution is summarized in Table I. However, it was not possible to
collect tissue samples from 5/37 patients with CRC. A total of 32
paired tumor and adjacent healthy tissues were collected from the
freshly resected colon tissue of patients with CRC who underwent an
elective colon cancer surgery. Histological grading system was used
for grades of malignancy of CRC based on the highest poorly
differentiated clusters (29).
 | Table I.Demographic features of patients
group with colorectal cancer and control group. |
Table I.
Demographic features of patients
group with colorectal cancer and control group.
|
| Demographic
features |
|---|
|
|
|
|---|
|
| Age (years) | Gender |
|
|---|
|
|
|
|
|
|---|
| Groups | Mean ± standard
deviation | P-value | Male | Female | P-value |
|---|
| Patient group
(n=37) | 55.97±12.63 | 0.992a | 15 (40.5%) | 22 (59.5%) | 0.594b |
| Healthy control
group (n=37) | 55.94±11.84 | 15 (40.5%) | 22 (59.5%) |
The present study was reviewed and approved by the
Medical Faculty Ethics Committee of the University of Hamadan
(reference no. 3482, 2014; Hamadan, Iran). All samples were
collected from Bistoon Hospital (Kermanshah, Iran) from March to
November 2016. Written informed consent was obtained from all
participants. No patients had received chemotherapy or radiotherapy
prior to the surgical procedure. Healthy subjects without any
evidence of CRC following a colonoscopy were included as a control
group. As illustrated in Table I, all
patients and healthy individuals were matched for demographic
variables.
Small tissue samples were immediately stored in
liquid nitrogen for subsequent methylation assays, while other
samples were fixed in formalin for the routine histological
examination. Blood samples were obtained from 37 patients with CRC
and 37 healthy individuals prior to surgery. For each subject, 5 ml
of blood was collected in an EDTA vacutainer tube. Plasma was
isolated using centrifugation at 2,500 × g and 4°C for 15
min. All samples were then transferred into polypropylene tubes and
stored at −80°C for use in subsequent experiments.
DNA extraction from tissue and plasma
samples
Total genomic DNA was purified from 200 µl plasma
and 20 mg tissue samples. The samples were dissolved in lysis
buffer (Qiagen GmbH, Hilden, Germany) containing protease, followed
by DNA extraction using a QIAamp DNA Blood Mini kit (Qiagen GmbH,
Hilden, Germany; cat. no. 51104) according to the manufacturer's
instructions. DNA concentration and purity were determined using a
NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Inc.,
Wilmington, DE, USA). Purified DNA was eluted in buffer AE (Qiagen
GmbH) and stored at −20°C for use in subsequent experiments.
Bisulfite modification
DNA treatment with bisulfite leads to the
deamination of unmethylated cytosines, while leaving methylated
cytosines unchanged. The resulting sequence differences from the
conversion of deaminated cytosine into uracil and subsequently into
thymine make it possible to analyze the methylation patterns of a
DNA sequence (30,31). A successful bisulfite alteration
depends on essential factors such as similar bisulfite
concentration, temperature, clean and entirely denatured DNA,
freshly provided bisulfite and a valid pH status (32). In the present study, DNA extracted
from tissue and plasma samples was modified with sodium bisulfite
treatment using an EpiTect Fast DNA Bisulfite kit (Qiagen GmbH;
cat. no. 59826) according to the manufacturer's instructions. The
reaction consisted of bisulfite mix and DNA protection buffer from
the kit, in addition to 500 ng template and distilled water to make
a total volume of 140 µl. The bisulfite treatment was performed
using an Applied Biosystem VeritiThermal Cycler (Applied
Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with
the following thermocycling conditions: Initial denaturation step
at 95°C for 5 min; incubation 60°C for 15 min; second denaturation
step at 95°C for 5 min; incubation 60°C for 15 min; and hold at
20°C for 10 min. The procedure included the following steps:
Preparation of DNA from the sample; bisulfite-mediated conversion
of unmethylated cytosines (bisulfite thermocycling program);
binding of the converted single-stranded DNA to the membrane of a
MinElute DNA spin column; washing with wash buffer (Qiagen GmbH);
sulfonation of membrane-bound DNA; washing with desulfonation
buffer (Qiagen GmbH) of the membrane-bound DNA to remove the
sulfonation agent; and elution of the pure converted DNA from the
spin column. A poly-A carrier RNA was added to improve binding of
small quantities of DNA to the spin column membrane. Following
bisulfite treatment, the quality and quantity of DNA were evaluated
by the aforementioned protocol. Purified DNA was eluted in buffer
AE and stored at −20°C for use in subsequent experiments.
MethyLight method
In the present study, the MethyLight assay was
carried out using two primers and a TaqMan probe, which were
specifically annealed to the fully methylated DNA at gene of
interest and one assay was carried out using two primers and a
TaqMan probe, which were specifically annealed to ALU-C4, a
consensus DNA sequence as reference gene for normalization. The
MethyLight method can be used in DNA methylation analysis, to
detect methylated DNA sequences in presence of the unmethylated DNA
(33,34).
