Cancer has been described as a set of diseases
driven by progressive genetic abnormalities, including mutations in
tumor suppressor genes, oncogenes and chromosomal abnormalities,
and by aberrant epigenetic alterations (1). Epigenetic alterations identified in
cancer include global DNA hypomethylation, particularly in
repetitive regions, but also in the intronic and the coding regions
of genes. These alterations may result in the reactivation of
transposons, the loss of chromosomal stability and imprinting
patterns. Another epigenetic modification is gene-specific DNA
hypermethylation, particularly in promoter regions of tumor
suppressor genes, deregulation of histone modification patterns and
consequently alterations in gene expression. Additionally, small
non-coding RNA (ncRNA) deregulation has been studied in detail in
various types of cancer in recent years (1). All these alterations drive the
transformation of wild-type cells into highly malignant tumor
consisting of neoplastic cells with metastatic potential and
unlimited proliferation capacities (1,2).
Currently, three major classes of small regulatory
RNAs have been identified: microRNAs, small interfering RNAs
(siRNAs) and piRNAs. The least well investigated class of ncRNAs,
piRNAs were originally identified in 2006 as ncRNAs that interacted
with PIWI proteins, which are a subclass of the conserved Argonaute
family of proteins (3–6). The main characteristics of piRNAs are
that they are single-stranded ncRNAs, with an average length of
24–32 nucleotides exhibiting highly conserved functions across
species (3,7). Notably, unlike other ncRNAs, piRNAs are
generated from a small number of long single-stranded RNA
precursors, transcribed from distinct transposons referred to as
‘piRNA clusters’ by a Dicer-independent mechanism (8,9). However,
certain piRNAs are also encoded in 3′untranslated regions of
intergenic non-coding transcripts and protein-coding genes
(10). In Drosophila, piRNAs
are primarily derived from intergenic repetitive regions in the
genome, including transposable elements. Two models of piRNA
biogenesis have been suggested in Drosophila: The primary
pathway and the secondary ‘ping-pong cycle’ (11). Certain characteristics of the
biogenesis of piRNAs have also been described in other organisms
(3,4).
Cloning of piRNAs in Drosophila revealed that this group of
small ncRNAs also included repeat-associated siRNAs, which were
previously identified in plants (12)
and trypanosomes (13).
On the basis of phylogenetic analysis, Argonaute
proteins may be divided into two subclasses: The AGO subfamily,
based on Arabidopsis thaliana Ago1 proteins, and the PIWI
subfamily, related to Drosophila melanogaster Piwi proteins.
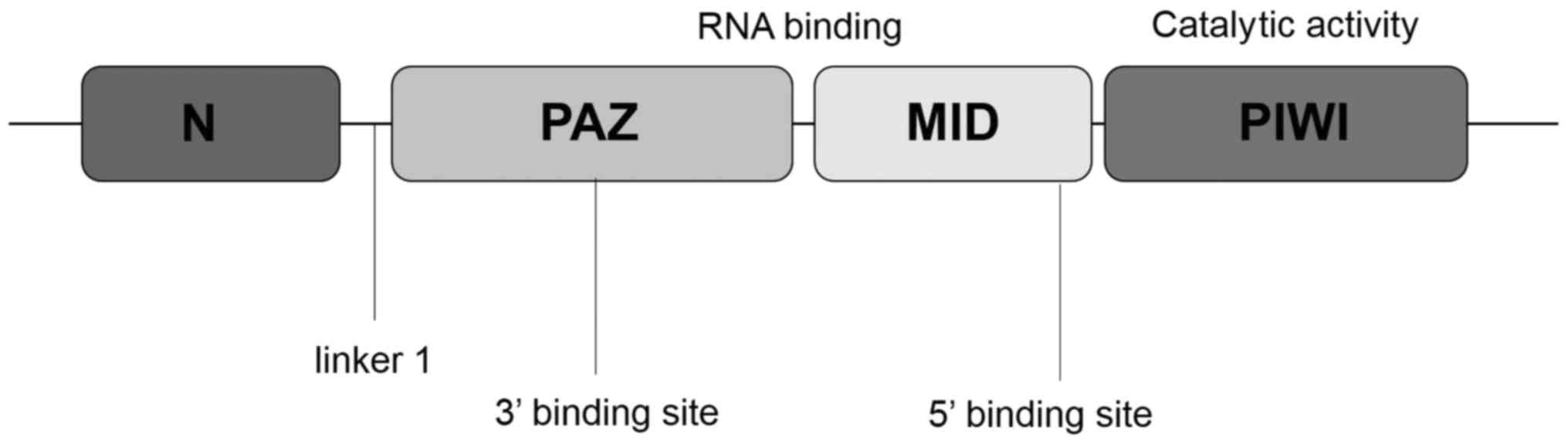
The AGO and PIWI subclasses of Argonaute are composed of three
characteristic domains: the PIWI-Argonaute-Zwille (PAZ) domain, the
middle (MID) domain and the PIWI domain (14) (Fig. 1).
The PAZ domain of Argonaute proteins recognizes the 3′ end of the
RNA, which, in the case of piRNAs, is invariably modified with a
2′-O-methyl group (15). The
MID domain, which is located between the PAZ domain and the PIWI
domain, is similar to the glucose/galactose-binding protein and the
Lac repressor (16). The primary
function of the MID domain is to provide a binding pocket for the
phosphorylated 5′ end of guide strand RNA. The PIWI domain, which
is unique to the AGO protein superfamily, adopts a classical RNase
H fold, and three residues within the PIWI domain form a catalytic
triad (generally Asp-Asp-His) (14,16). A
previous study has revealed that PIWI proteins, but not AGO, are
arginine-methylated by protein arginine methyltransferase 5 and, as
a consequence, symmetric dimethylarginines (sDMAs) at their
N-termini are formed (17). Several
members of the Tudor-domain-containing proteins (Tudor family
proteins) specifically bind to sDMAs and serve a crucial role in
PIWI function (11,18,19).
Current understanding of PIWI protein function has
primarily been the result of loss-of-function studies carried out
in mice, D. melanogaster, C. elegans and zebrafish.
These studies have indicated that the PIWI-piRNA system is involved
in germline development, primarily spermatogenesis and maintenance
of germline and somatic stem cells (20,21).
Further studies have indicated that the piRNA-PIWI signaling
pathway serves a crucial role in transposon repression, epigenetic
regulation and translation control (20). The epigenetic role of PIWI proteins in
germ and stem cell regulation has been the subject of study in a
number of organisms: In Drosophila mutants lacking
PIWI genes, the inhibition of germline stem cell renewal and
depletion of gametes in males and females were observed (22–24).
Homozygous Miwi, Mili and Miwi2 knockout male
mice exhibited arrested spermatogenesis, apoptosis of germ cells
and decreased testis size (25–27).
PIWI-piRNA complexes are involved in maintaining genomic integrity
in germline stem cells and have been demonstrated to be critical
for silencing transposon regions in the genome by clustering at
these elements and by methylating DNA (28).
