Introduction
Cervical cancer (CC) is one of the most common solid
tumors in females worldwide, and has a high mortality rate
(1–3)
due to asymptomatic development of the disease delaying diagnosis
(4,5).
CC is a gynecological malignancy associated with oncogenic human
papillomavirus (HPV) infection (6,7). In
addition to HPV infection, other factors affect the development of
CC, including immunological disorders and genetic malfunctions such
as point mutations, deletions, amplifications and rearrangements of
DNA (6). Previous studies have
suggested that epigenetic changes may significantly impact cervical
carcinogenesis (6–9).
Epigenetic alterations are heritable traits that
impact the regulation of gene expression without altering the DNA
sequence (10). These traits control
genetic and transcriptional activity during growth, differentiation
or organism adaptation to environmental changes (6). One epigenetic mechanism of DNA
methylation consists of cytosine methylation in
cytosine-phosphate-guanine (CpG) dinucleotide islands, located in
the promoter region of numerous genes (6,11,12). During malignant transformation, CpG
islands become hypermethylated, silencing the expression of
suppressor genes and leading to a loss in the control of cell
proliferation (13,14). By contrast, the hypomethylation of
oncogenes increases cell division and enhances the metastasis of
cancer cells (14). The process of
methylation has been well characterized in recent years, but the
underlying mechanism of demethylation, particularly during
carcinogenesis, remains to be elucidated (6,15,16). Ten-eleven translocation (TET) proteins
have an important role in DNA demethylation, with reduced
expression observed in various tumors (6,13,17–22).
The TET protein family includes TET1, TET2 and TET3
(10,23). The TET1 and TET3 proteins use the CXXC
zinc motif to bind to5-methylcytosine (5-mC) in CpG islands
(16,17,24). The
TET proteins have been revealed to catalyze the oxidation of 5-mC
to 5-hydroxymethylcytosine (5-hmC) (25). Subsequently, 5-hmC is oxidized to
5-formylocytosine (5-fC) and 5-carboxycytosine (5-caC), eventually
converting 5-mC to cytosine (13,17,26). This
transformation may contribute to unlocking the promoter regions of
suppressor genes and facilitating the development of cancer
(8,13,16). Low
TET expression levels are correlated with decreased 5-hmC levels in
malignant tissues and with clinicopathological features in various
primary cancer tissues (13,17,20,22,27).
However, little is understood regarding the levels of TET
expression in cervical cancerous and non-cancerous tissue.
Therefore, the present study evaluated the expression levels of
TET1, TET2 and TET3 transcripts in cervical cancerous (n=80) and
non-cancerous (n=41) tissues. Furthermore, the TET1, TET2 and
TET3transcript levels were compared in patient groups stratified by
clinicopathological variables in primary CC and non-cancerous
cervical tissues.
Materials and methods
Patients and tissue samples
Primary CC tissue samples were collected following
surgical resection between June 2013 and August 2015 from 80 female
Caucasian patients, which is representative of the female Polish
population. Patients were treated at the Department of Radiotherapy
and Gynecological Oncology Greater Cancer Center (Poznań, Poland).
Non-cancerous cervical tissues were obtained from 41 women with
uterine fibroids undergoing uterine surgical resection in the
Division of Gynecological Surgery, Poznań University of Medical
Sciences (Poznań, Poland).
At the time of surgery, the mean age of patients in
the cancer and control groups was 58.6±11.4 and 49.9±9.0 years,
respectively. Of the 80 females in the study group, 11 patients
were classified as <45 years, 48 were aged 45–60 and 21 were
aged >60 years. Among the 41 women in the control group, 12
patients were classified as <45 years of age, 24 were aged 45–60
and 5 were >60 years. Among the 80 patients with CC, 4 patients
were classified as stage I, 26 as stage II, 43 as stage III and 7
as stage IV, based on the International Federation of Gynecology
and Obstetrics (FIGO) classification system and World Health
Organization (28). Cancerous and
non-cancerous cervical tissue samples were obtained following
protocol approval by the Local Ethics Committee of Poznań
University of Medical Sciences. Oral and written informed consent
were obtained from all participants in the study. A portion of the
tissue sample was immediately snap-frozen in liquid nitrogen and
stored at −80°C until RNA isolation. The remaining portion was used
for histopathological assessment, which was performed by an
experienced pathologist (Greater Poland Cancer Centre, Poznań,
Poland).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis of TET transcript
levels
Frozen tissue was homogenized and total RNA was
isolated according to the protocol of Chomczyński and Sacchi
(29). RNA quality was determined
spectrophotometrically using a BioPhotometer® from
Eppendorf AG (Hamburg, Germany) and 2% agarose gel electrophoresis.
RNA samples were reverse transcribed to cDNA using Moloney Murine
Leukemia Virus (M-MLV) reverse transcriptase (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) according to the
manufacturer's protocol. RT-qPCR was performed using a Light
Cycler1480 real-time PCR detection system (Roche Diagnostics GmbH,
Mannheim, Germany) using EvaGreen® (Solis BioDyne,
Tartu, Estonia) as the detection dye. The thermal cycling
conditions were as follows: 15 min activation, followed by 40
cycles consisting of 10 sec denaturation at 95°C, 10 sec annealing
at 58°C, 10 sec at 72°C. The transcript levels for patients and
controls were quantified by the relative quantification method
using a calibrator, which is a standard curve described in the
Relative Quantification Manual, Roche Diagnostics GmbH (Mannheim,
Germany). The calibrator was prepared as a cDNA mix from all
samples. For amplification, 1 µl (total 20 µl) of cDNA using 9 µl
of 5X HOT FIREPol® EvaGreen® qPCR Mix Plus
(no ROX) (Solis BioDyne) was used. Primer sequences are presented
in Table I. A total of 1 µl of 10 µM
primer was used per reaction. All analyses included a negative
control without cDNA, each experiment was repeated three times for
all samples. The quantity of TET1, TET2 and TET3 transcripts in
each sample was corrected by the measurement of porphobilinogen
deaminase cDNA levels and expressed as a multiple of the copies in
the calibrator.
 | Table I.Primers used in reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I.
