Introduction
Liver cancer is the fifth most frequently diagnosed
type of cancer worldwide in males; however, it is the second most
common cause of cancer-associated mortality. In women, liver cancer
is the seventh most commonly diagnosed type of cancer and the sixth
leading cause of cancer-associated mortality (1). Hepatocellular carcinoma (HCC) is the
most common form of liver cancer, and accounts for ~70–85% of all
cases (1). Patients with HCC
typically exhibit few early symptoms, and thus, have low early
diagnostic rates. Cases are usually confirmed at a late stage,
which is past the most opportunistic time for surgery (2). Due to poor prognosis, metastasis and
recurrence are likely to occur following surgery, the 5-year
survival rate for patients with HCC is 30–40% (3). Current efforts are focused on
identifying reliable biomarkers to predict HCC occurrence and
development.
The N-myc downstream-regulated gene 1 (NDRG1; also
termed Drg1, cap43, RTP, Rit2 and PROXY-1) belongs to the NDRG gene
family. The NDRG1 gene is positioned at chromosome location 8q24.2
(4) or 8q24.3 (5). The NDRG1 gene has 60,085 bp, including
16 exons and 15 introns, and encodes a 2997-bp RNA with a 1182-bp
region that translates into the NDRG1 protein (4). The NDRG1 protein is ~42,835 Da in
length, and consists of 394 amino acids (6,7). In normal
liver tissue, the NDRG1 protein is generally expressed in biliary
epithelial cells but not in hepatocytes (8). In addition, NDRG1 protein staining is
not affected by the condition of the liver (such as hepatitis or
cirrhosis) or the type of infecting hepatitis virus (9).
In hepatoma cells, NDRG1 expression has been
observed to be significantly increased compared with normal
hepatocytes, and NDRG1 protein was generally expressed in the
cytoplasm and membrane, but rarely in the nucleus (8–10).
However, a previous in vitro study revealed that DNA damage
increased the levels of NDRG1 nuclear expression (11). Previously, NDRG1 has been defined as
an anti-tumor gene in several cancers due to its involvement in
tumor invasion, metastasis and proliferation (12–16).
However, theories regarding the function of NDRG1 vary across
different studies. For example, a previous study stated that NDRG1
suppressed tumor growth in HCC (17).
By contrast, other studies have suggested that NDRG1 exerted a
stimulating effect on HCC (18,19).
Therefore, the present study utilized a series of experiments in
order to clarify the function of the NDRG1 gene in different HCC
cell lines in vitro, and used a polymerase chain reaction
(PCR) array test to determine downstream associated genes that may
also be regulated by NDRG1. The present study identified that
knockdown of NDRG1 expression resulted in the upregulation of
several genes, including integrin β3 (ITGB3).
ITGB3, also termed platelet glycoprotein III and
cluster of differentiation 61, belongs to the integrin family.
Within this family of cell surface receptors and adhesion
molecules, ITGB3 regulates cellular proliferation, migration, cell
survival and cell morphology, performs a main role in the processes
of cell adhesion and movement, and can affect tumor growth and
metastasis (20). However, the exact
role of ITGB3 during tumor growth and metastasis requires
additional studies. The integrin αvβ3 and αIIbβ3 are members of the
ITGB3 family. Integrin αvβ3 is mainly expressed on the surface of
endothelial cells, smooth muscle cells, monocytes and platelets
(21). Integrin αvβ3 has been
observed to affect tumor angiogenesis and is also strongly
expressed in malignant tumor angiogenic endothelial cells (22). Integrin αvβ3 could promote tumor cell
growth and angiogenesis in melanoma and breast cancer (23,24) and
blocking integrin αvβ3 resulted in reduced proliferation and
invasion in ovarian cancer (25).
Furthermore, integrin αIIbβ3 is mainly expressed in platelets and
megakaryocytes, and it has been observed to regulate platelet
interacting with tumor cells, which might help to tumor metastatic
spread (20). Currently, correlation
studies between NDRG1 and ITGB3 have not been reported. In the
present, study it was shown that knockdown of NDRG1 expression
resulted in the upregulation of ITGB3. However, the regulatory
mechanism between NDRG1 gene expression and ITGB3 function requires
additional investigation.
Materials and methods
Tissue samples
The tissue microarray (Hliv-HCC150cs-01) was
purchased from Shanghai Outdo Biotech Co., Ltd. (Shanghai, China),
and included 75 specimens of cancerous HCC tissue and corresponding
adjacent degenerative tissue. Table I
describes the patient characteristics of the study population.
 | Table I.Characteristics of the study
population (n=75). |
Table I.
Characteristics of the study
population (n=75).
| Categorical
variables | Patients, n
(%) |
|---|
| Gender |
|
|
Female | 11 (14.7) |
|
Male | 64 (85.3) |
| Age, years (mean ±
standard deviation) | 52.5±10.5 |
| AJCC cancer
staging |
|
| I | 23 (30.7) |
| II | 26 (34.7) |
|
III | 23 (30.7) |
| IV | 3 (4.0) |
Cell lines and culture
The human HCC BEL7402 and SMMC7721 cell lines, the
human colon carcinoma SW480 and SW620 cell lines, a cell line with
a high metastatic potential (MHCC-97H) and a cell line with a low
metastatic potential (MHCC-97L) were all obtained from the Cell
Bank of Type Culture Collection of Chinese Academy of Sciences
(Shanghai, China). The BEL7402, SMMC7721, SW480 and SW620 cell
lines were maintained in Roswell Park Memorial Institute
(RPMI)-1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). The MHCC-97H and MHCC-97L cell lines were maintained in
Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% (v/v) fetal bovine serum
(Gibco; Thermo Fisher Scientific, Inc.) and 1% antibiotic solution
(Gibco™ Penicillin-Streptomycin; Thermo Fisher Scientific, Inc.)
and incubated at 37°C in a humidified incubator under 5%
CO2. The detach BEL7402 cells mimic the metastatic
cancer cells in the vascular system. Access method of attach and
detach BEL7402 cells was used according to our previous description
(26).
Immunohistochemistry (IHC)
The IHC staining method was used according to a
previous description (27).
Plasmid construction and
transfection
Three candidate NDRG1 knockdown plasmids were
purchased from Shanghai Genechem Co., Ltd. (Shanghai, China). The
BEL7402 cells were transfected with the short hairpin (sh) RNA
plasmid using the jetPRIME® transfection reagent
(Polyplus-transfection, Illkirch, France) according to the
manufacturer's protocol. The shRNA sequence was
5′-CTCTAAACAACCCTGAGAT-3′, and was designed according to the
sequence provided under the GenBank accession number NM_001258432.1
(28). The shRNA sequence was
inserted into the eukaryotic expression vector GV102 (GeneChem Co.,
Ltd., Shanghai, China). The manufacturer's protocol was followed
for the transfection, using non-transfected and untreated cells as
controls. Total RNA was extracted 48 h post-transfection, and total
protein was extracted 72 h post-transfection. Western blot analysis
and quantitative (q) PCR were used to validate the effects of NDRG1
downregulation in transfected HCC cells.
Western blot analysis
Experiments were conducted at least three times.