The functions of PIWI in the germline have been
extensively studied. The expression of the human PIWI protein
PIWIL1 has been described primarily in germ cells and hematopoietic
stem cells (29). PIWIL1 has been
detected in human cluster of differentiation (CD)34+
hematopoietic progenitor cells, but not in well-differentiated cell
populations (29). However, several
lines of evidence have indicated that the human PIWI proteins
PIWIL1 and PIWIL2 are aberrantly expressed in various types of
cancer (30,31). Preliminary studies suggest that
overexpression and ectopic expression of PIWIL1 is associated with
several types of tumor (31,32). Primarily on the basis of
immunohistochemical studies, the increased expression of PIWIL1 has
been detected in breast (33,34), esophageal (35), pancreas (36), gastric (37) and endometrial (38) carcinoma. In the majority of cases,
increased levels of PIWIL1 were markedly associated with an
advanced histological tumor grade, advanced clinical stage and a
poorer clinical outcome for patients. Positive staining of PIWIL1
in colorectal cancer tissue has been identified to be a marker of
poor prognosis for patients with colorectal cancer (39). Increased expression of PIWIL1 in
hepatocellular carcinoma (HCC) was positively correlated with tumor
size and metastasis and negatively correlated with survival rates
(40). Liu et al (41) detected an increased level of PIWIL1
expression in high-grade squamous intraepithelial lesions and
cervical cancer, compared with in wild-type cervical tissue. The
alterations in PIWIL1 levels were associated with advanced
pathological stage and cisplatin resistance of cancer. In
vitro and in vivo studies demonstrated that PIWIL1
upregulation contributes to increased tumorigenesis, resistance to
chemotherapeutic drugs, acquisition of self-renewal abilities and
elevated expression of stem-cell-related transcription markers
including octamer-binding protein 4 (OCT4), homeobox protein NANOG
(NANOG) and Polycomb complex protein BMI1 (41). The levels of PIWIL1 protein and
transcript were significantly upregulated in intratumor tissue from
patients with non-small cell lung cancer (NSCLC), compared with in
peritumor tissue (42). In addition,
using gain-of-function and loss-of-function strategies, a positive
association between the expression of PIWIL1 and proliferation of
the NSCLC cell line A549 was identified (42). Cao et al (34) demonstrated that PIWIL1 affects the
cell cycle by regulating the expression level of transforming
growth factor-β receptors, cyclin-dependent kinase (CDK) 4, CDK6
and CDK8 in breast cancer (34).
PIWIL1 overexpression in colon cancer cell lines promoted
proliferation and induced global DNA methylation in vitro
(43).
Previous studies have indicated that PIWIL2 may also
serve an important role in tumor development. Increased expression
of PIWIL2 was identified in breast, cervical, gastric, ovarian,
prostate and colorectal cancer (37,38,44–49).
PIWIL2 expression has been observed in various stages of breast
cancer, and its expression was associated with increased expression
of the estrogen receptor and proliferation marker Ki-67, as well as
cancer progression (38,50). Increased levels of PIWIL2 have also
been observed in testicular seminomas, but not in testicular
non-seminoma tumors (51,52). Immunohistochemical analysis of
prostate cancer tissues revealed increased expression of PIWIL2 in
cancer cells, as compared with in non-tumorous adjacent tissues
(48). Using an in vitro model
of prostate cancer cell lines, it was demonstrated that silencing
the expression of PIWIL2 significantly decreased cell
invasion and migration, downregulated the expression of neuronal
(N-)cadherin, protein TWIST (TWIST) and vimentin, and upregulated
the expression of epithelial (E-)cadherin, matrix
metalloproteinase-9 and factors associated with EMT (48). Increased expression of PIWIL2 in
colorectal cancer tissue was significantly associated with a
decreased degree of differentiation and invasion, and reduced
overall survival time (12 months median survival period, vs. 28
months for patients with low PIWIL2) (49). Significantly increased expression of
PIWIL2 was observed in primary tumor of colon cancer and lymph node
metastasis compared with in non-tumorous colon tissue. An increased
level of PIWIL2 was associated with a decreased degree of
differentiation of the tumor and invasion, and a lower 5-year
overall survival rate (56.6 vs. 84.3%) (51,53).
PIWIL2 knockdown in colon cancer cells significantly
decreased proliferation, migration and colony formation, increased
apoptosis in vitro, and decreased tumor cell proliferation
in vivo (53). It was
suggested that PIWIL2 acts as an oncogene by inhibiting
apoptosis and promoting cell proliferation through the signal
transducer and activator of transcription/B-cell lymphoma
extra-large signaling pathway (51).
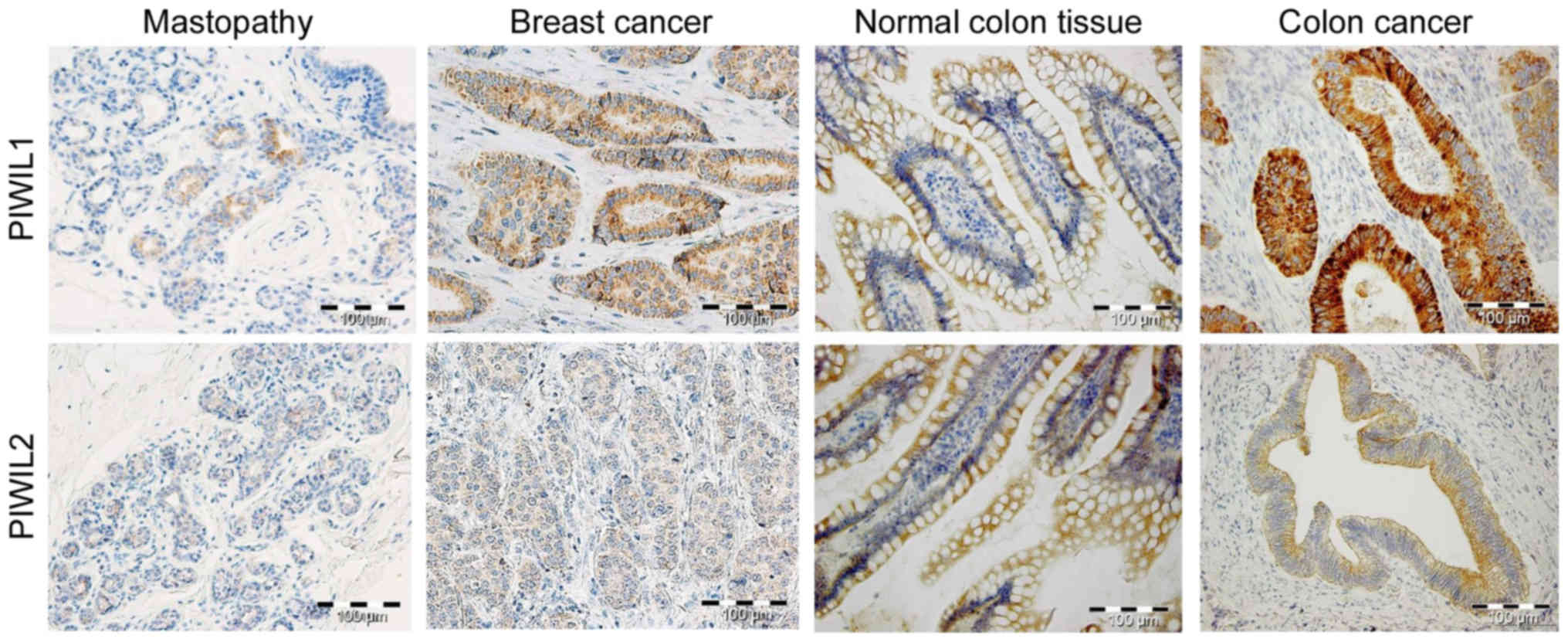
Our recent study demonstrated a decreased level of PIWIL2 in colon
cancer tissue compared with in non-tumorous adjacent tissue
(Fig. 2) (54). Furthermore, a marked negative
association between PIWIL1 and PIWIL2 was observed in wild-type
colorectal tissue (Fig. 2).