Primer and probe design
The primers and probes were designed using Beacon
Designer™ (version 8.13; www.premierbiosoft.com/molecular_beacons; Premier
Biosoft International, Palo Alto, CA, USA). The primers and probes
were designed to specifically amplify fully methylated
bisulfite-converted DNA, a region within the promoter CpG island in
the gene of interest, at the vicinity of the annotated
transcription start site provided by the University of California
Santa Cruz Genome Browser (genome.ucsc.edu) and the Eukaryotic Promoter Database
(EPD; epd.vital-it.ch/human/human_database.php). EPD is an
annotated non-redundant collection of eukaryotic POL II promoters
for which the transcription start site has been determined
experimentally. The MethyLight assay exhibited an amplicon of 94 bp
(deposited at GenBank; accession no. NM 015087). The software
designed primers with a melting temperature (Tm) of
61.8–62.8°C and probes with a Tm of 71.4°C for SPG20
gene promoter sequence. In the MethyLight assay, ALU-C4, a
consensus DNA sequence, was used as a reference gene and an
Alu-based MethyLight control reaction was designed to normalize
input DNA. The primers and probes used in the present study were
according to those previously reported for ALU-C4 sequences as a
reference gene (illustrated in Table
II). The primers and probes to the ALU-C4 reference gene were
used to bind a bisulfite-converted DNA region of the sequence
independently of its methylation status and lack of any CpG in this
region; the amplicon size was 98 bp (35). The ALU-C4 reaction has previously been
demonstrated to be less susceptible to normalization errors caused
by cancer-associated aneuploidy and copy number changes, compared
with the single-copy genes that previously applied in another study
(34,36). All primers and probes sequences used
in the present study are illustrated in Table II. All primers and probes were
purchased from and underwent high performance liquid chromatography
purification at Metabion (Steinkirchen, Germany).
 | Table II.Primer and probe sequences used for
MethyLight polymerase chain reaction. |
Table II.
Primer and probe sequences used for
MethyLight polymerase chain reaction.
| Primers and
probes | Sequence
(5′-3′) |
|---|
| SPG20 |
|
| Forward
primer |
5′-GGTGATGAATTCGTTGGT |
|
| AAGACGC-3′ |
| Reverse
primer |
5′-ACCTCGCTCCCGCCACAAAA-3′ |
|
Probe | 6FAM
5′-CAACACGCCGCCGCCGCAACCTAA-3′TAMRA |
| ALU-C4 |
|
| Forward
primer |
5′-GGTTAGGTATAGTGGTTTATATTTGTAATTTTAGTA-3′ |
| Reverse
primer |
5′-ATTAACTAAACTAATCTTAAACTCCTAACCTCA-3′ |
|
Probe | 6FAM
5′-CCTACCTTAACCTCCC-3′TAMRA |
MethyLight quantitative polymerase
chain reaction (qPCR)
The qPCR reactions were performed in 96-well plates
on a 7500 Real-Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.), using the EpiTect® MethyLight PCR +
ROX Vial kit (Qiagen GmbH; cat. no. 59496) according to the
manufacturer's protocol. The reactions were performed in a volume
of 20 µl that included: 2X EpiTect MethyLight Master mix (without
ROX) 10 µl containing (HotStarTaq Plus DNA Polymerase, EpiTect
Probe PCR buffer, deoxyadenosine triphosphate, deoxycytidine
triphosphate, deoxyguanosine triphosphate and thymidine
triphosphate of ultrapure quality); 2 µl 10X primer-probe mix
containing 200 nM probe and 400 nM forward and reverse primers; 0.4
µl 50X ROX Dye Solution (passive reference dye for normalization of
fluorescent signals); 2 µl (20 ng) bisulfite-treated DNA template;
and 5.6 µl RNase-free water. Cycle conditions were as follows:
Initial PCR activation step at 95°C for 5 min, 50 cycles at 95°C
for 15 sec (denaturation step) and 60°C for 1 min (combined
annealing/extension step with fluorescence data collection). Each
plate included standard curves for the SPG20 and ALU-C4 sequence,
DNA samples, a methylated human bisulfite-converted DNA (EpiTect
PCR Control DNA; cat. no. 59655; Qiagen GmbH) as a positive
control, unmethylated human bisulfite-converted DNA (EpiTect PCR
Control DNA; cat. no. 59665; Qiagen GmbH) as a negative control,
unconverted human genomic DNA from healthy human blood samples,
which was not amplified, and a no template control, including the
entire ingredients of the reaction except the template DNA. The
standard curve was plotted using the Cq value (13,14,18,19,34,37)
of the methylated human bisulfite-converted DNA at various
concentrations vs. the log value of input DNA concentration. An
acceptable MethyLight PCR result was obtained, and is demonstrated
by the PCR efficiency (96–102%), correct slope value (−3.65 - −3.2)
and R2=0.9985. In addition, to establish the standard
curve, commercial methylated human bisulfite-converted DNA (EpiTect
PCR Control DNA; cat. no. 59655; Qiagen GmbH) was used as a fully
methylated control to calculate the percentage of methylated
reference (PMR) of the samples. PMR is the degree of methylation of
each sample relative to the fully methylated control.
Chemiluminescence immunoassay
All of the plasma samples were measured to assess
the CEA concentration using a Liaison® chemiluminescence
analyzer with supporting reagents (Diasorin, Saluggia, Italy). The
quantitative determination of CEA in plasma samples was performed
using the Liaison CEA kit (cat. no. 314311; Diasorin) according to
the manufacturer's protocol.
Statistical analysis
The qPCR MethyLight PCR data were analyzed using
software from ABI 7500 SDS version 1.3.1. The level of methylated
DNA (the PMR) was calculated for all samples using the following
formula: [(SPG20/ALU)sample/(SPG20/ALU)positive
control]x100 (20,35,38,39). A
specific threshold was applied to classify the samples as positive
(threshold ≤ PMR) or negative (threshold > PMR) methylation
status. The optimal cut off point was the highest PMR value
obtained from healthy samples (17).