Total protein was extracted using a radio immunoprecipitation assay
lysis buffer (Beyotime Institute of Biotechnology, Haimen, China)
and separated using SDS-PAGE (12% separating gel and 5% stacking
gel). A total of 20 µg of protein was loaded into each well. The
target proteins were then transferred onto polyvinylidene fluoride
membranes (EMD Millipore, Billerica, MA, USA). Subsequent to being
blocked with 5% non-fat milk for 2 h at room temperature, the
membranes were incubated with a primary anti-human NDRG1 antibody
(catalog no. PA5-18109; dilution, 1:1,000; Pierce, Thermo Fisher
Scientific, Inc.) or anti-human ITGB3 antibody (catalog no. 13,166;
dilution 1:1,000; Cell Signaling Technology, Inc., Danvers, MA,
USA) at 4°C overnight. The membranes were then incubated with a
secondary peroxidase-conjugated affiniPure rabbit anti-goat IgG
antibody (cat. no. ZB2306; dilution, 1:10,000; Zhong Shan Jin Qiao
Inc., Beijing, China) for NDRG1 and goat anti-rabbit IgG antibody
(cat. no. ZB2301; dilution, 1:10,000; Zhong Shan Jin Qiao Inc.) for
integrin β3 at room temperature for 1 h. Rabbit anti-GAPDH
(dilution, 1:1,000; Hangzhou Goodhere Biotechnology Co., Ltd.,
Hangzhou, China) was used as the control. An enhanced
chemiluminescence method was used to visualize western blot results
(cat. no. WBLUC0100; EMD Millipore). Protein band densitometry was
measured using Image J2x (National Institutes of Health, Bethesda,
MA, USA).
qPCR
Experiments were conducted at least three times.
BEL7402 cells transfected with empty plasmid vector
(BEL7402vec) and BEL7402 cells were as negative
controls. Total RNA was extracted from transfected and
non-transfected BEL7402 cells using the Quick-RNA extraction
reagent (Ambion; Thermo Fisher Scientific, Inc., USA). qPCR was
performed using the SYBR Green Premix Ex Taq™ (catalog no. RR420Q;
Takara Bio, Inc., Otsu, Japan) following the manufacturer's
protocol, using 35 cycles of 94°C for 30 sec and 60°C for 30 sec.
The primers for NDRG1 were as follows: Forward,
5′-AACCCACACAGTCACCCTGC-3′ and reverse, 5′-ACTCCACCACGGCATCCACT-3′.
The primers for GAPDH were as follows: Forward,
5′-GAGAAGTATGACAACAGCCTCAA-3′ and reverse,
5′-TGAGTCCTTCCACGATACCAA-3′. The 2−∆∆Cq method was used
as the normalization method for analysis (29).
PCR array assay
The RT2 Profiler™ PCR Array Human Tumor
Metastasis kit was purchased from Qiagen, Inc. (catalog no. 330231;
Valencia, CA, USA). The procedures for RNA purification,
complementary DNA first strand synthesis and qPCR were conducted
according to the manufacturer's protocols. The primers for 84 genes
known to be involved in metastasis were provided. The reaction
system for qPCR was prepared according to the manufacturer's
protocol, using 40 cycles of 95°C for 15 sec and 60°C for 60 sec.
Briefly, 20 µl of the total reaction mixture was added to each well
of a 96-well plate containing 5 µg RNA. Data analysis was completed
using the Web-based PCR Array Data Analysis Software on the Qiagen
website (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php).
Cellular proliferation test
A cell suspension of 100 µl (20,000 cells) was
inoculated in each well of a 96-well plate. Subsequently, cells
were cultured at 37°C in a humidified incubator with 5%
CO2 overnight. Cellular proliferation was assessed using
a Cell Counting Kit-8 assay (CCK-8; Dojindo Molecular Technologies,
Inc., Kumamoto, Japan) assay according to the manufacturer's
protocol. In briefly, 10 µl of the CCK-8 assay solution was added
to each well and incubated at 37°C in a humidified incubator with
5% CO2 for 2 h. Absorbance values were then measured at
450 nm with a spectrophotometer.
Wound healing assay
A total of 1×106 cells were seeded into
35-mm dishes, and when they reached 90% confluence, a scratch was
created with a 200-µl pipette tip. Subsequently, cells were
cultured in a serum-free RPMI-1640 medium at 37°C in a humidified
incubator with 5% CO2 for the next 48 h. Micrographs
were captured at 0 and 48 h. Three separate studies were conducted,
and Image-Pro Plus version 6.0 (Media Cybernetics, Inc., Rockville,
MD, USA) was used for data analysis.
Transwell migration and Matrigel
invasion assays
For Matrigel invasion assay, 1×105 cells
were seeded in the upper chamber with Matrigel (NO. 356,234; BD
Biosciences, USA) in serum-free medium for 20 h as previously
described (30). Transwell migration
assay was conducted without Matrigel and cultured for 6 h as
previously described (30).
Subsequently, the membrane was swabbed to remove the cells in the
upper chamber, and the membrane was stained with 0.5% crystal
violet dissolved in methanol. Image-Pro Plus version 6.0 was used
to count the number of cells that adhered to the membrane of the
inserts. A total of three replicates were performed.
Statistical analysis
All data were processed using the SPSS 19.0
statistical software program (IBS SPSS, Armonk, NY, USA).
Comparisons between 2 groups were performed using an independent
sample t-test and comparisons among >2 groups were performed
using one-way analysis of variance. Correlation analyses were
conducted using the Spearman's rank correlation coefficient
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
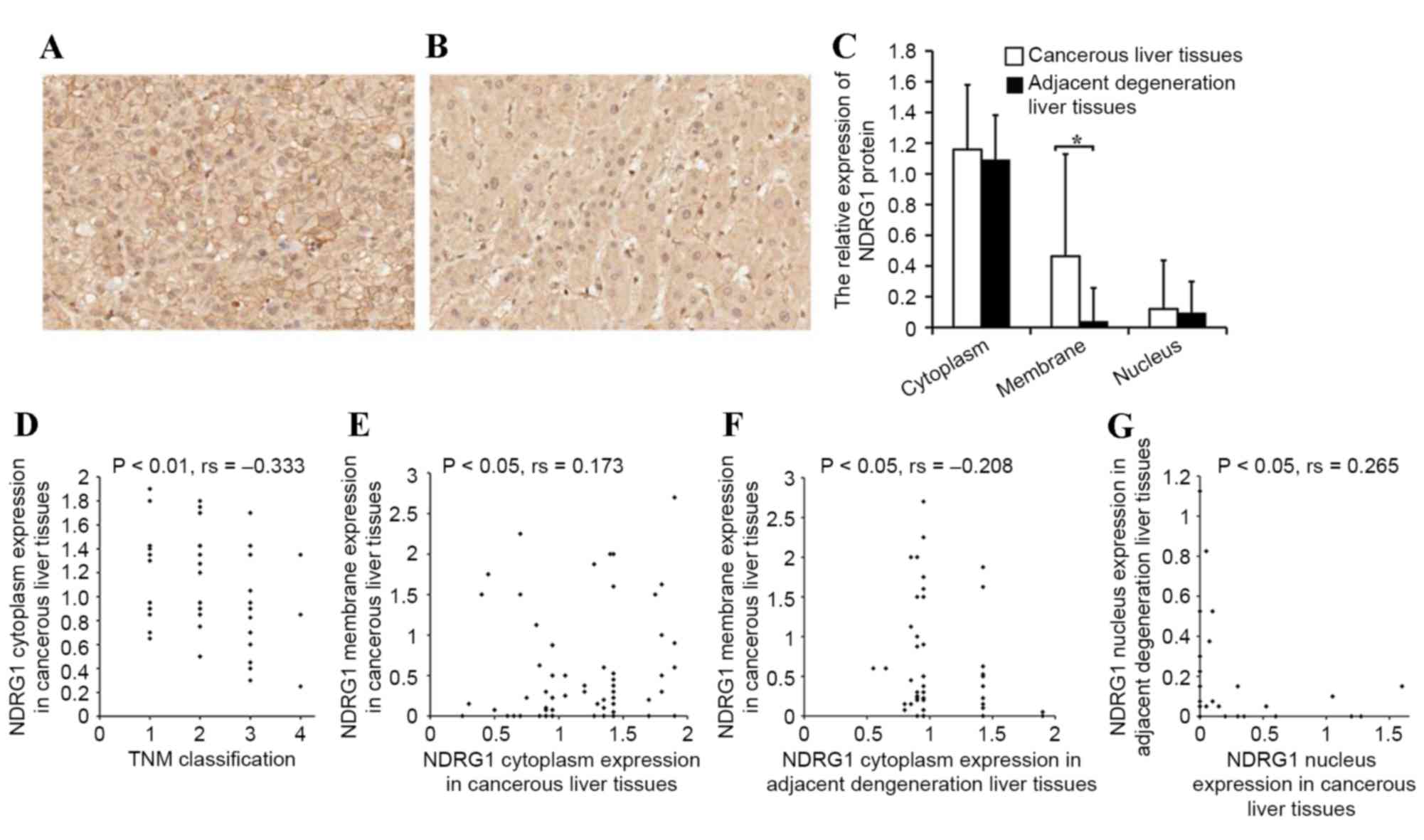
NDRG1 protein expression in HCC
tissue
The present study analyzed the characteristics of
NDRG1 protein expression in HCC tissue. The NDRG1 protein was
primarily expressed in the cytoplasm, membrane and nucleus of
cancerous cells and the corresponding adjacent degenerative cells.