Similarly, Nikpour et al (55)
reported the absence of PIWIL2 expression in several bladder
carcinoma cell lines and bladder cancer tissues. These authors
suggested that that ectopic expression of PIWIL2 is not essential
for the pathogenesis of human bladder carcinoma (55). The contradictory results indicate
possible reciprocal regulation between PIWIL1 and PIWIL2 in colon
cancer (54).
With regard to other members of the PIWI protein
family, PIWIL4 has been identified to be overexpressed in human
cervical cancer tissue, as compared with in non-tumorous adjacent
tissue (56). Furthermore, the
expression of PIWIL4, but not of PIWIL1, PIWIL2 or PIWIL3, was
significantly increased in renal cell carcinoma (57). PIWIL2 and PIWIL4 mRNA
were expressed at an increased level in various breast cancer cell
lines compared with in mammary epithelial cells, whereas PIWIL1 and
PIWIL3 transcripts were undetectable (19). In the case of gastric cancer,
expression of four members of the PIWI protein family was markedly
increased in tumor tissue compared with in non-tumorous adjacent
tissue (37). Increased expression
was associated with advanced clinical tumor-node-metastasis (TNM)
classification, advanced T-stage and lymph node metastasis, but
only PIWIL1 and PIWIL2 levels were associated with poorer overall
survival (37).
piRNAs have not been extensively studied in cancer;
however, a limited number of preliminary studies suggest that
piRNAs are altered in cancer. A specific piRNA-651 has been
demonstrated to be aberrantly overexpressed in various tumors
compared with in wild-type tissues (58,59). In
NSCLC, a significant increase in piRNA-651 levels was identified to
be associated with cancer progression (58). Furthermore, the upregulation of
piRNA-651 in A549 lung cancer cells caused a significant increase
in cell viability and metastasis, and was associated with the
upregulation of cyclin D1 and CDK4 in vivo and in
vitro. Inhibition of piRNA-651 in gastric cancer cell lines was
associated with decreased cellular proliferation (59). The level of piRNA-823 was positively
associated with tumor lymph node metastasis and distant metastasis
(60). Conversely, upregulated
expression of piRNA-651 was observed in gastric, colon, lung and
breast cancer tissues (58–60). Increased levels of piRNA-4987, −20365,
−20485 and −20582 were observed in breast tumors compared with in
non-cancerous tissues, and were associated with lymph node
metastasis (60). In spite of these
discrepancies, the detection of piRNAs in blood and cancer tissues
may be a valid biomarker for identifying circulating or cancer stem
cells within the tumor. Recently, Martinez et al (7) described somatic and malignant expression
patterns of numerous piRNAs. They detected that 273 piRNAs of the
~20,000 known piRNAs are expressed in somatic non-malignant
tissues; in corresponding tumor tissues, they detected a
significantly increased number of piRNAs, and expression patterns
were identified to be specific to malignancies and their clinical
features (7).
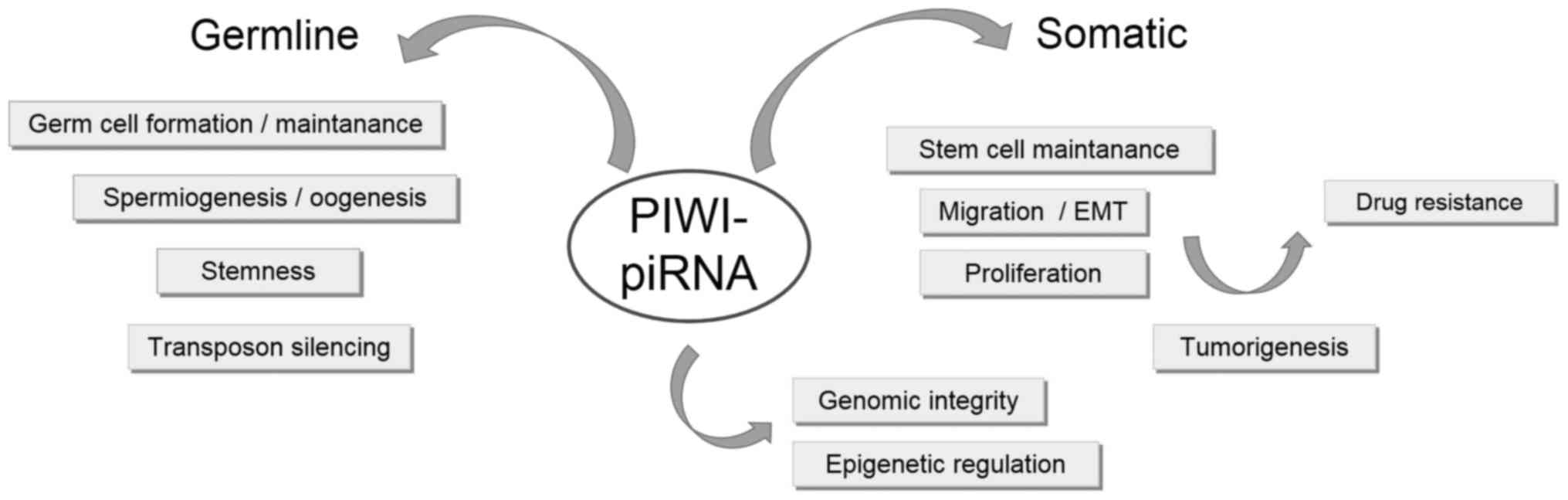
A PIWI and piRNA model of function in non-tumorous
and cancer tissue has been proposed (Fig.
3). piRNAs, along with abundant expression of PIWI, in germline
stem cells regulate transposon silencing through DNA methylation
during spermatogenesis. In this way, germline cells (expressing
PIWI at an increased level) develop normally into somatic tissues
in which PIWI proteins are absent (28,61).
However, in cancer cells, PIWI and piRNAs exhibit increased
expression, which results in aberrant DNA methylation, silencing of
tumor suppressor genes and an abnormal ‘stem-like’ state of cancer
cells (28). Furthermore, the
hypothesis that PIWIL1 and PIWIL2 in cancer contribute to
tumorigenesis by transcriptionally silencing tumor suppressor genes
through epigenetic mechanisms is supported by observation of their
orthologs in mice and D. melanogaster (62–66). In
mice, Mili and Miwi2 mutants fail to establish de
novo DNA methylation of transposon sequences, which is required
for transcriptional silencing of transposons in the genome
(62,63). It has been suggested that piRNAs acts
as a guide for directing transposon-specific DNA methylation
(64). In D. melanogaster,
Piwi is localized in the nucleus and such subcellular localization
determines its function during transposon silencing (66,67).
Additionally, PIWI proteins, by suppressing the expression of
particular transposons, may be involved in genomic instability, one
of the most common alterations to occur in cancer (67). Despite the growing attention on the
PIWI-piRNA signaling pathway, only a limited number of studies have
described the underlying molecular mechanism by which PIWI proteins
contribute to tumorigenesis (28,61).
Cancer consists of a population of genetically and
epigenetically heterogeneous individual cells, exhibiting distinct
molecular and phenotypic characteristics, and proliferative
potential. Heterogeneity of the tumor cells within the cancer led
to the concept of the existence of CSCs, which present epigenetic
alterations and signaling pathways characteristic of stem cells,
including self-renewal capacity, rapid proliferation and
multilineage differentiation (68,69).