Statistical data analysis was carried
out using SPSS software (version 16.0; SPSS, Inc., Chicago, IL,
USA)
Fisher's exact test and Pearson's χ2 test
were used to measure the association between clinicopathological
characteristics and classified variables. Where appropriate,
Mann-Whitney U test, independent samples t-test and one-way (by
Tukey test) analysis of variance were used to compare categorical
and continuous variables. P<0.05 was considered to indicate a
statistically significant difference. A Receiver Operating
Characteristics (ROC) curve was generated using PMR values. The
percentile of the highest PMR value across all adjacent healthy
tissues or healthy plasma samples were used as a cut-off point of
PMR in tissue and plasma samples, respectively.
Results
DNA methylation of plasma samples
To identify potential DNA methylation biomarkers
that are appropriate for the detection of CRC, SPG20
promoter-methylated DNA was investigated in plasma and tissue
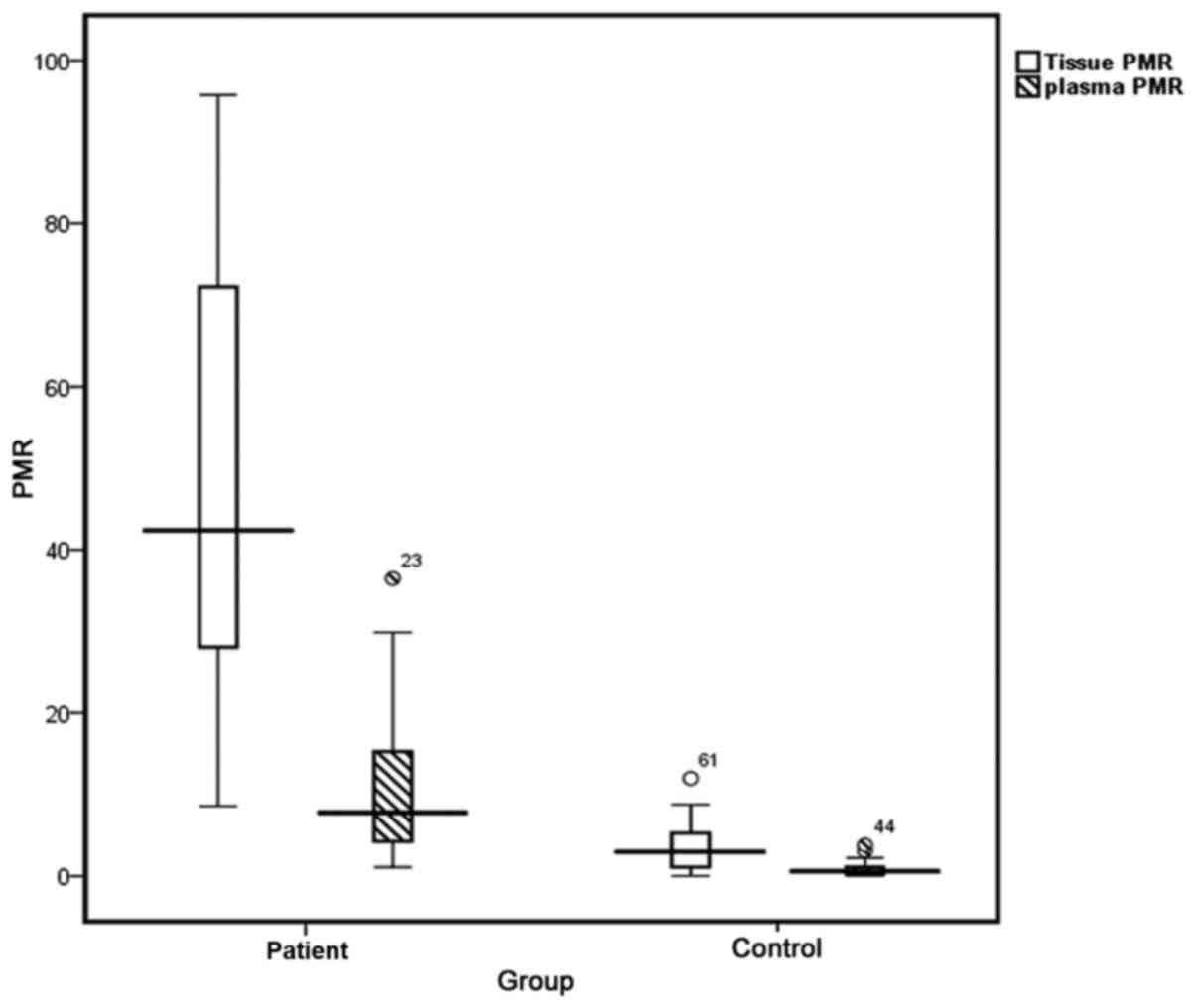
samples using the MethyLight method. As illustrated in Table III, plasma specimens from patients
with CRC and the healthy control group exhibited median PMR values
of 7.7 (95% CI, 4.15–15.28) and 0.59 (95% CI, 0.14–1.12),
respectively. The highest PMR value obtained from the healthy
plasma samples was considered as the threshold of methylation
status; samples with a PMR >3.52 were considered to be positive.