The NDRG1 protein immunoreactivities differed in various
subcellular locations; cytoplasm immunoreactivity was the
strongest, followed by that in the membrane, and was weakest within
the nucleus. No significant difference was identified between NDRG1
expression in the cytoplasm or nucleus of cancerous cells compared
with that of the corresponding adjacent degenerative cells.
However, membrane expression in cancer cells was significantly
higher compared with the expression of NDRG1 in adjacent
degenerative cells (P<0.05; Fig.
1A-C). There was also a negative correlation between the NDRG1
cytoplasm staining in cancerous cells and tumor-node-metastasis
(TNM) stage (P<0.01; rs=−0.333; Fig. 1D) (31).
In other words, increased NDRG1 cytoplasm expression was observed
in tumors with earlier TNM stages compared with tumors with later
TNM stages. A positive correlation was observed between NDRG1
membrane expression and cytoplasmic expression in cancer cells
(P<0.05; rs=0.173; Fig.
1E). A negative correlation was also identified between
membrane expression in cancerous cells and cytoplasmic expression
in adjacent degenerative cells (P<0.05; rs=−0.208;
Fig. 1F). A positive correlation was
also observed between NDRG1 nucleus expression in adjacent
degenerative cells and cancer cells (P<0.05;
rs=0.265; Fig. 1G).
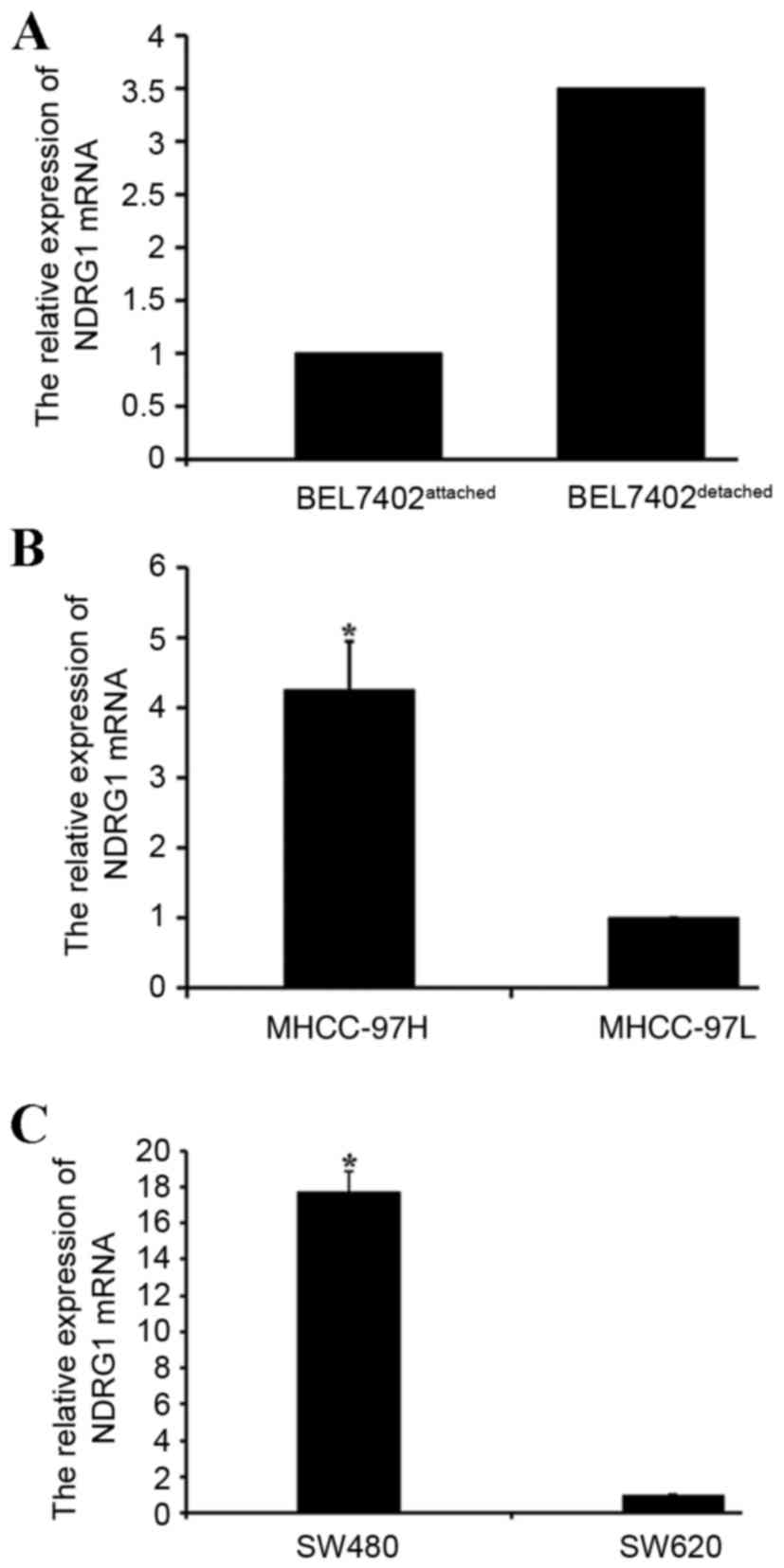
NDRG1 expression levels in different
cell culture models
The present study analyzed the expression of NDRG1
mRNA in different cell culture models using qPCR. The results
revealed an increase in NDRG1 messenger (m)RNA expression in
detached human HCC BEL7402 cells compared with that in attached
BEL7402 cells (Fig. 2A). The NDRG1
mRNA expression levels were also higher in MHCC-97H cells compared
with those in MHCC-97L cells (P<0.05; Fig. 2B). The NDRG1 mRNA expression levels in
colon carcinoma SW620 cells were lower when compared with those in
colon carcinoma SW480 cells (P<0.05; Fig. 2C).