Although the CSC hypothesis is still evolving, there is evidence to
support this model of cancer development and progression (70). Research based on the CSC model has
focused on a particular subset of cells that may be explicitly
targeted for more effective therapies for cancer (70,71). As
numerous so-called cancer/testis antigens (CTAs) that are
transiently expressed in developing germ cells have also been
identified in various tumors in humans, these may be targets for
immunotherapy (71). Due to the
restricted expression of PIWI in testis and in various types of
tumors, PIWIs may be CTAs. Currently, the most widely used method
to identify CSCs is through their expression of particular
cell-surface markers called CDs; an example is the antigen CD133,
also known as prominin-1, originally classified as a marker of
hematopoietic endothelial progenitor cells and neural stem cells
(72–74). CD133 is also a marker for
organ-specific stem cells and CSCs, and several types of tumor,
including brain, hepatocellular, colon, pancreas and prostate
(75–79). However, markers expressed exclusively
by CSCs currently identified are not specific for cancer stem
cells. Therefore, characterization of the genetic and epigenetic
alterations that occur in CSCs may provide important insights into
the processes of cancer development and metastasis.
To date, certain key signaling pathways have been
identified, which may be aberrantly regulated in CSCs, and thus may
represent potential targets for cancer diagnostics and therapies
(80,81). A number of studies have indicated that
several fundamental signaling pathways, including Wnt/β-catenin,
Notch and Hedgehog, serve critical roles in normal stem cells and
CSCs (81). Furthermore, it has been
identified that transcription factors OCT4, SRY-box 2 (SOX2),
NANOG, Krüppel-like factor 4 (KLF4), c-MYC and LIN28 are
responsible for the regulation of pluripotency and self-renewal of
embryonic stem cells (ESCs). Expression of these factors, known as
ESC markers, is restricted to pluripotent stem cells, downregulated
during embryonic development and undetectable in adult wild-type
tissues (82). However, alterations
in the expression of ESC-associated proteins have been demonstrated
in a number of types of cancer: OCT4, also known as POU domain,
class 5, transcription factor 1, is expressed in ESCs and adult
stem cells, and has been proposed to be associated with the
pluripotency, proliferative potential and self-renewal of ESCs and
germ cells (83). The transcription
factor NANOG, a downstream target of OCT4, which contributes to
cell fate determination of the pluripotent inner cell mass during
embryonic development, is also specifically expressed in human
pluripotent ESCs (82).
Patients with co-expression of OCT4 and NANOG have
been demonstrated to exhibit significantly worse overall survival
and poor prognosis of several malignancies, including oral
(84), glioma (85), gastric (86), rectal (87) and hepatocellular (88) cancer. Overexpression of NANOG was
markedly associated with poor prognosis, lymph node metastasis and
Dukes' classification of colorectal cancer (89). SOX2, together with OCT4 and NANOG,
serves a crucial role in the maintenance of ESC pluripotency.
Previous studies have demonstrated that SOX2 is involved in
promoting tumorigenesis, proliferation and dedifferentiation of
human lung squamous cell carcinoma and breast cancer (90,91). In
ovarian cancer, SOX2 expression increases the expression of CSC
markers, the potential to form tumor spheres and in vivo
tumor-initiating capability. Furthermore, SOX2-expressing cells
display enhanced apoptosis resistance in response to conventional
chemotherapies (92). In pancreatic
carcinoma, alterations in SOX2 were identified to be associated
with the invasion and metastatic potential of tumors, suggesting
that SOX2 is involved in later events of carcinogenesis (93). In rectal cancer, the increased levels
of CD133, OCT4 and SOX2 were significantly associated with tumor
recurrence and decreased disease-free survival time (87). Yin et al (88) demonstrated that pluripotent stem cell
genes are associated with HCC progression and poor prognosis.
Expression of SOX2 and LIN28 in HCC was correlated with an
increased tumor size, whereas an increased expression level of
c-MYC was associated with vascular invasion (88). An increased level of KLF4 was
associated with the aggressiveness of HCC, vascular invasion and
cancer differentiation (88).
Together with OCT4, SOX2 and C-MYC, KLF4 is a pivotal factor in the
generation of induced pluripotent cells and is involved in the
epigenetic reprogramming of a somatic genome; KLF4 is required to
maintain the cell morphology of mammary epithelial cells (94). Notably, Tiwari et al (95) observed that downregulation of KLF4
induces EMT through alterations in the expression of key genes
involved in EMT, including those encoding N-cadherin, vimentin,
β-catenin, vascular endothelial growth factor A and c-Jun
N-terminal kinase 1 (95).
PIWI proteins, due to restricted expression during
embryonic development and aberrant expression in various types of
cancer, have been suggested to act as oncogenes or constitute a
marker for CSCs. Reactivated expression of PIWI in cancer and
association with certain already defined ESC-associated proteins
indicates the participation of these proteins in the process of
tumor growth (28). Positive
associations between PIWIL1 and OCT4 mRNA levels, as
well as PIWIL2 and SOX2, in colon cancer tissues have
been identified (54). Thus, the
expression of various CSC markers in various types of cancer has
been extensively studied; however, the functional characteristics
of these markers, co-expression with transcription factors, other
signaling pathways and epigenetic mechanisms require further
study.
The alterations in cell motility and adhesiveness,
as well as the adaptation to the new microenvironment, observed
during tumor growth appear to be crucial in determining the
metastatic potential and invasiveness of cancer cells. EMT is a
critical process enabling the migration, invasion and metastasis of
tumor cells from the primary tumor to distant organs. EMT is
characterized by long-lasting morphological and molecular
alterations in epithelial cells as a result of transdifferentiation
towards a mesenchymal cell type (96). During this process, epithelial cells
acquire fibroblast-like properties, and demonstrate decreased
adhesion and increased motility (96). A number of transcription factors have
been implicated in the control of EMT, including zinc-finger
protein SNAI1 (SNAI1), TWIST, zinc-finger protein SLUG (SLUG) and
zinc-finger E-box-binding homeobox (ZEB) 1 and 2, which directly
repress mediators of epithelial cell adhesion, including E-cadherin
and components of adherens junctions. In addition, the self-renewal
capacity of CSCs appears to be essential for EMT during early steps
of metastasis (97–99).
A number of studies have indicated that metastatic
cancer cells, which have presumably undergone EMT, may exhibit a
CSC-like phenotype: For instance, overexpression of SNAI1, a
central transcription factor that regulates EMT, induces a CSC-like
phenotype in colorectal cancer cells by directly repressing
epithelial markers, including E-cadherin, and by regulating
mesenchymal markers (100). SNAI1
upregulation led to increased cell migration and increased
metastasis in vivo, and was associated with a more
aggressive phenotype, increased rates of distant metastases and
poorer clinical outcomes in breast and ovarian carcinoma, and HCC
(100). Epithelial breast cancer
cells that undergo EMT induced by SNAI1 factor, exhibit
CD44-positive and CD24-negative expression, as well as expression
of stem-like genes, including NANOG, KLF4 and
transcription factor 4 (100).