Based on this threshold, 30/37 (81.1%) patient's plasma samples
exhibited a positive methylation status. The PMR values were
significantly higher in plasma samples from patients with CRC, as
compared with those from control subjects (P<0.05). No
significant differences were identified in the frequency of SPG20
hypermethylation status (positive or negative) in plasma samples
from patients with CRC according to tumor type, size and location,
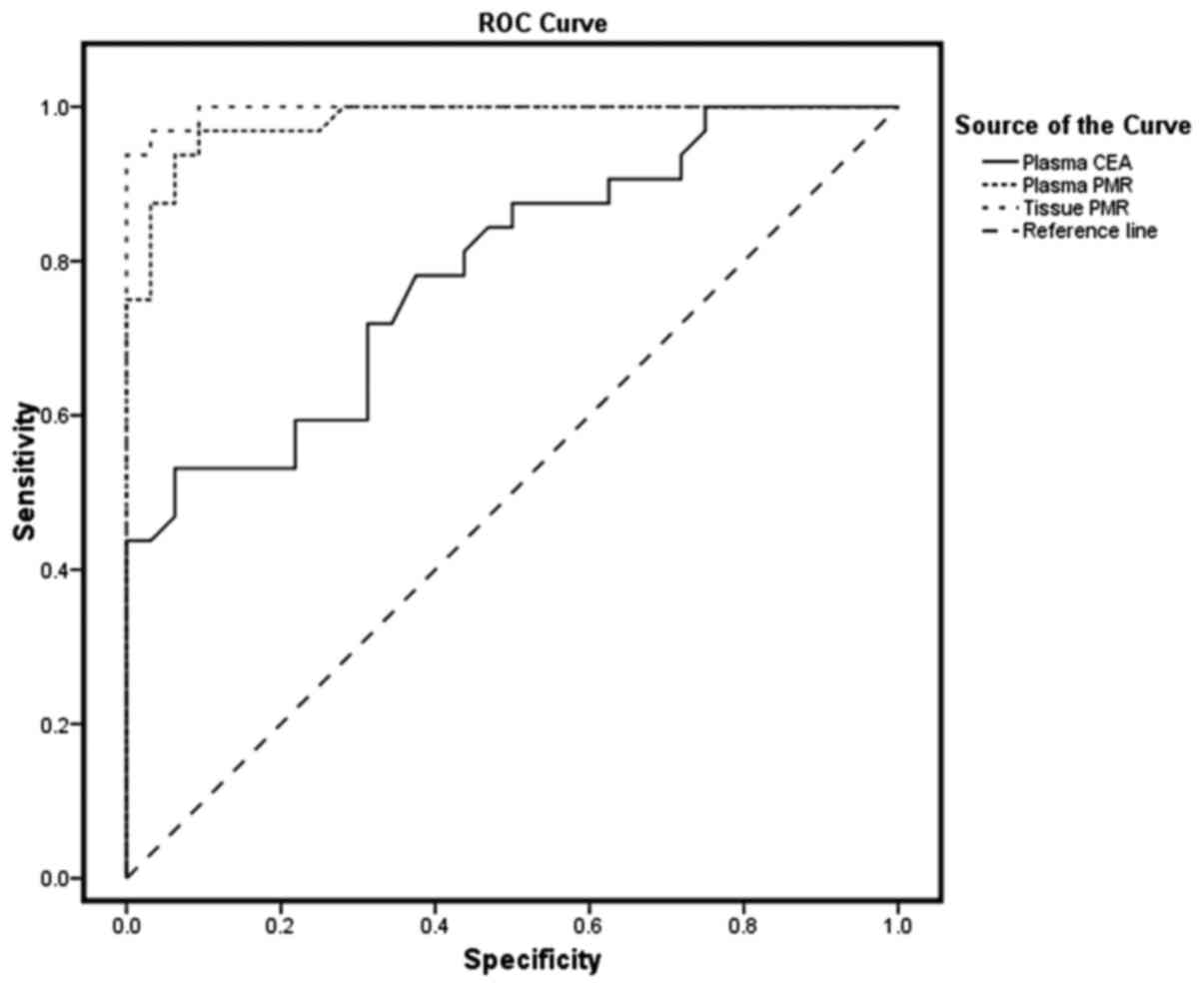
histological grade of differentiation (29), age, and gender (Table IV). Using the ROC curve, the AUC was
identified as 0.984 in plasma samples, exhibiting a sensitivity of
81.1% and specificity of 96.9% (Fig.
1).
 | Table III.SPG20 gene PMR value of plasma,
tissue samples and plasma level of CEA in patients with CRC and in
the control group. |
Table III.
SPG20 gene PMR value of plasma,
tissue samples and plasma level of CEA in patients with CRC and in
the control group.
|
|
|
|
| Percentiles |
|
|---|
|
|
|
|
|
|
|
|---|
| Parameter | No. of
patients | Minimum | Maximum | 25th | 50th | 75th | P-value |
|---|
| Tissue PMR |
|
|
|
|
|
|
<0.05a |
|
Tumor | 32 | 8.61 | 95.81 | 27.69 | 42.39 | 72.26 |
|
| Healthy
adjacent | 32 | 0.01 | 11.5 | 0.07 | 2.97 | 5.29 |
|
| Plasma PMR |
|
|
|
|
|
|
<0.05a |
|
Patient | 37 | 1.1 | 36.49 | 4.15 | 7.70 | 15.2 |
|
|
Control | 37 | 0.01 | 3.52 | 0.14 | 0.59 | 1.12 |
|
| Plasma CEA |
|
|
|
|
|
|
<0.05a |
|
Patient | 37 | 0.45 | 72.10 | 2.10 | 4.80 | 9.16 |
|
|
Control | 37 | 0.34 | 4.87 | 0.63 | 0.99 | 2.84 |
|
 | Table IV.Frequency of SPG20 hypermethylation
status in plasma DNA of patients with CRC and clinicopathological
parameters. |
Table IV.