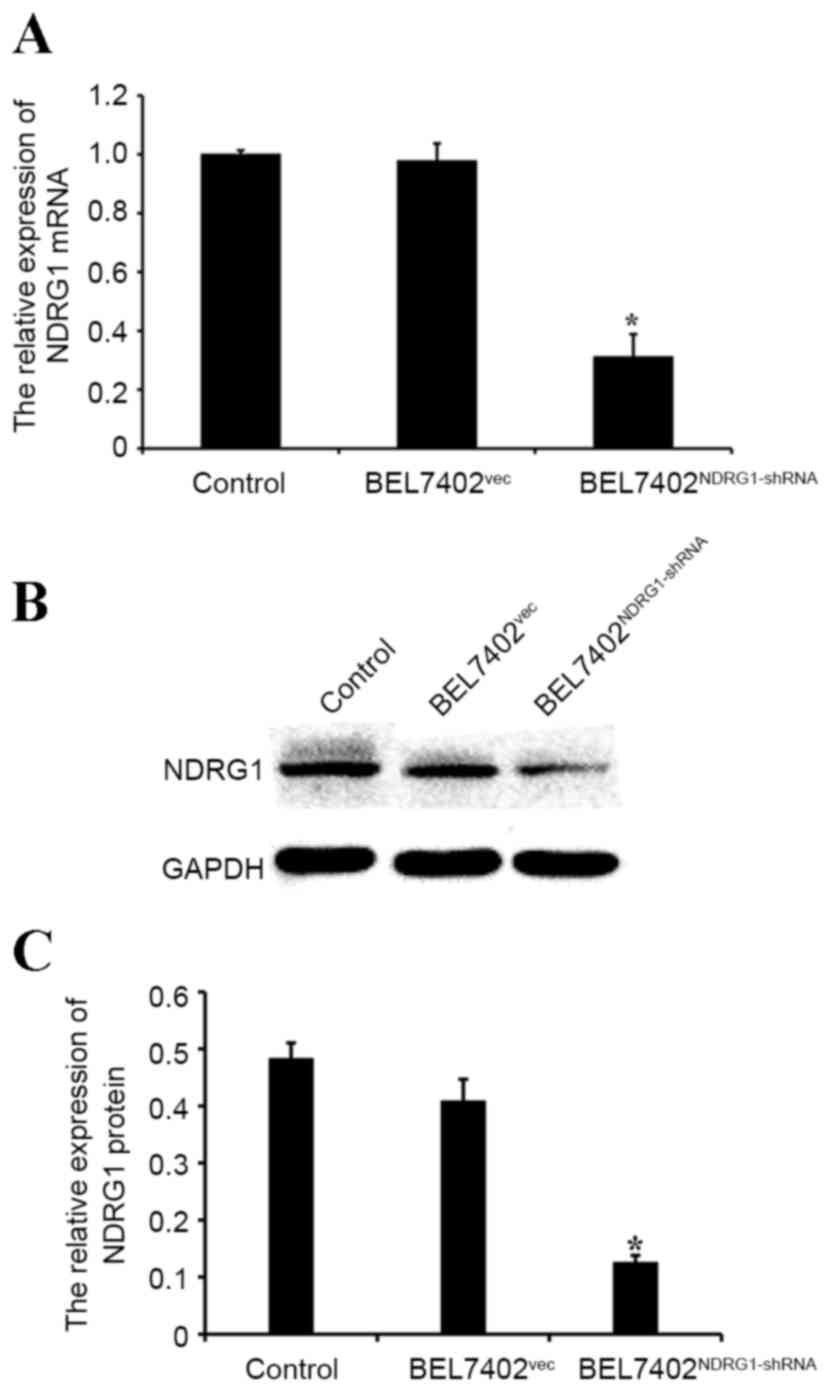
NDRG1 expression is significantly
downregulated in NDRG1-shRNA-transfected BEL7402 cells
To investigate NDRG1 expression following the
knockdown of the NDRG1 gene with shRNA, the present study performed
a western blot analysis and qPCR on transfected and control cells.
The western blot analysis and qPCR demonstrated that NDRG1
expression was significantly decreased in the BEL7402 cells
transfected with NDRG1-shRNA compared with that in the control
cells (P<0.05; Fig. 3A-C).
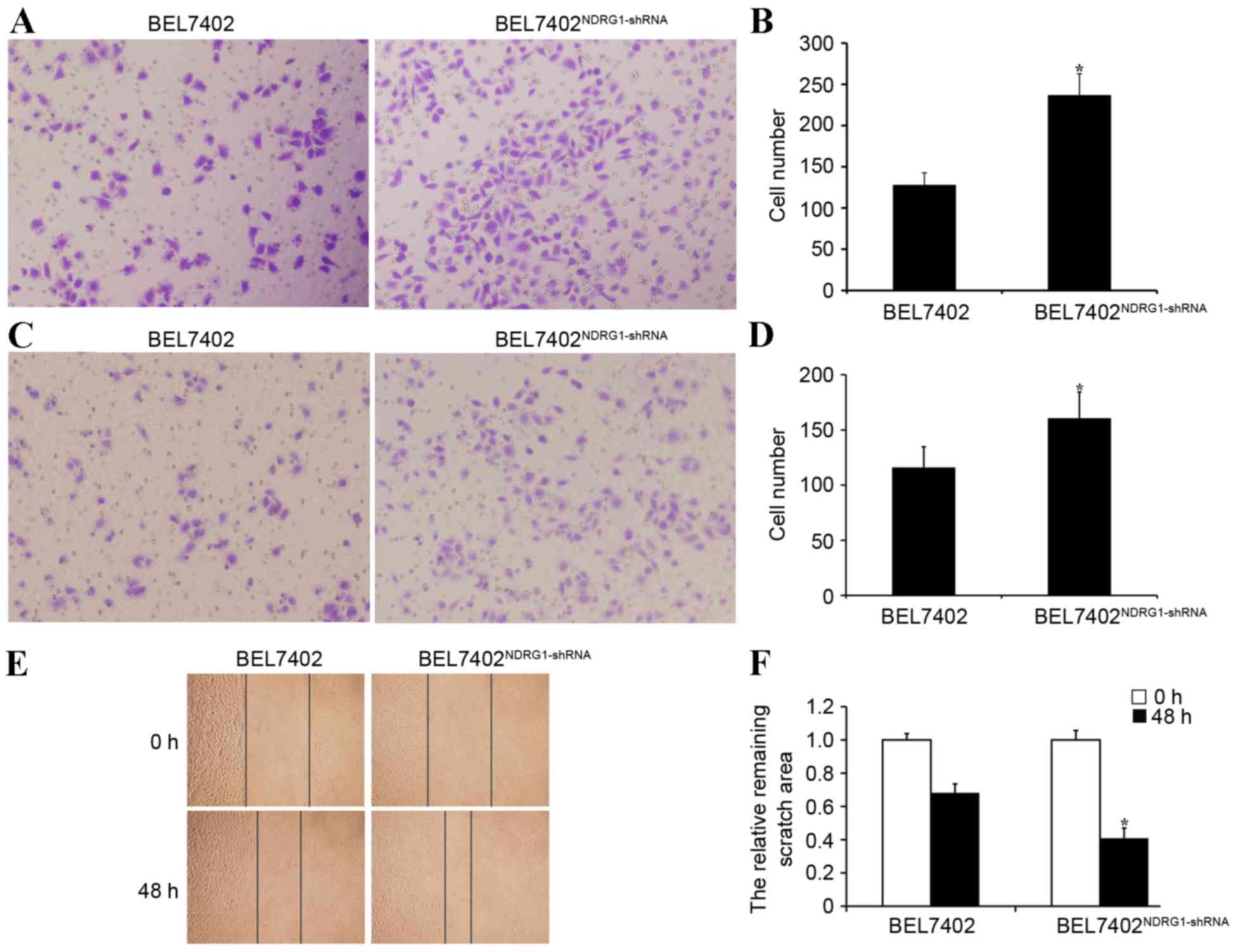
NDRG1 inhibits invasion and metastasis
in BEL7402 cells
Transwell migration assay demonstrated that
NDRG1-shRNA BEL7402 cells were significantly more capable of
motility compared with BEL7402 cells transfected with empty plasmid
vector (P<0.05; Fig. 4A and B).
The Matrigel invasion assay revealed that the BEL7402 cells had a
significantly higher invasive ability with NDRG1 knockdown
(P<0.05), indicating that NDRG1 may inhibit HCC cell invasion
in vitro (Fig. 4C and D). The
wound healing assay also showed that NDRG1-shRNA BEL7402 cells were
significantly more capable of movement compared with BEL7402 cells
transfected with empty plasmid vector (P<0.05; Fig. 4E and F). Therefore, NDRG1 had the
ability to inhibit HCC cell migration and invasion in
vitro.