Ectopic expression of OCT4 and NANOG in lung carcinoma was
demonstrated to increase the CD133-positive cell subpopulation,
activate SLUG, promote EMT and enhance drug resistance (101). Furthermore, double knockdown of
OCT4 and NANOG in A549 lung cancer cells suppressed
the expression of SLUG, reversed the EMT process, inhibited
tumorigenic and metastatic capacities and significantly increased
the survival time of transplanted nude mice (101). In nasopharyngeal carcinoma,
increased expression of OCT4 and NANOG was significantly associated
with a decreased rate of survival (36.1% 5-year survival rate,
compared with 76.7%) (102). OCT4
and NANOG were primarily located at the invasive front of tumors,
and were significantly associated with increased levels of various
aggressive clinical factors, and advanced TNM classification and
clinical stage; furthermore, these proteins were positively
associated with EMT-associated markers (102). The limited effectiveness of standard
anticancer therapies has been attributed to the existence of
heterogeneous highly drug-resistant populations of CSCs that are
responsible for initiation, development and tumor metastasis, as
well as response to treatment. Chen et al (103) identified stem-like cancer cells of
the colon cancer cell line HCT116 that co-express CD133 and CD44
markers. Cells with increased expression of CD133 and CD44 were
undifferentiated, with self-renewal and epithelial lineage
differentiation capabilities in vitro, and increased
expression of CSC and EMT markers (103). Furthermore,
CD133+/CD44+ cells represented increased
invasive abilities in vitro and increased tumorigenic
properties in vivo (103). In
addition, alterations in the expression of EMT-associated genes
driven by KLF4 downregulation was demonstrated by Tiwari et
al (95), as aforementioned.
Previous studies have identified a direct connection
between stem cell self-renewal and cancer development and invasion
(68–70). However, the potential association
between CSCs and the PIWI-piRNA signaling pathway remains
unexplored. Evidence exists concerning the roles of PIWI proteins
in various types of tumors; however, experimental reports, as
described, often appear to be contradictory. PIWI proteins in
complex with piRNA have been demonstrated to be involved in
epigenetic regulation in germline cells by transposon element
regulation and in somatic tissue in the activation of gene
expression by promoting euchromatic histone modifications and
transcription of piRNAs (106,107).
Although the number of piRNAs expressed in somatic tissue is
significantly lower than in germline cells, tissue specificity
associated with the expressed piRNAs has been demonstrated
(7).
Aberrant expression of PIWI and piRNAs that target
mRNA transcripts may serve a driving role through the degradation
or inhibition of tumor suppressor genes or oncogenes (108). Another example of the piRNA-mediated
influence on tumorigenesis is by mutagenic retrotranspositions and
genomic instability initiation (107,108).
PIWI-piRNA complexes contribute to cancer development through
aberrant DNA methylation resulting in genomic silencing and
promoting a stem-like state of cancer cells (28,61,108,109).
A number of studies indicate that stem-like cancer cells represent
the cells that have undergone EMT and acquired metastatic
capabilities. Our previous study indicates a reciprocal regulation
between PIWI proteins and complex signaling network linking markers
characterized for CSCs (54) and
transcription factors involved in the EMT process; however, further
research is required to elucidate the underlying molecular
mechanisms.
The present review was partly supported by the
Leading National Research Center (KNOW, 2014-2018) of Wrocław
Center for Biotechnology and WroVasc Project-Integrated
Cardiovascular Centre, co-financed by the European Regional
Development Fund within the Innovative Economy Operational
Programme 2007–2013 (POIG 1.1–3).
|
1
|
Kanwal R and Gupta S: Epigenetic
modifications in cancer. Clin Genet. 81:303–311. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kristensen LS, Nielsen HM and Hansen LL:
Epigenetics and cancer treatment. Eur J Pharmacol. 625:131–142.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aravin A, Gaidatzis D, Pfeffer S,
Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ,
Kuramochi-Miyagawa S, Nakano T, et al: A novel class of small RNAs
bind to MILI protein in mouse testes. Nature. 442:203–207.
2006.PubMed/NCBI
|
|
4
|
Girard A, Sachidanandam R, Hannon GJ and
Carmell MA: A germline-specific class of small RNAs binds mammalian
Piwi proteins. Nature. 442:199–202. 2006.PubMed/NCBI
|
|
5
|
Grivna ST, Beyret E, Wang Z and Lin H: A
novel class of small RNAs in mouse spermatogenic cells. Genes Dev.
1:1709–1714. 2006. View Article : Google Scholar
|
|
6
|
Watanabe T, Takeda A, Tsukiyama T, Mise K,
Okuno T, Sasaki H, Minami N and Imai H: Identification and
characterization of two novel classes of small RNAs in the mouse
germline: Retrotransposon-derived siRNAs in oocytes and germline
small RNAs in testes. Genes Dev. 1:1732–1743. 2006. View Article : Google Scholar
|
|
7
|
Martinez VD, Vucic EA, Thu KL, Hubaux R,
Enfield KS, Pikor LA, Becker-Santos DD, Brown CJ, Lam S and Lam WL:
Unique somatic and malignant expression patterns implicate
PIWI-interacting RNAs in cancer-type specific biology. Sci Rep.
5:104232015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suzuki R, Honda S and Kirino Y: Piwi
expression and function in cancer. Front Genet. 3:2042012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luteijn MJ and Ketting RF:
PIWI-interacting RNAs: From generation to transgenerational
epigenetics. Nat Rev Genet. 14:523–534. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Robine N, Lau NC, Balla S, Jin Z, Okamura
K, Kuramochi-Miyagawa S, Blower MD and Lai EC: A broadly conserved
pathway generates 3′UTR-directed primary piRNAs. Curr Biol.
19:2066–2076. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ku HY and Lin H: PIWI proteins and their
interactors in piRNA biogenesis, germline development and gene
expression. Natl Sci Rev. 1:205–218. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Llave C, Kasschau KD, Rector MA and
Carrington JC: Endogenous and silencing associated small RNAs in
plants. Plant Cell. 14:1605–1619. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Djikeng A, Shi H, Tschudi C and Ullu E:
RNA interference in Trypanosoma brucei: Cloning of small
interfering RNAs provides evidence for retroposon-derived 24–26
nucleotide RNAs. RNA. 7:1522–1530. 2001.PubMed/NCBI
|
|
14
|
Farazi TA, Juranek SA and Tuschl T: The
growing catalog of small RNAs and their association with distinct
Argonaute/Piwi family members. Development. 135:1201–1214. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Simon B, Kirkpatrick JP, Eckhardt S,
Reuter M, Rocha EA, Andrade-Navarro MA, Sehr P, Pillai RP and
Carlopamgno T: Recognition of 2′-O-methylated 3′-end of piRNA by
the PAZ domain of a Piwi protein. Structure. 19:172–180. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jinek M and Doudna JA: A three dimensional
view of the molecular machinery of RNA interference. Nature.
457:405–412. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kirino Y, Kim N, de Planell-Saguer M,
Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA and
Mourelatos Z: Arginine methylation of Piwi proteins catalysed by
dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol.