Frequency of SPG20 hypermethylation
status in plasma DNA of patients with CRC and clinicopathological
parameters.
|
|
| Hypermethylation
status in plasma |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
characteristic | No. of
patients | Positive (%) | Negative (%) | P-value |
|---|
| Age |
|
|
| 0.339 |
| <50
years | 11 | 8/11 (72.7) | 3/11 (22.3) |
|
| >50
years | 26 | 22/26 (84.6) | 4/26 (15.4) |
|
| Gender |
|
|
| 0.283 |
|
Female | 22 | 18/22 (81.8) | 4/22 (18.2) |
|
Male | 15 | 12/15 (80) | 3/15 (20) |
|
| Tumor type |
|
|
| 0.330 |
|
Adenocarcinoma | 21 | 18/21 (85.7) | 3/21 (14.3) |
|
Mucinous | 11 | 9/11 (81.8) | 2/11 (18.2) |
|
| Histological grade
of differentiation |
|
|
| 0.672 |
|
Poorly | 7 | 6/7 (85.7) | 1/7 (14.3) |
|
|
Moderately | 9 | 8/9 (88.9) | 1/9 (11.1) |
|
|
Well | 16 | 13/16 (81.2) | 3/16 (18.8) |
| Tumor location |
|
|
| 0.476 |
|
Distal | 8 | 7/8 (87.5) | 1/8 (12.5) |
|
Proximal | 24 | 20/24 (83.3) | 4/24 (16.7) |
| Tumor size |
|
|
| 0.164 |
| <50
mm | 13 | 10/13 (76.9) | 3/13 (23.1) |
| ≥50
mm | 19 | 17/19 (89.4) | 2/19 (10.6) |
|
DNA methylation of tissue samples
As depicted in Table
III, median PMR values of 42.39 (95% CI, 27.69–72.26) and 3.61
(95% CI, 1.07–5.29) were identified in tumor and adjacent healthy
tissues, respectively (Fig. 2). The
highest PMR value obtained from adjacent healthy tissues was
considered to be the threshold; samples with a PMR >11.5 were
interpreted to be positive for methylation status. Based on this
threshold, positive methylation status was detected in 30/32 tumor
tissue samples (93.8%). As illustrated in Table III, the PMR value was significantly
higher in the tumor tissues compared with in the adjacent healthy
tissues (P<0.05). Tissue samples exhibited an AUC of 0.996,
representing a sensitivity of 93.8% and specificity of 99.96% for
this methylation marker. Results from PMR values indicate that
there is no statistically significant association between the
tissue and plasma samples from patients with CRC and the
clinicopathological characteristics (Table V).
 | Table V.Association between PMR value of
plasma, tissue samples and plasma level of CEA and
clinicopathological characteristics of patients with CRC. |
Table V.
Association between PMR value of
plasma, tissue samples and plasma level of CEA and
clinicopathological characteristics of patients with CRC.
|
|
| Tissue PMR | Plasma PMR | Plasma CEA |
|---|
|
|
|
|
|
|
|---|
| Clinicopathological
characteristic | No. of
patients | Mean ± standard
deviation | P-value | Mean ± standard
deviation | P-value | Mean ± standard
deviation | P-value |
|---|
| Tumor type |
|
| 0.432a |
| 0.155a |
| 0.161a |
|
Adenocarcinoma | 21 | 50.96±28.2 |
| 12.88±9.9 |
| 5.43±5.0 |
|
|
Mucinous | 11 | 42.91±24.0 |
| 8.01±6.53 |
| 12.18±20.6 |
|
| Histological grade
of differentiation |
|
| 0.991b |
| 0.144b |
| 0.163b |
|
Poorly | 7 | 49.32±33.1 |
| 9.48±8.3 |
| 3.86±3.0 |
|
|
Moderately | 9 | 47.45±24.3 |
| 16.30±12.4 |
| 3.08±2.3 |
|
|
Well | 16 | 45.18±26.9 |
| 9.1±6.41 |
| 12.08±17.1 |
|
| Tumor location |
|
| 0.799a |
| 0.151a |
| 0.193a |
|
Distal | 8 | 50.35±25.8 |
| 7.15±4.0 |
| 16.73±23.4 |
|
|
Proximal | 24 | 47.50±27.5 |
| 12.56±10.0 |
| 4.75±4.1 |
|
| Tumor size |
|
| 0.645a |
| 0.561a |
| 0.131a |
| <50
mm | 13 | 45.51±28.7 |
| 10.04±8.8 |
| 3.58±2.6 |
| ≥50
mm | 19 | 50.06±25.9 |
| 12.00±9.4 |
| 10.60±16.0 |
|
CEA of plasma samples
The performance of the CEA tumor marker was compared
with PMR values derived from plasma samples. Median CEA levels were
demonstrated to be 4.83 (95% CI, 2.1–9.16) and 1.24 (95% CI,
0.63–2.84) in samples from patients with CRC and the healthy
control group, respectively.
Discussion
In the present study, the PMR values for the SPG20
promoter were measured in plasma and tissue samples obtained from
patients with CRC, and in plasma samples from healthy individuals.
The MethyLight assay was used to determine the methylation status
of the samples. Because of its high specificity, sensitivity and
reproducibility, the MethyLight assay is able to measure a small
volume of a DNA template, making it an ideal tool to effectively
quantify DNA methylation in clinical samples (33,34). In
the MethyLight assay, ALU-C4, a consensus DNA sequence, was used as
a reference gene, demonstrating less local cancer-associated
genomic alteration compared with single-copy genes such as β-actin,
GAPDH, myogenic differentiation 1 and collagen type II α 1 chain
(34,36). The PMR values in plasma samples from
patients with CRC were significantly higher compared with samples
from healthy individuals. Median PMR values for patients' plasma
samples were 12 times higher compared with samples from healthy
patients.