NDRG1 inhibits BEL7402 cellular
proliferation
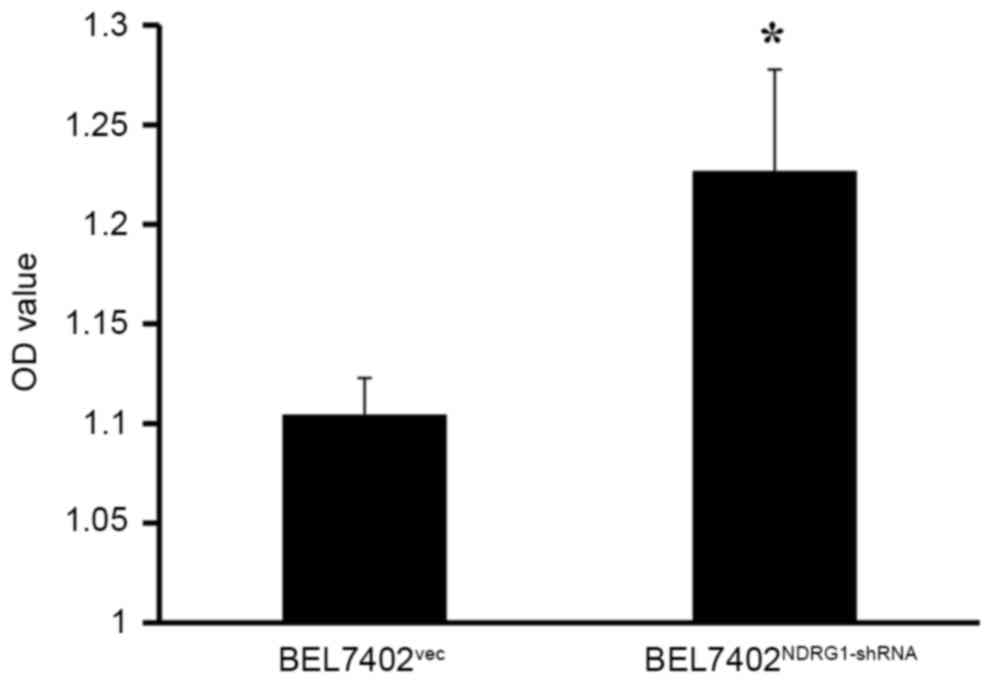
The CCK-8 assay was performed to detect the
proliferation of BEL7402 cells at a 48 h following transfection
with NDRG1-shRNA. The results demonstrated that BEL7402 cellular
proliferation was enhanced due to NDRG1 knockdown. Therefore, NDRG1
may inhibit BEL7402 cellular proliferation at a level that was
statistically significant (P<0.05; Fig. 5).
Downstream molecular candidates of
NDRG1 inhibit tumor metastasis
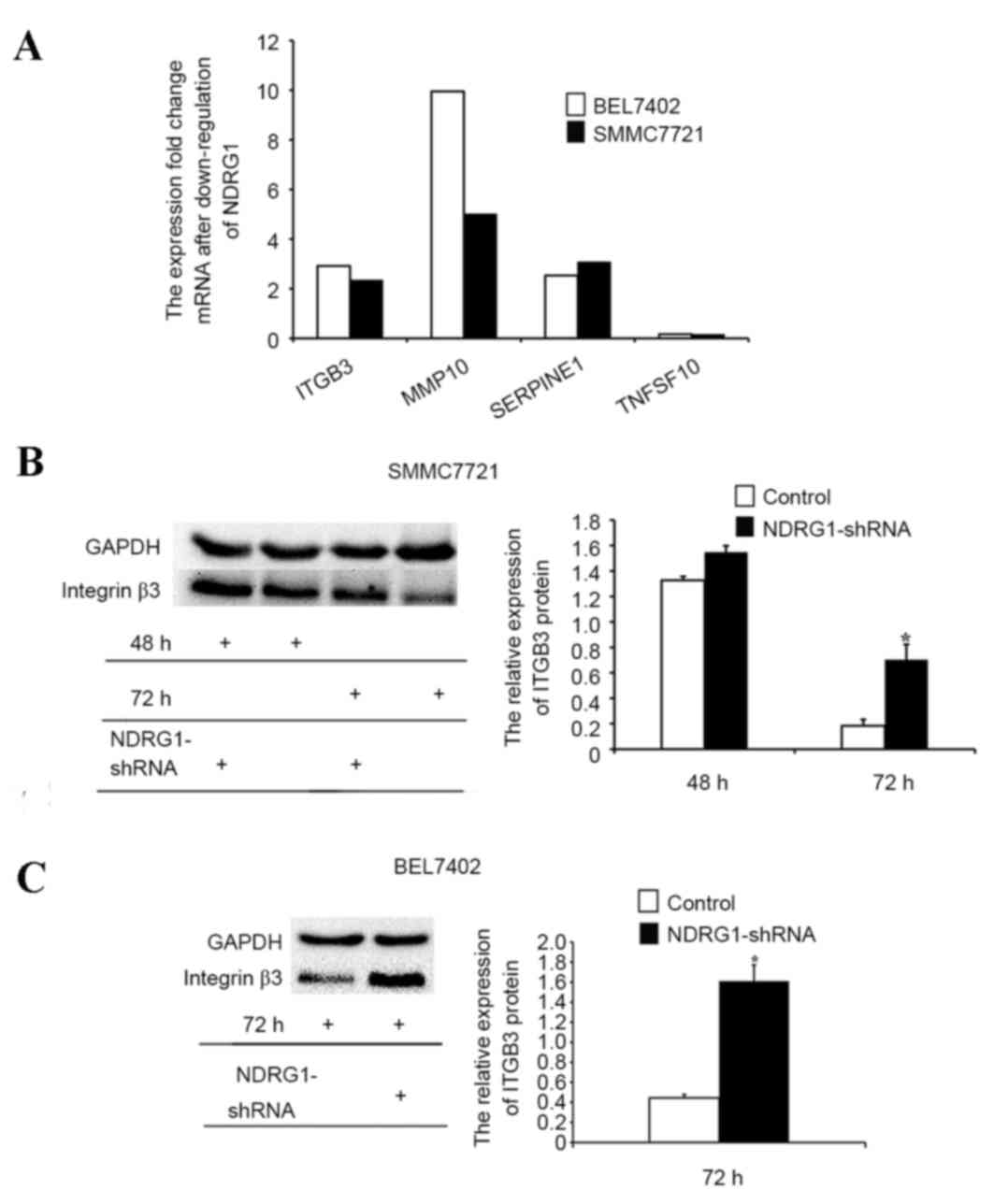
The PCR array assay was used to determine the
downstream gene candidates of NDRG1 that inhibited tumor
metastasis. The results demonstrated that NDRG1 knockdown
upregulated ITGB3, MMP10 and SERPINE1 and downregulated TNFSF10 in
both BEL7402 and SMMC7721 cell lines (Fig. 6A). Furthermore, western blot analysis
was used to determine ITGB3 protein expression. The results
demonstrated that ITGB3 protein expression was increased in
NDRG1-downregulated BEL7402 and SMMC7721 cells compared with that
in the control cells, indicating that the NDRG1 gene may suppress
tumor metastasis by regulating ITGB3 expression (P<0.05;
Fig. 6B and C).
Discussion
Currently, the function of the NDRG1 gene is
controversial (17–19); therefore, the present study
investigated the role of NDRG1 in HCC as well as its molecular
mechanism. Firstly, NDRG1 expression was detected in HCC tissue and
cells. The present results demonstrated that NDRG1 was expressed in
cancerous liver cells and adjacent degenerative liver cells. A
previous study observed that NDRG1 was expressed in 6% of cirrhosis
and benign liver lesions subsequent to staining (10). These results indicated that NDRG1 may
participate in the full progression of the occurrence and
development of HCC, from liver cell degeneration to malignant
changes. Although NDRG1 stained stronger on the membrane of liver
cancerous cells compared with that in the adjacent degenerative
liver cells, there was no staining difference in the cytoplasm or
nucleus. As mentioned previously, the expression of the NDRG1 gene
was always negative in normal liver cells (8–10,18). Therefore, the levels of NDRG1
expression in liver cell membrane may estimate the degree of injury
in cells or the extent of cell cancerization.