11:652–658. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vagin VV, Wohlschlegel J, Qu J, Jonsson Z,
Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ and Aravin
AA: Proteomic analysis of murine Piwi proteins reveals a role for
arginine methylation in specifying interaction with Tudor family
members. Genes Dev. 23:1749–1762. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hashim A, Rizzo F, Marchese G, Ravo M,
Taralllo R, Nassa G, Giurato G, Santamaria G, Cordella A,
Cantarella C and Weisz A: RNA sequencing identifies specific
PIWI-interacting small non-coding RNA expression patterns in breast
cancer. Oncotarget. 5:9901–9910. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Juliano C, Wang J and Lin H: Uniting
germline and stem cells: The function of Piwi proteins and the
piRNA pathway in diverse organisms. Annu Rev Genet. 45:447–469.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thomson T and Lin H: The biogenesis and
function PIWI proteins and piRNAs: Progress and prospect. Annu Rev
Cell Dev Biol. 25:355–376. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sasaki T, Shiiohama A, Minoshima S and
Shimizu N: Identification of eight members of the Argonaute family
in the human genome. Genomics. 82:323–330. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Williams RW and Rubin GM: Argonautel is
required for efficient RNA interference in Drosophila
embryos. Proc Natl Acad Sci USA. 99:6889–6894. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Carmell MA, Xuan Y, Yhang MQ and Hannon
HJ: The Argonaute family: Tentacles that reach into RNAi,
developmental control, stem cell maintenance, and tumorigenesis.
Genes Dev. 16:2733–2742. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Carmell MA, Girard A, van de Kant HJ,
Bourc'his D, Bestor TH, de Rooij DG and Hannon GJ: MIWI2 is
essential for spermatogenesis and repression of transposons in the
mouse male germline. Dev Cell. 12:503–514. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kuramochi-Miyagawa S, Kimura T, Ijiri TW,
Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et
al: Mili, a mammalian member of piwi family gene, is essential for
spermatogenesis. Development. 131:839–849. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Deng W and Lin H: Miwi, a murine homolog
of piwi, encodes a cytoplasmic protein essential for
spermatogenesis. Dev Cell. 2:819–830. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Siddiqi S and Matushansky I: Piwis and
piwi-interacting RNAs in the epigenetics of cancer. J Cell Biochem.
113:373–380. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sharma AK, Nelson MC, Brandt JE, Wessman
M, Mahmud N, Weller KP and Hoffman R: Human CD34(+) stem cells
express the HIWI gene, a human homologue of the Drosophila
gene piwi. Blood. 97:426–434. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tan Y, Liu L, Liao M, Zhang C, Hu S, Zou
M, Gu M and Li X: Emerging roles for PIWI proteins in cancer. Acta
Biochim Biophys Sin (Shanghai). 47:315–324. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qiao D, Zeeman AM, Deng W, Looijenga LH
and Lin H: Molecular characterization of hiwi, a human member of
the piwi gene family whose overexpression is correlated with
seminomas. Oncogene. 21:3988–3999. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Taubert H, Greither T, Kaushal D, Würl P,
Bache M, Bartel F, Kehlen A, Lautenschläger C, Harris L, Kraemer K,
et al: Expression of the stem cell self-renewal gene Hiwi and risk
of tumour-related death in patients with soft-tissue sarcoma.
Oncogene. 15:1098–1100. 2007. View Article : Google Scholar
|
|
33
|
Wang DW, Wang ZH, Wang LL, Song Y and
Zhang GZ: Overexpression of hiwi promotes growth of human breast
cancer cells. Asian Pac J Cancer Prev. 15:7553–7558. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cao J, Xu G, Lan J, Huang Q, Tang Z and
Tian L: High expression of piwi-like RNA mediated gene silencing 1
is associated with poor prognosis via regulating transforming
growth factor-β receptors and cyclin-dependent kinases in breast
cancer. Mol Med Rep. 13:2829–2835. 2016.PubMed/NCBI
|
|
35
|
He W, Wang Z, Wang Q, Fan Q, Shou C, Wang
J, Giercksky KE, Nesland JM and Suo Z: Expression of HIWI in human
esophageal squamous cell carcinoma is significantly associated with
poorer prognosis. BMC Cancer. 9:4262009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Grochola LF, Greither T, Taubert H, Möller
P, Knippschild U, Udelnow A, Henne-Bruns D and Würl P: The stem
cell-associated Hiwi gene in human adenocarcinoma of the pancreas:
Expression and risk of tumor-related death. Br J Cancer.
99:1083–1088. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Y, Liu Y, Shen X, Zhang X, Chen X,
Yang C and Gao H: The PIWI protein acts as a predictive marker for
human gastric cancer. Int J Clin Exp Pathol. 5:315–325.
2012.PubMed/NCBI
|
|
38
|
Liu JJ, Shen R, Chen L, Ye Y, He G, Hua K,
Jarjoura D, Nakano T, Ramesh GK, Shapiro CL, et al: Piwil2 is
expressed in various stages of breast cancers and has the potential
to be used as a novel biomarker. Int J Clin Exp Pathol. 3:328–337.
2010.PubMed/NCBI
|
|
39
|
Zeng Y, Qu LK, Meng L, Liu CY, Dong B,
Xing XE, Wu J and Shou CC: HIWI expression profile in cancer cells
and its prognostic value for patients with colorectal cancer. Chin
Med J (Engl). 124:2144–2149. 2011.PubMed/NCBI
|
|
40
|
Zhao YM, Zhou JM, Wang LR, He HW, Wang XL,
Tao ZH, Sun HC, Wu WZ, Fan J, Tang ZY and Wang L: HIWI is
associated with prognosis in patients with hepatocellular carcinoma
after curative resection. Cancer. 118:2708–2717. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu W, Gao Q, Chen K, Xue X, Li M, Chen Q,
Zhu G and Gao Y: Hiwi facilitates chemoresistance as a cancer stem
cell marker in cervical cancer. Oncol Rep. 32:1853–1860. 2016.
|
|
42
|
Wang Y, Liu J, Wu G and Yang F:
Manipulations in HIWI levels exerts influence on the proliferation
of human non-small cell lung cancer cells. Exp Ther Med.
11:1971–1976. 2016.PubMed/NCBI
|
|
43
|
Yang L, Bi L, Liu Q, Zhao M, Cao B, Li D
and Xiu J: Hiwi promotes the proliferation of colorectal cancer
cells via upregulating global DNA methylation. Dis Markers.
2015:3830562015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ye Y, Yin DT, Chen L, Zhou Q, Shen R, He
G, Yan Q, Tong Z, Issekutz AC, Shapiro CL, et al: Identification of
Piwil2-like (PL2L) proteins that promote tumorigenesis. PLoS One.
5:e134062010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu X, Sun Y, Guo J, Ma H, Li J, Dong B,
Jin G, Zhang J, Wu J, Meng L and Shou C: Expression of hiwi gene in
human gastric cancer was associated with proliferation of cancer
cells. Int J Cancer. 118:1922–1929. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
He G, Chen L, Ye Y, Xiao Y, Hua K,
Jarjoura D, Nakano T, Barsky SH, Shen R and Gao JX: Piwil2
expressed in various stages of cervical neoplasia is a potential
complementary marker for p16. Am J Transl Res. 2:156–169.
2010.PubMed/NCBI
|
|
47
|
Chen C, Liu J and Xu G: Overexpression of
PIWI proteins in human stage III epithelial ovarian cancer with
lymph node metastasis. Cancer Biomark. 13:315–321. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Yang Y, Zhang X, Song D and Wei J: Piwil2
modulates the invasion and metastasis of prostate cancer by
regulating the expression of matrix metalloproteinase-9 and
epithelial-mesenchymal transitions. Oncol Lett. 10:1735–1740.