Results from ROC curve analysis demonstrated that
there was a high AUC of 0.984 for plasma samples, indicating a
sensitivity of 81.1% and a specificity of 96.9% to discriminate
carcinoma from normal status. To the best of our knowledge, no
studies have previously investigated the methylation status of
SPG20 in plasma samples. Thus, the results of the current study
were compared with studies performed on other genes. In a previous
study by Warren et al (39),
septin 9 (SEPT9) promoter methylation analysis was performed in 50
plasma samples from patients with CRC and 90 plasma samples from
healthy individuals, demonstrating a sensitivity of 90% and
specificity of 88% with a 12% false-positive rate for CRC
detection. In a previous study, Grutzmann et al (40) analyzed SEPT9 methylation in plasma,
representing 72% sensitivity and 90% specificity for CRC detection.
Kalmar et al (41) applied the
PMR threshold value for SEPT9 methylation and reported a
sensitivity of 8.3, 30.8 and 88.2% for plasma derived from healthy
subjects, and patients with adenoma and CRC, respectively.
Blood-based methylation analysis on numerous genes, including human
mutL homolog 1, helicase-like transcription factor (42) and runt related transcription factor 3
(43), has revealed sensitivities
between 34–90% and specificities between 69–100% for CRC
diagnosis.
In the present study, the tissue sample results
demonstrated 93.8% sensitivity and 96.9% specificity based on a
putative threshold value. A significant difference was identified
in the median PMR values of tumor tissues, compared with adjacent
healthy tissues. Consistent with a study conducted by Lind et
al (18), promoter
hypermethylation of SPG20 was analyzed in tumor, adenoma and
healthy tissue samples. However, it was revealed that the SPG20
promoter was methylated in 91, 75 and 2% of the tumor, adenoma and
healthy tissue samples, respectively. In addition, an AUC of 0.947,
a sensitivity of 88% and specificity of 100% were identified
(18). Notably, results from the
present study were comparable with those reported by Zhang et
al (44), whereby the SPG20
promoter methylated DNA with methyl specific PCR was analyzed in
tissue and stool samples from patients with CRC. They demonstrated
that there is 85.4% sensitivity and 96.9% specificity, and 88%
sensitivity and 100% specificity in tissue and stool samples,
respectively. In the present study, the association between SPG20
methylation status and clinicopathological characteristics of
patients with CRC was evaluated. However, no significant
association was identified between these characteristics. Lind
et al (17) assessed the
promoter methylation status of specific genes, including
cannabinoid receptor interacting protein 1 (CNRIP1), fibrillin 1
(FBN1), internexin neuronal intermediate filament protein α (INA),
mal T-cell differentiation protein (MAL), synuclein α (SNCA) and
SPG20, in CRC, adenoma, and healthy mucosa tissue samples.
Consistent with the results of the current study, the results
indicated that the genes were methylated in tumor (65–94%), adenoma
(35–91%) and healthy mucosa samples (0–5%). In addition, it was
reported that no significant difference was identified in the
methylation status of these genes with clinicopathological
characteristics of patients with CRC (17). Bethge et al (19) assessed CNRIP1, FBN1, INA, MAL, SNCA
and SPG20 genes in 97 cancer cell lines, demonstrating that SNCA
and SPG20 were methylated in 97 and 92% of the tumor samples,
respectively. In the present study, it was demonstrated that SPG20
methylation exhibited a significantly higher sensitivity (81.1%)
compared with the CEA tumor marker (48.6%). The preoperative CRC
detection rates of CEA were 30–50% in patients with CRC (45–47).
Currently, colonoscopy is an invasive method used
for CRC diagnosis with poor patient compliance. Furthermore,
non-invasive methods, including FOBT and CEA exhibit low
specificity and sensitivity (48).
Therefore, the development of an acceptable diagnostic method with
minimal invasiveness, compared with conventional approaches, is
required. In contrast to genetic examination that requires multiple
parallel assessments to identify the position of a mutation in a
desired gene, promoter methylation occurs in a specific sequence of
the gene that can be easily examined using a one step-cost
effective analysis. In conclusion, the detection of SPG20 gene
methylated DNA using the MethyLight method was revealed to be a
highly specific and sensitive epigenetic biomarker for CRC. In
addition, the promoter methylation status of SPG20 was demonstrated
to exhibit satisfactory sensitivity and specificity in plasma
samples, indicating the significance of SPG20 methylation as a
noninvasive biomarker for CRC. To the best of our knowledge, this
is the first study to investigate the promoter methylation status
of SPG20, as a biomarker, in CRC plasma samples. However, further
studies with larger sample sizes are required in order to evaluate
the specific role of SPG20 methylation as a biomarker for CRC in
plasma samples.
Acknowledgements
The present study was part of a PhD thesis of N.A.
Rezvani, which was supported by the Department of Research and
Technology at the Hamadan University of Medical Sciences (grant no.
9304101836). The authors would like to thank the staff of the
Medical Genetics Laboratory, Reference Laboratory and Kermanshah
University of Medical Sciences for their assistance during the
present study.