A previous study demonstrated that high NDRG1
expression in patients with HCC was associated with a short overall
survival rate and a poor prognosis. High NDRG1 expression was
detected in poorly differentiated HCC and high TNM stages (10). However, when the present study
considered NDRG1 expression in different subcellular localizations,
it identified that there was a negative correlation between the
cytoplasmic expression of NDRG1 in cancerous liver cells and the
TNM stage, which indicates that higher expression levels were
observed in smaller sized tumors. In our previous study, it was
revealed that the cytoplasmic expression of NDRG1 may be associated
with lymph node metastasis (27).
The expression of NDRG1 may therefore become a
marker of the degree of invasiveness for local tumors, although the
NDRG1 expression observed in the nucleus of the cancerous and
degenerative liver cells in the present study may be coincidental.
However, there was a positive correlation between the expression of
NDRG1 in the nucleus of cancerous liver cells and that in the
adjacent degenerative liver cells, indicating that the regulating
factor of NDRG1 nuclear expression acts equally on cancerous and
degenerative liver cells. However, WoLF PSORT (an advanced protein
subcellular localization prediction tool) and amino acid sequence
analysis demonstrated that the NDRG1 protein lacked the motifs used
for localization in nucleus (32).
Thus, NDRG1 nucleus localization may rely on protein
phosphorylation. Several phosphorylation sites have been identified
in the C-terminal site of the NDRG1 protein (33).
Reports have shown that serum- and
glucocorticoid-induced protein kinase 1 could phosphorylate NDRG1
on the amino acids Thr328, Ser330, Thr346, Thr356 and Thr366
(34,35). Glycogen synthesis kinase 3β (GSK3β)
was shown to be able to phosphorylate Ser342, Ser352 and Ser362 of
NDRG1 (36). These results are
valuable to understanding the role of NDRG1, since protein
phosphorylation is reversible and affects NDRG1 subcellular
localization in the cell (37). In
addition, NDRG1 can localize in the nucleus subsequent to binding
to the 70-kDa heat shock cognate protein (Hsc70) (38). Hsc70 is a molecular chaperone that
mediates mast cell transport between the cytoplasm and nucleus
(39).
NDRG1 was revealed to inhibit the proliferation and
invasion of BEL7402 cells, which facilitated the hypothesis that
NDRG1 expression levels may be lower in a cell line with a high
metastatic potential compared with those in a cell line with a low
metastatic potential. However, higher expression of NDRG1 mRNA was
detected in MHCC-97H cells compared with that in MHCC-97L cells. In
addition, higher expression of NDRG1 mRNA was detected in detached
BEL7402 cells compared with that in attached BEL7402 cells. Thus,
these findings were contrary to the expected results, and suggest
that NDRG1 overexpression may be the compensatory mechanism. To
clarify the expression trend of different metastatic potential
cells lines, the present study detected the NDRG1 mRNA expression
of colon carcinoma SW480 and SW620 cells. SW480 cells and SW620
cells were respectively derived from primary tumor and metastatic
lymph nodes of the same person, which may make clear the expression
of NDRG1 in primary and metastatic tissues. The result showed that
NDRG1 expression was lower in SW620 compared with SW480 cells,
which was contrary to the results of HCC cell lines. Maybe the
NDRG1 expression was tissue specific. It has been reported that the
level of RNA polymerase II bound to the NDRG1 promoter was lower in
SW620 cells compared with SW480 cells, which reduced histone H4
acetylation, and enhanced histone H3 Ser10 phosphorylation
(40). The unique histone
modifications may be the possible mechanism for the different
expression of NDRG1 in cell lines of different metastatic potential
(40).
A previous study identified that NDRG1 inhibited the
proliferation and invasion of cancer cells. In prostate cancer
cells, the aberrant methylation of NDRG1 CpG islands caused
downregulation of the NDRG1 promotor, leading to accelerated
cellular proliferation and invasion of cancer cells (41). To illustrate the function of NDRG1 in
HCC, the present study performed a series of in vitro
experiments. Knockdown of the NDRG1 gene increased the
proliferation of BEL7402 cells, indicating that NDRG1 inhibits the
proliferation of hepatoma cells in vitro, which is
consistent with the results of a previous study (17). However, a study in Hep3B and HepG2
cells revealed that the downregulation of NDRG1 decreased cell
growth (19). A previous study
indicated that NDRG1 directly interacted with GSK3β and Nur77 (also
known as NR4A1, nuclear receptor subfamily 4 group A member 1)
(42). This interaction could prevent
the degradation of β-catenin, and consequently regulate
β-catenin-relevant downstream signaling pathways to promote the
growth of Hep3B and HepG2 cells (42). A previous study demonstrated that
NDRG1 induced G0/G1-phase cell cycle arrest
in HCC cell lines, and may incorporate cell cycle regulators such
as p21 and cyclin-dependent kinase 4 in the NDRG1-induced cell
cycle arrest (17). The suppression
of NDRG1 inhibited tumor growth by inducing extensive cellular
senescence in HCC cells (17,43).
To confirm the controversial function of NDRG1 in
the metastasis of HCC, NDRG1 was knocked down using NDRG1-shRNA in
BEL7402 cells in the present study, demonstrating that NDRG1
downregulation promotes HCC cell invasion and metastasis. This
result indicates that NDRG1 inhibits the invasion and metastasis of
HCC cells. A previous study demonstrated that NDRG1 overexpression
maintained membrane E-cadherin and β-catenin levels while
inhibiting transforming growth factor (TGF)-β-stimulated cellular
migration and invasion (44), and
this finding indirectly confirmed the results of the present study.
A previous study demonstrated a potential mechanism by which NDRG1
effectively inhibited rat prostate cancer AT6.1 cells from
metastasizing to the lungs (45).
This mechanism may also regulate the cell structural protein actin.
Therefore, NDRG1 could affect the formation and regulation of actin
filaments by inhibiting the rho-associated protein kinase
1/phosphorylated myosin light chain 2 signaling pathway (46). A recent study has also demonstrated
that NDRG1 could suppress tumor metastasis by inhibiting the focal
adhesion kinase/paxillin signaling pathway (47).
To investigate the mechanism by which NDRG1 is
involved in the proliferation and metastasis of HCC, the present
study screened the downstream gene candidates regulated by NDRG1
that inhibited tumor metastasis. The results demonstrated that, in
the BEL7402 and SMMC7721 cell lines, a decrease in NDRG1 expression
led to an increase in ITGB3 expression. The expression levels of
the ITGB3 protein were detected using western blot analysis, and
were consistent with the PCR array results. A previous study has
demonstrated that integrin αvβ3 could increase adhesion and
invasion of HCC (48). ITGB3 could
enhance the TGF-β1/H2O2/HOCl-inducing
invasive ability of non-metastatic HCC cells via the TGF-β1
signaling pathway (49).
In addition, reducing the levels of integrin αvβ3
suppressed the cathepsin B-induced proliferation of HCC (50). These previous studies confirmed the
findings of the present study. Integrin signaling induced
cytoskeleton organization change and contraction, which
consequently promoted cellular migration. A potential mechanism by
which this happens could be that activated integrin αvβ3 was not
effective for the primary tumor growth, but it could collaborate
with platelets in order to promote tumor cell metastasis from the
bloodstream (51). To the best of our
knowledge, the association between NDRG1 and ITGB3 has not been
studied to date. The role of NDRG1 on the inhibition of tumor
metastasis and cellular proliferation was considered to occur via
the regulation of ITGB3. The microarray results from colon cancer
tissue demonstrated a negative correlation between NDRG1 expression
in the nucleus of cancerous cells and ITGB3 stromal expression in
adjacent degenerative cells. A negative correlation was also
revealed between NDRG1 expression in the nucleus of cancerous cells
and ITGB3 expression in cancer tissue. These results suggest that
NDRG1 is involved in regulating ITGB3 expression at the
transcriptional level (data not shown).