2015.PubMed/NCBI
|
|
49
|
Oh SJ, Kim SM, Kim YO and Chang HK:
Clinicopathologic implications of PIWIL2 expression in colorectal
cancer. Korean J Pathol. 46:318–323. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lee JH, Jung C, Javadian-Elyaderani P,
Schweyer S, Schütte D, Shoukier M, Karimi-Busheri F, Weinfeld M,
Rasouli-Nia A, Hengstler JG, et al: Pathway of proliferation and
apoptosis driven in breast cancer stem cells by stem cell protein
piwil2. Cancer Res. 70:4569–4579. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Lee JH, Schütte D, Wulf G, Füzesi L,
Radzun HJ, Schweyer S, Engel W and Nayernia K: Stem-cell protein
Piwil2 is widely expressed in tumors and inhibits apoptosis through
activation of Stat3/Bcl-XL pathway. Hum Mol Genet. 15:201–211.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gainetdinov IV, Skvortsova YV, Stukacheva
EA, Bychenko OS, Kondratieva SA, Zinovieva MV and Azhikina TL:
Expression profiles of PIWIL2 short isoforms differ in testicular
germ cell tumors of various differentiation subtypes. PLoS One.
9:e1125282014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Li D, Sun X, Yan D, Huang J, Luo Q, Tang H
and Peng Z: Piwil2 modulates the proliferation and metastasis of
colon cancer via regulation of matrix metallopeptidase 9
transcriptional activity. Exp Biol Med (Maywood). 237:1231–1240.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Litwin M, Dubis J, Arczyńska K, Piotrowska
A, Frydlewicz A, Karczewski M, Dzięgiel P and Witkiewicz W:
Correlation of HIWI and HILI expression with cancer stem cell
markers in colorectal cancer. Anticancer Res. 35:3317–3324.
2015.PubMed/NCBI
|
|
55
|
Nikpour P, Forouzandeh-Moghaddam M, Ziaee
SA, Dokun OY, Schulz WA and Mowla SJ: Absence of PIWIL2 (HILI)
expression in human bladder cancer cell lines and tissues. Cancer
Epidemiol. 33:271–275. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Su C, Ren ZJ, Wang F, Liu M, Li X and Tang
H: PIWIL4 regulates cervical cancer cell line growth and is
involved in down-regulating the expression of p14ARF and p53. FEBS
Lett. 586:1356–1362. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Al-Janabi O, Wach S, Nolte E, Weigelt K,
Rau TT, Stöhr C, Legal W, Schick S, Greither T, Hartmann A, et al:
Piwi-like 1 and 4 gene transcript levels are associated with
clinicopathological parameters in renal cell carcinoma. Biochim
Biophys Acta. 1842:686–690. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Li D, Luo Y, Gao Y and Yang Y, Wang Y, Xu
Y, Tan S, Zhang Y, Duan J and Yang Y: piR-651 promotes tumour
formation in non-small cell lung carcinoma through the upregulation
of cyclin D1 and CDK4. Int J Mol Med. 38:927–936. 2016.PubMed/NCBI
|
|
59
|
Cheng J, Guo JM, Xiao BX, Miao Y, Jaing Z,
Zhou H and Li QN: piRNA, the new non-coding RNA, is aberrantly
expressed in human cancer cells. Clinica Chim Acta. 412:1621–1625.
2011. View Article : Google Scholar
|
|
60
|
Huang G, Hu H, Xue X, Shen S, Gao E, Guo
G, Shen X and Zhang X: Altered expression of piRNAs and their
relation with clinicopathologic features of breast cancer. Clin
Transl Oncol. 15:563–568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Siddigi S, Terry M and Matushansky I: Hiwi
mediated tumorigenesis is associated with DNA hypermethylation.
PLoS One. 7:e337112012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Aravin AA, Sachidanandam R, Bourc'his D,
Schaefer C, Pezic D, Toth KF, Bestor T and Hannon GJ: A piRNA
pathway primed by individual transposons is linked to de novo DNA
methylation in mice. Mol Cell. 26:785–799. 2008. View Article : Google Scholar
|
|
63
|
Kuramochi-Miyagawa S, Watanabe T, Gotoh K,
Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri
TW, et al: DNA methylation of retrotransposon genes is regulated by
Piwi family members MILI and MIWI2 in murine fetal testes. Genes
Dev. 22:908–917. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Brennecke J, Malone CD, Aravin AA,
Sachidanandam R, Stark A and Hannon GJ: An epigenetic role for
maternally inherited piRNAs in transposon silencing. Science.
322:1387–1392. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Saito K, Nishida KM, Mori T, Kawamura Y,
Miyoshi K, Nagami T, Siomi H and Siomi MC: Specific association of
Piwi with rasiRNAs derived from retrotransposon and heterochromatic
regions in the Drosophila genome. Genes Dev. 20:2214–2222.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Saito K: The epigenetic regulation of
transposable elements by PIWI-interacting RNAs in
Drosophila. Genes Genet Syst. 88:9–17. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
van Wolfswinkel JC and Ketting RF: The
role of small non-coding RNAs in genome stability and chromatin
organization. J Cell Sci. 123:1825–1839. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Lobo NA, Shimono Y, Qian D and Clarke MF:
The biology of cancer stem cells. Annu Rev Cell Dev Biol.
23:675–699. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Pardal R, Clarke MF and Morrison SJ:
Applying the principles of stem-cell biology to cancer. Nat Rev
Cancer. 3:895–902. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Islam F, Gopalan V, Smith RA and Lam AK:
Translational potential of cancer stem cells: A review of the
detection of cancer stem cells and their roles in cancer recurrence
and cancer treatment. Exp Cell Res. 335:135–147. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yao J, Caballero OL, Yung WK, Weinstein
JN, Riggins GJ, Strausberg RL and Zhao Q: Tumor subtype-specific
cancer-testis antigens as potential biomarkers and
immunotherapeutic targets for cancers. Cancer Immunol Res.
2:371–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Yin AH, Miraglia S, Zanjani ED,
Almeida-Porada G, Ogawa M, Leary AG, Olweus J, Kearney J and Buck
DW: AC133, a novel marker for human hematopoietic stem and
progenitor cells. Blood. 90:5002–5012. 1997.PubMed/NCBI
|
|
73
|
Salven P, Mustjoki S, Alitalo R, Alitalo K
and Rafii S: VEGFR-3 and CD133 identify a population of
CD34+lymphatic/vascular endothelial precursor cells. Blood.
101:168–172. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Uchida N, Buck DW, He D, Reitsma MJ, Masek
M, Phan TV, Tsukamoto AS, Gage FH and Weissman IL: Direct isolation
of human central nervous system stem cells. Proc Natl Acad Sci USA.
97:14720–14725. 2009. View Article : Google Scholar
|
|
75
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
76
|
Suetsugu A, Nagaki M, Aoki H, Motohashi T,
Kunisada T and Moriwaki H: Characterization of CD133+
hepatocellular carcinoma cells as cancer stem/progenitor cells.
Biochem Biophys Res Commun. 351:820–824. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
O'Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Hermann PC, Huber SL, Herrler T, Aicher A,
Ellwart JW, Guba M, Bruns CJ and Heeschen C: Distinct populations
of cancer stem cells determine tumor growth and metastatic activity
in human pancreatic cancer. Cell Stem Cell. 1:313–323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Collins AT, Berry PA, Hyde C, Stower MJ
and Maitland NJ: Prospective identification of tumorigenic prostate
cancer stem cells. Cancer Res. 65:10946–10951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Klonisch T, Wiechec E, Hombach-Klonisch S,
Ande SR, Wesselborg S, Schulze-Osthoff K and Los M: Cancer stem
cell markers in common cancers-therapeutic implications. Trends Mol
Med. 14:445–460. 2008. View Article : Google Scholar
|
|
81
|
Takebe N, Miele L, Harris PJ, Jeong W,
Bando H, Kahn M, Yang SX and Ivy SP: Targeting Notch, Hedgehog, and
Wnt pathways in cancer stem cells: Clinical update. Nat Rev Clin
Oncol. 12:445–464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Wang J, Rao S, Chu J, Shen X, Levasseur
DN, Theunissen TW and Orkin SH: A protein interaction network for
pluripotency of embryonic stem cells. Nature. 444:364–368. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Niwa H, Miyazaki J and Smith AG:
Quantitative expression of Oct-3/4 defines differentiation,
dedifferentiation or self-renewal of ES cells. Nature Genet.