Glossary
Abbreviations
Abbreviations:
|
SPG20
|
spastic paraplegia 20
|
|
CRC
|
colorectal cancer
|
|
CEA
|
carcinoembryonic antigen
|
|
EPD
|
eukaryotic promoter database
|
|
Tm
|
melting temperature
|
|
SEPT9
|
septin 9
|
|
CNRIP1
|
cannabinoid receptor interacting
protein 1
|
|
FBN1
|
fibrillin 1
|
|
INA
|
internexin neuronal intermediate
filament protein α
|
|
MAL
|
mal, T-cell differentiation
protein
|
|
SNCA
|
synuclein α
|
|
FOBT
|
fecal occult blood test
|
|
ROC
|
receiver-operating characteristics
|
|
PMR
|
percent methylated reference
|
|
AUC
|
area under the ROC curve
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pawa N, Arulampalam T and Norton JD:
Screening for colorectal cancer: Established and emerging
modalities. Nat Rev Gastroenterol Hepatol. 8:711–722. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stigliano V, Sanchez-Mete L, Martayan A
and Anti M: Early-onset colorectal cancer: A sporadic or inherited
disease? World J Gastroenterol. 20:12420–12430. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mohebbi M, Nourijelyani K, Mahmoudi M,
Moghadaszadeh B, Mohammad K, Zeraati H and Fotouhi A: Time of
occurrence and age distribution of digestive tract cancers in
northern Iran. Iranian J Publ Health. 37:8–19. 2008.
|
|
6
|
Smith RA, Cokkinides V and Eyre HJ:
American cancer society guidelines for the early detection of
cancer, 2006. CA Cancer J Clin. 56:11–25; quiz 49–50. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dawson MA and Kouzarides T: Cancer
epigenetics: From mechanism to therapy. Cell. 150:12–27. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Irizarry RA, Ladd-Acosta C, Wen B, Wu Z,
Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al:
The human colon cancer methylome shows similar hypo-and
hypermethylation at conserved tissue-specific CpG island shores.
Nat Genet. 41:178–186. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rakyan VK, Down TA, Balding DJ and Beck S:
Epigenome-wide association studies for common human diseases. Nat
Rev Genet. 12:529–541. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chan AO, Broaddus RR, Houlihan PS, Issa
JP, Hamilton SR and Rashid A: CpG island methylation in aberrant
crypt foci of the colorectum. Am J Pathol. 160:1823–1830. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fleischhacker M and Schmidt B: Circulating
nucleic acids (CNAs) and cancer-a survey. Biochim Biophys Acta.
1775:181–232. 2007.PubMed/NCBI
|
|
12
|
Shirahata A and Hibi K: Serum vimentin
methylation as a potential marker for colorectal cancer. Anticancer
Res. 34:4121–4125. 2014.PubMed/NCBI
|
|
13
|
deVos T, Tetzner R, Model F, Weiss G,
Schuster M, Distler J, Steiger KV, Grützmann R, Pilarsky C,
Habermann JK, et al: Circulating methylated SEPT9 DNA in plasma is
a biomarker for colorectal cancer. Clin Chem. 55:1337–1346. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Herbst A, Rahmig K, Stieber P, Philipp A,
Jung A, Ofner A, Crispin A, Neumann J, Lamerz R and Kolligs FT:
Methylation of NEUROG1 in serum is a sensitive marker for the
detection of early colorectal cancer. Am J Gastroenterol.
106:1110–1118. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lee BB, Lee EJ, Jung EH, Chun HK, Chang
DK, Song SY, Park J and Kim DH: Aberrant methylation of APC, MGMT,
RASSF2A, and Wif-1 genes in plasma as a biomarker for early
detection of colorectal cancer. Clin Cancer Res. 15:6185–6191.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lind GE, Kleivi K, Meling GI, Teixeira MR,
Thiis-Evensen E, Rognum TO and Lothe RA: ADAMTS1, CRABP1, and NR3C1
identified as epigenetically deregulated genes in colorectal
tumorigenesis. Cell Oncol. 28:259–272. 2006.PubMed/NCBI
|
|
17
|
Lind GE, Danielsen SA, Ahlquist T, Merok
MA, Andresen K, Skotheim RI, Hektoen M, Rognum TO, Meling GI, Hoff
G, et al: Identification of an epigenetic biomarker panel with high
sensitivity and specificity for colorectal cancer and adenomas. Mol
Cancer. 10:852011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lind GE, Raiborg C, Danielsen SA, Rognum
TO, Thiis-Evensen E, Hoff G, Nesbakken A, Stenmark H and Lothe RA:
SPG20, a novel biomarker for early detection of colorectal cancer,
encodes a regulator of cytokinesis. Oncogene. 30:3967–3978. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bethge N, Lothe RA, Honne H, Andresen K,
Trøen G, Eknæs M, Liestøl K, Holte H, Delabie J, Smeland EB and
Lind GE: Colorectal cancer DNA methylation marker panel validated
with high performance in Non-Hodgkin lymphoma. Epigenetics.
9:428–436. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bakowska JC, Jupille H, Fatheddin P,
Puertollano R and Blackstone C: Troyer syndrome protein spartin is
mono-ubiquitinated and functions in EGF receptor trafficking. Mol
Biol Cell. 18:1683–1692. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Eastman SW, Yassaee M and Bieniasz PD: A
role for ubiquitin ligases and Spartin/SPG20 in lipid droplet
turnover. J Cell Biol. 184:881–894. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsang HT, Edwards TL, Wang X, Connell JW,
Davies RJ, Durrington HJ, O'Kane CJ, Luzio JP and Reid E: The
hereditary spastic paraplegia proteins NIPA1, spastin and spartin
are inhibitors of mammalian BMP signalling. Hum Mol Genet.
18:3805–3821. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hooper C, Puttamadappa SS, Loring Z,
Shekhtman A and Bakowska JC: Spartin activates
atrophin-1-interacting protein 4 (AIP4) E3 ubiquitin ligase and
promotes ubiquitination of adipophilin on lipid droplets. BMC Biol.