In conclusion, as a key signaling molecule
regulating tumor proliferation and metastasis, NDRG1 is likely to
become the target of anti-tumor metastasis or to be a biomarker
observing the prognosis and metastasis of patients with tumors. In
order to be considered as a drug target, additional studies are
required on NDRG1 in order to identify the molecular mechanism
behind its anti-tumor activity. The present study identified that
NDRG1 inhibited the proliferation and metastasis of HCC by
regulating ITGB3 expression at the transcriptional level. However,
the association between NDRG1 and ITGB3 requires additional
investigation through in vitro and in vivo
experiments.
Acknowledgements
The present study was supported by grants from the
Natural Science Foundation of China (grant no. 30873025), the
Natural Science Foundation of Shandong Province (grant nos.
ZR2010HM033 and ZR2014HM016) and the Science and Technology
Development of Shandong Province (grant no. 2016GSF201118).
Glossary
Abbreviations
Abbreviations:
|
NDRG1
|
N-myc downstream-regulated gene 1
|
|
HCC
|
hepatocellular carcinoma
|
|
ITGB3
|
integrin β3
|
|
CCK-8
|
Cell Counting Kit-8
|
|
MMP10
|
matrix metalloprotease 10
|
|
SERPINE1
|
serine protease inhibitor clade E
member 1
|
|
TNFSF10
|
tumor necrosis factor receptor
superfamily member 10
|
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bruix J and Llovet JM: Prognostic
prediction and treatment strategy in hepatocellular carcinoma.
Hepatology. 35:519–524. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aravalli RN, Steer CJ and Cressman EN:
Molecular mechanisms of hepatocellular carcinoma. Hepatology.
48:2047–2063. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
van Belzen N, Dinjens WN, Eussen BH and
Bosman FT: Expression of differentiation-related genes in
colorectal cancer: Possible implications for prognosis. Histol
Histopathol. 13:1233–1242. 1998.PubMed/NCBI
|
|
5
|
Thierry-Mieg D and Thierry-Mieg J:
AceView: A comprehensive cDNA-supported gene and transcripts
annotation. Genome Biol. 7:(Suppl 1). S12.1–14. 2006. View Article : Google Scholar
|
|
6
|
van Belzen N, Dinjens WN, Diesveld MP,
Groen NA, van der Made AC, Nozawa Y, Vlietstra R, Trapman J and
Bosman FT: A novel gene which is up-regulated during colon
epithelial cell differentiation and down-regulated in colorectal
neoplasms. Lab Invest. 77:85–92. 1997.PubMed/NCBI
|
|
7
|
Zhou D, Salnikow K and Costa M: Cap43, a
novel gene specifically induced by Ni2+ compounds.
Cancer Res. 58:2182–2189. 1998.PubMed/NCBI
|
|
8
|
Sibold S, Roh V, Keogh A, Studer P, Tiffon
C, Angst E, Vorburger SA, Weimann R, Candinas D and Stroka D:
Hypoxia increases cytoplasmic expression of NDRG1, but is
insufficient for its membrane localization in human hepatocellular
carcinoma. FEBS Lett. 581:989–994. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Akiba J, Ogasawara S, Kawahara A, Nishida
N, Sanada S, Moriya F, Kuwano M, Nakashima O and Yano H: N-myc
downstream regulated gene 1 (NDRG1)/Cap43 enhances portal vein
invasion and intrahepatic metastasis in human hepatocellular
carcinoma. Oncol Rep. 20:1329–1335. 2008.PubMed/NCBI
|
|
10
|
Chua MS, Sun H, Cheung ST, Mason V,
Higgins J, Ross DT, Fan ST and So S: Overexpression of NDRG1 is an
indicator of poor prognosis in hepatocellular carcinoma. Mod
Pathol. 20:76–83. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kurdistani SK, Arizti P, Reimer CL, Sugrue
MM, Aaronson SA and Lee SW: Inhibition of tumor cell growth by
RTP/rit42 and its responsiveness to p53 and DNA damage. Cancer Res.
58:4439–4444. 1998.PubMed/NCBI
|
|
12
|
Lee JC, Chung LC, Chen YJ, Feng TH and
Juang HH: N-myc downstream-regulated gene 1 downregulates cell
proliferation, invasiveness, and tumorigenesis in human oral
squamous cell carcinoma. Cancer Lett. 355:242–252. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kim-Fuchs C, Winterhalder S, Winter A,
Malinka T, Born D, Schäfer S, Stroka D, Gloor B, Candinas D and
Angst E: The silencing of N-myc downstream-regulated gene-1 in an
orthotopic pancreatic cancer model leads to more aggressive tumor
growth and metastases. Dig Surg. 31:135–142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu ZY, Xie WB, Yang F, Xiao LW, Wang XY,
Chen SY and Li ZG: NDRG1 attenuates epithelial-mesenchymal
transition of nasopharyngeal cancer cells via blocking Smad2
signaling. Biochim Biophys Acta. 1852:1876–1886. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chang X, Xu X, Ma J, Xue X, Li Z, Deng P,
Zhang S, Zhi Y, Chen J and Dai D: NDRG1 expression is related to
the progression and prognosis of gastric cancer patients through
modulating proliferation, invasion and cell cycle of gastric cancer
cells. Mol Biol Rep. 41:6215–6223. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hosoya N, Sakumoto M, Nakamura Y, Narisawa
T, Bilim V, Motoyama T, Tomita Y and Kondo T: Proteomics identified
nuclear N-myc downstream-regulated gene 1 as a prognostic tissue
biomarker candidate in renal cell carcinoma. Biochim Biophys Acta.
1834:2630–2639. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Akiba J, Murakami Y, Noda M, Watari K,
Ogasawara S, Yoshida T, Kawahara A, Sanada S, Yasumoto M, Yamaguchi
R, et al: N-myc downstream regulated gene1/Cap43 overexpression
suppresses tumor growth by hepatic cancer cells through cell cycle
arrest at the G0/G1 phase. Cancer Lett. 310:25–34. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cheng J, Xie HY, Xu X, Wu J, Wei X, Su R,
Zhang W, Lv Z, Zheng S and Zhou L: NDRG1 as a biomarker for
metastasis, recurrence and of poor prognosis in hepatocellular
carcinoma. Cancer Lett. 310:35–45. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yan X, Chua MS, Sun H and So S: N-Myc
down-regulated gene 1 mediates proliferation, invasion, and
apoptosis of hepatocellular carcinoma cells. Cancer Lett.
262:133–142. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Desgrosellier JS and Cheresh DA: Integrins
in cancer: Biological implications and therapeutic opportunities.
Nat Rev Cancer. 10:9–22. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tadokoro S, Tomiyama Y, Honda S, Kashiwagi
H, Kosugi S, Shiraga M, Kiyoi T, Kurata Y and Matsuzawa Y: Missense
mutations in the beta(3) subunit have a different impact on the
expression and function between alpha(IIb)beta(3) and
alpha(v)beta(3). Blood. 99:931–938. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Varner JA and Cheresh DA: Tumor
angiogenesis and the role of vascular cell integrin alphavbeta3.
Important Adv Oncol. 69–87. 1996.PubMed/NCBI
|
|
23
|
Brooks PC, Strömblad S, Klemke R, Visscher
D, Sarkar FH and Cheresh DA: Antiintegrin alpha v beta 3 blocks
human breast cancer growth and angiogenesis in human skin. J Clin
Invest. 96:1815–1822. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Petitclerc E, Strömblad S, von Schalscha
TL, Mitjans F, Piulats J, Montgomery AM, Cheresh DA and Brooks PC:
Integrin alpha(v)beta3 promotes M21 melanoma growth in human skin
by regulating tumor cell survival. Cancer Res. 59:2724–2730.