24:372–376. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
84
|
Chiou SH, Yu CC, Huang CY, Lin SC, Liu CJ,
Tsai TH, Chou SH, Chien CS, Ku HH and Lo JF: Positive correlations
of Oct4 and Nanog in oral cancer stem-like cells and high grade
oral squamous cell carcinoma. Clin Cancer Res. 14:4085–4095. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Guo Y, Liu S, Wang P, Zhao S, Wang F, Bing
L, Zhang Y, Ling EA, Gao J and Hao A: Expression profile of
embryonic stem cell-associated genes Oct4, Sox2 and Nanog in human
glioma. Histopathology. 59:763–775. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Matsuoka J, Yashiro M, Sakuari K, Kubo N,
Tanaka H, Muguruma K, Sawada T, Ohira M and Hirakawa K: Role of the
stemness factors Sox2, Oct3/4, and Nanog in gastric carcinoma. J
Surg Res. 174:130–135. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Saigusa S, Tanaka K, Toiyama Y, Yokoe T,
Okugawa Y, Ioue Y, Miki C and Kusunoki M: Correlation of CD133,
OCT4, and Sox2 in rectal cancer and their association with distant
recurrence after chemoradiotherpy. Ann Surg Oncol. 16:3488–3498.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Yin X, Li YW, Jin JJ, Zhou Y, Ren ZG, Qiu
SJ and Zhang BH: The clinical and prognostic implications of
pluripotent stem cell gene expression in hepatocellular carcinoma.
Oncol Lett. 5:1155–1162. 2013.PubMed/NCBI
|
|
89
|
Meng HM, Zheng P, Wang XY, Liu C, Sui HM,
Wu SJ, Zhou J, Ding YQ and Li J: Overexpression of Nanog predicts
tumor progression and poor prognosis in colorectal cancer. Cancer
Biol Ther. 9:295–302. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Sholl LM, Barletta JA, Yeap BY, Chirieac
LR and Hornick JL: Sox2 protein expression is an independent poor
prognostic indicator in stage I lung adenocarcinoma. Am J Surg
Pathol. 34:1193–1198. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lengerke C, Fehm T, Kurth R, Neubauer H,
Scheble V, Müller F, Schneider F, Petersen K, Wallwiener D, Kanz L,
et al: Expression of the embryonic stem cell marker SOX2 in
early-stage breast carcinoma. BMC Cancer. 11:422011. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Bareiss PM, Paczulla A, Wang H, Schairer
R, Wiehr S, Kohlhofer U, Rothfuss OC, Fischer A, Perner S, Staebler
A, et al: SOX2 expression associates with stem cell state in human
ovarian carcinoma. Cancer Res. 73:5544–5555. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Sanada Y, Yoshida K, Ohara M, Oeda M,
Konishi K and Tsutani Y: Histopathological evaluation of stepwise
progression of pancreatic carcinoma with immunohistochemical
analysis of gastric epithelial transcription factor SOX2:
Comparison of expression patterns between invasive components and
cancerous or nonneoplastic intraductal components. Pancreas.
32:164–170. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Tiwari N, Meyer-Schaller N, Arnold P,
Antoniadis H, Pachkov M, van Nimwegen E and Christofori G: Klf4 is
a transcriptional regulator of genes critical for EMT, including
Jnk1 (Mapk8). PLoS One. 8:e573292013. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Thiery JP, Aclogue H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wu CY, Tsai YP, Wu MZ, Teng SC and Wu KJ:
Epigenetic reprogramming and post-transcriptional regulation during
the epithelial-mesenchymal transition. Trends Genet. 28:454–463.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Esteban MA, Bao X, Zhuang Q, Zhou T, Qin B
and Pei D: The mesenchymal-to-epithelial transition in somatic cell
reprogramming. Curr Opin Genet Dev. 22:423–428. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging role axis of evil in the war
on cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Fan F, Samuel S, Evans KW, Lu J, Xia L,
Zhou Y, Sceusi E, Tozzi F, Ye XC, Mani SA and Ellis LM:
Overexpression of Snail induces epithelial-mesenchymal transition
and a cancer stem cell-like phenotype in human colorectal cancer
cells. Cancer Med. 1:5–16. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
101
|
Chiou SH, Wang ML, Chou YT, Chen CJ, Hong
CF, Hsieh WJ, Chang HT, Chen YS, Lin TW, Hsu HS and Wu CW:
Coexpression of Oct4 and Nanog enhances malignancy in lung
adenocarcinoma by inducing cancer stem cell-like properties and
epithelial-mesenchymal transdifferentiation. Cancer Res.
70:10433–10444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Luo W, Li S, Peng B, Ye Y, Deng X and Yao
K: Embryonic stem cell markers SOX2, OCT4 and Nanog expression and
their correlations with epithelial-mesenchymal transition in
nasopharyngeal carcinoma. PLoS One. 8:e563242013. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Chen KL, Pan F, Jiang H, Chen JF, Pei L,
Xie FW and Liang HJ: Highly enriched CD133(+)CD44(+) stem-like
cells with CD133(+)CD44(high) metastatic subset in HCT116 colon
cancer cells. Clin Exp Metastasis. 28:751–763. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Zhanga H, Renb Y, Xuc H, Pengd D, Duane C
and Liua C: The expression of stem cell protein Piwil2 and piR-932
in breast cancer. Surgical Oncol. 22:217–223. 2013. View Article : Google Scholar
|
|
105
|
Botchkina IL, Rowehl RA, Rivadeneira DE,
Karpeh MS Jr, Crawford H, Dufour A, Ju J, Wang Y, Leyfman Y and
Botchkina GI: Phenotypic subpopulations of metastatic colon cancer
stem cells: Genomic analysis. Cancer Genomic Proteomics. 6:19–30.
2009.PubMed/NCBI
|
|
106
|
Zou AE, Zheng H, Saad MA, Rahimy M, Ku J,
Kuo SZ, Honda TK, Wang-Rodriguez J, Xuan Y, Korrapati A, et al: The
non-coding landscape of head and neck squamous cell carcinoma.
Oncotarget. 7:51211–51222. 2016.PubMed/NCBI
|
|
107
|
Watanabe T and Lin H: Posttranscriptional
regulation of gene expression by Piwi proteins and piRNAs. Mol
Cell. 56:18–27. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Moyano M and Stefani G: piRNA involvement
in genome stability and human cancer. J Hematol Oncol. 8:382015.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Ng KW, Anderson C, Marshall EA, Minatel
BC, Enfield KS, Saprunoff HL, Lam WL and Martinez VD:
Piwi-interacting RNAs in cancer: Emerging functions and clinical
utility. Mol Cancer. 15:52016. View Article : Google Scholar : PubMed/NCBI
|