8:722010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ciccarelli FD, Proukakis C, Patel H, Cross
H, Azam S, Patton MA, Bork P and Crosby AH: The identification of a
conserved domain in both spartin and spastin, mutated in hereditary
spastic paraplegia. Genomics. 81:437–441. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Connell JW, Lindon C, Luzio JP and Reid E:
Spastin couples microtubule severing to membrane traffic in
completion of cytokinesis and secretion. Traffic. 10:42–56. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sagona AP and Stenmark H: Cytokinesis and
cancer. FEBS Lett. 584:2652–2661. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Heresbach D, Manfredi S, D'Halluin PN,
Bretagne JF and Branger B: Review in depth and meta-analysis of
controlled trials on colorectal cancer screening by faecal occult
blood test. Eur J Gastroenterol Hepatol. 18:427–433. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lao VV and Grady WM: Epigenetics and
colorectal cancer. Nat Rev Gastroenterol Hepatol. 8:686–700. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bonetti Reggiani L, Barresi V, Bettelli S,
Domati F and Palmiere C: Poorly differentiated clusters (PDC) in
colorectal cancer: What is and ought to be known. Diagn Pathol.
11:312016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Susan JC, Harrison J, Paul CL and Frommer
M: High sensitivity mapping of methylated cytosines. Nucleic Acids
Res. 22:2990–2997. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Grunau C, Clark SJ and Rosenthal A:
Bisulfite genomic sequencing: Systematic investigation of critical
experimental parameters. Nucleic Acids Res. 29:E65–E75. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Clark SJ: Studying mammalian DNA
methylation: Bisulfite modification. CRC Press; Boca Raton: 2004,
View Article : Google Scholar
|
|
33
|
Ogino S, Kawasaki T, Brahmandam M, Cantor
M, Kirkner GJ, Spiegelman D, Makrigiorgos GM, Weisenberger DJ,
Laird PW, Loda M and Fuchs CS: Precision and performance
characteristics of bisulfite conversion and real-time PCR
(MethyLight) for quantitative DNA methylation analysis. J Mol
Diagn. 8:209–217. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Eads CA, Danenberg KD, Kawakami K, Saltz
LB, Blake C, Shibata D, Danenberg PV and Laird PW: MethyLight: A
high-throughput assay to measure DNA methylation. Nucleic Acids
Res. 28:E322000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Weisenberger DJ, Campan M, Long TI, Kim M,
Woods C, Fiala E, Ehrlich M and Laird PW: Analysis of repetitive
element DNA methylation by MethyLight. Nucleic Acids Res.
33:6823–6836. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jackson K, Yu MC, Arakawa K, Fiala E, Youn
B, Fiegl H, Müller-Holzner E, Widschwendter M and Ehrlich M: DNA
hypomethylation is prevalent even in low-grade breast cancers.
Cancer Biol Ther. 3:1225–1231. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pfaffl MW: Quantification strategies in
real-time polymerase chain reactionApplied Microbiology. Filion M:
Caister Academic Press; Norfolk, UK: pp. 53–61. 2012
|
|
38
|
Widschwendter M, Siegmund KD, Müller HM,
Fiegl H, Marth C, Müller-Holzner E, Jones PA and Laird PW:
Association of breast cancer DNA methylation profiles with hormone
receptor status and response to tamoxifen. Cancer Res.
64:3807–3813. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Warren JD, Xiong W, Bunker AM, Vaughn CP,
Furtado LV, Roberts WL, Fang JC, Samowitz WS and Heichman KA:
Septin 9 methylated DNA is a sensitive and specific blood test for
colorectal cancer. BMC Med. 9:1332011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Grutzmann R, Molnar B, Pilarsky C,
Habermann JK, Schlag PM, Saeger HD, Miehlke S, Stolz T, Model F,
Roblick UJ, et al: Sensitive detection of colorectal cancer in
peripheral blood by septin 9 DNA methylation assay. PLoS One.
3:e37592008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kalmar A, Toth K, Wasserkort R, Sipos F,
Wichmann B, Valcz G, Tulassay Z and Molnar B: The detection of
methylated Septin 9 in tissue and plasma of colorectal neoplasia
and its relationship to the amount of free circulating DNA. Cancer
Res. 74:1850. 2014. View Article : Google Scholar
|
|
42
|
Leung WK, To KF, Man EP, Chan MW, Bai AH,
Hui AJ, Chan FK and Sung JJ: Quantitative detection of promoter
hypermethylation in multiple genes in the serum of patients with
colorectal cancer. Am J Gastroenterol. 100:2274–2279. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tan SH, Ida H, Lau QC, Goh BC, Chieng WS,
Loh M and Ito Y: Detection of promoter hypermethylation in serum
samples of cancer patients by methylation-specific polymerase chain
reaction for tumour suppressor genes including RUNX3. Oncol Rep.
18:1225–1230. 2007.PubMed/NCBI
|
|
44
|
Zhang H, Song YC and Dang CX: Detection of
hypermethylated spastic paraplegia-20 in stool samples of patients
with colorectal cancer. Int J Med Sci. 10:230–234. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yamashita K and Watanabe M: Clinical
significance of tumor markers and an emerging perspective on
colorectal cancer. Cancer Sci. 100:195–199. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yamashita K, Waraya M, Kim MS, Sidransky
D, Katada N, Sato T, Nakamura T and Watanabe M: Detection of
methylated CDO1 in plasma of colorectal cancer; a PCR study. PLoS
One. 9:e1135462014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang SY, Lin M and Zhang HB: Diagnostic
value of carcinoembryonic antigen and carcinoma antigen 19-9 for
colorectal carcinoma. Int J Clin Exp Pathol. 8:9404–9409.
2015.PubMed/NCBI
|
|
48
|
Koga Y, Yamazaki N and Matsumura Y: Fecal
biomarker for colorectal cancer diagnosis. Rinsho Byori.
63:361–368. 2015.(In Japanese). PubMed/NCBI
|