1999.PubMed/NCBI
|
|
25
|
Landen CN, Kim TJ, Lin YG, Merritt WM,
Kamat AA, Han LY, Spannuth WA, Nick AM, Jennnings NB, Kinch MS, et
al: Tumor-selective response to antibody-mediated targeting of
alphavbeta3 integrin in ovarian cancer. Neoplasia. 10:1259–1267.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kong Q, Wu G, Han L, Zhang Z, Du J, Sun W
and Cao L: A transfection method of PS-asODNs targeting ANGPTL4 in
multicellular structures of hepatocarcinoma cell line. Cancer Gene
Ther. 22:285–290. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Song Y, Lv L, Du J, Yue L and Cao L:
Correlation of N-myc downstream-regulated gene 1 subcellular
localization and lymph node metastases of colorectal neoplasms.
Biochem Biophys Res Commun. 439:241–246. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Homo sapiens N-myc downstream regulated 1
(NDRG1), transcript variant 3, mRNA. NCBI Reference Sequence:
NM_001258432.1. https://www.ncbi.nlm.nih.gov/nuccore/386643031
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cao L, Han L, Zhang Z, Li J, Qu Z, Du J,
Liang X, Liu Y, Liu H, Shi Y, et al: Involvement of
anoikis-resistance in the metastasis of hepatoma cells. Exp Cell
Res. 315:1148–1156. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Edge SB and Compton CC: The American joint
committee on cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Horton P, Park KJ, Obayashi T, Fujita N,
Harada H, Adams-Collier CJ and Nakai K: WoLF PSORT: Protein
localization predictor. Nucleic Acids Res. 35:(Web Server issue).
W585–W587. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bandyopadhyay S, Wang Y, Zhan R, Pai SK,
Watabe M, Iiizumi M, Furuta E, Mohinta S, Liu W, Hirota S, et al:
The tumor metastasis suppressor gene Drg-1 down-regulates the
expression of activating transcription factor 3 in prostate cancer.
Cancer Res. 66:11983–11990. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Inglis SK, Gallacher M, Brown SG, McTavish
N, Getty J, Husband EM, Murray JT and Wilson SM: SGK1 activity in
Na+ absorbing airway epithelial cells monitored by
assaying NDRG1-Thr346/356/366 phosphorylation. Pflugers Arch.
457:1287–1301. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hoang B, Frost P, Shi Y, Belanger E,
Benavides A, Pezeshkpour G, Cappia S, Guglielmelli T, Gera J and
Lichtenstein A: Targeting TORC2 in multiple myeloma with a new mTOR
kinase inhibitor. Blood. 116:4560–4568. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Murray JT, Campbell DG, Morrice N, Auld
GC, Shpiro N, Marquez R, Peggie M, Bain J, Bloomberg GB, Grahammer
F, et al: Exploitation of KESTREL to identify NDRG family members
as physiological substrates for SGK1 and GSK3. Biochem J.
384:477–488. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Segawa T, Nau ME, Xu LL, Chilukuri RN,
Makarem M, Zhang W, Petrovics G, Sesterhenn IA, McLeod DG, Moul JW,
et al: Androgen-induced expression of endoplasmic reticulum (ER)
stress response genes in prostate cancer cells. Oncogene.
21:8749–8758. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lachat P, Shaw P, Gebhard S, van Belzen N,
Chaubert P and Bosman FT: Expression of NDRG1, a
differentiation-related gene, in human tissues. Histochem Cell
Biol. 118:399–408. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sugiki T, Taketomi Y, Kikuchi-Yanoshita R,
Murakami M and Kudo I: Association of N-myc downregulated gene 1
with heat-shock cognate protein 70 in mast cells. Biol Pharm Bull.
27:628–633. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li Q and Chen H: Transcriptional silencing
of N-Myc downstream-regulated gene 1 (NDRG1) in metastatic colon
cancer cell line SW620. Clin Exp Metastasis. 28:127–135. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ma W, Na M, Tang C, Wang H and Lin Z:
Overexpression of N-myc downstream-regulated gene 1 inhibits human
glioma proliferation and invasion via phosphoinositide 3-kinase/AKT
pathways. Mol Med Rep. 12:1050–1058. 2015.PubMed/NCBI
|
|
42
|
Lu WJ, Chua MS, Wei W and So SK: NDRG1
promotes growth of hepatocellular carcinoma cells by directly
interacting with GSK-3β and Nur77 to prevent β-catenin degradation.
Oncotarget. 6:29847–29859. 2015.PubMed/NCBI
|
|
43
|
Lu WJ, Chua MS and So SK: Suppressing
N-Myc downstream regulated gene 1 reactivates senescence signaling
and inhibits tumor growth in hepatocellular carcinoma.
Carcinogenesis. 35:915–922. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen Z, Zhang D, Yue F, Zheng M, Kovacevic
Z and Richardson DR: The iron chelators Dp44mT and DFO inhibit
TGF-β-induced epithelial-mesenchymal transition via up-regulation
of N-Myc downstream-regulated gene 1 (NDRG1). J Biol Chem.
287:17016–17028. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Bandyopadhyay S, Pai SK, Gross SC, Hirota
S, Hosobe S, Miura K, Saito K, Commes T, Hayashi S, Watabe M and
Watabe K: The Drg-1 gene suppresses tumor metastasis in prostate
cancer. Cancer Res. 63:1731–1736. 2003.PubMed/NCBI
|
|
46
|
Sun J, Zhang D, Zheng Y, Zhao Q, Zheng M,
Kovacevic Z and Richardson DR: Targeting the metastasis suppressor,
NDRG1, using novel iron chelators: Regulation of stress
fiber-mediated tumor cell migration via modulation of the
ROCK1/pMLC2 signaling pathway. Mol Pharmacol. 83:454–469. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wangpu X, Lu J, Xi R, Yue F, Sahni S, Park
KC, Menezes S, Huang ML, Zheng M, Kovacevic Z and Richardson DR:
Targeting the metastasis suppressor, N-Myc downstream regulated
gene-1, with novel Di-2-pyridylketone thiosemicarbazones:
Suppression of tumor cell migration and cell-collagen adhesion by
inhibiting focal adhesion kinase/paxillin signaling. Mol Pharmacol.
89:521–540. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tang NH, Chen YL, Wang XQ, Li XJ, Wu Y,
Zou QL and Chen YZ: N-terminal and C-terminal heparin-binding
domain polypeptides derived from fibronectin reduce adhesion and
invasion of liver cancer cells. BMC Cancer. 10:5522010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Feng XX, Liu M, Yan W, Zhou ZZ, Xia YJ, Tu
W, Li PY and Tian DA: β3 integrin promotes
TGF-β1/H2O2/HOCl-mediated induction of metastatic phenotype of
hepatocellular carcinoma cells by enhancing TGF-β1 signaling. PLoS
One. 8:e798572013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xu ZZ, Xiu P, Lv JW, Wang FH, Dong XF, Liu
F, Li T and Li J: Integrin alphavbeta3 is required for cathepsin
B-induced hepatocellular carcinoma progression. Mol Med Rep.
11:3499–3504. 2015.PubMed/NCBI
|
|
51
|
Weber MR, Zuka M, Lorger M, Tschan M,
Torbett BE, Zijlstra A, Quigley JP, Staflin K, Eliceiri BP, Krueger
JS, et al: Activated tumor cell integrin αβ3 cooperates with
platelets to promote extravasation and metastasis from the blood
stream. Thromb Res. 140:(Suppl 1). S27–S36. 2016. View Article : Google Scholar : PubMed/NCBI
|