Primers used in reverse
transcription-quantitative polymerase chain reaction analysis.
| Transcript | Forward (5′-3′) | Reverse (5′-3′) | Product size, bp | UCSC position
(GRCh37/hg19) of genes |
|---|
| TET1 |
ATACAATGGGCACCCTACCG |
GGGCTTGGGCTTCTACCAAA | 159 | chr10:70 320 117-70
454 239 |
| TET2 |
GCTGACAAACTCTACTCGG |
CTTCTGGCAAACTTACATCC | 188 | chr4:106 067 842-106
200 960 |
| TET3 |
CCCAAAGAGGAAGAAGTG |
GCAGTCAATCGCTATTTC | 129 | chr2:74 273 405-74
335 302 |
| PBGD |
GCCAAGGACCAGGACATC |
TCAGGTACAGTTGCCCATC | 160 | chr11:118 468 348–118
468 864 |
Statistical analysis
Statistical analysis was performed with STATISTICA
version 12 software (StatSoft, Inc., Tulsa, OK, USA) and Cytel
Studio version 10.0 (Cytel Software Corporation, Cambridge, MA,
USA). The data were presented as mean ± standard deviation and
median with range. For the comparison of variables with a normal
distribution, the unpaired t-test was used; in other cases, the
non-parametric Mann-Whitney U test or the Kruskal-Wallis test was
used to calculate statistically significant differences between the
compared mean values. P<0.05 was considered to indicate a
statistically significant difference.
Results
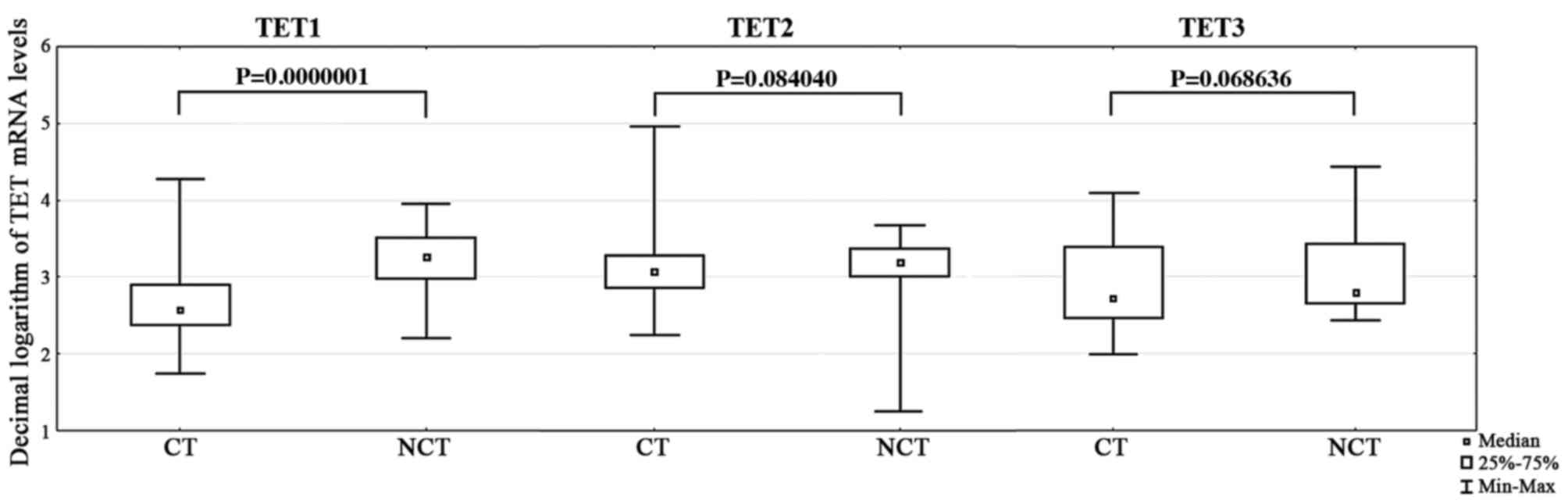
Significantly fewer TET1 transcripts (P=0.0000001)
were present in primary CC tissue from all patients, compared with
non-cancerous tissue from the control group (Table II; Fig.
1). However, there were no significant differences in TET2
(P=0.084040) and TET3 (P=0.068636) transcript levels between these
groups (Tables III and IV; Fig. 1).
Stratification of patients based on age, the FIGO classification
system, grade of differentiation and histological features was
performed to evaluate the differences in the TET1, TET2 and TET3
transcript levels between cancerous and non-cancerous tissues.
Significantly fewer TET1 transcripts were observed in stage I
(P=0.016), II (P<0.0001), III (P=0.00007), grade of
differentiationG1 (P=0.026), G2 (P=0.00006), G3 (P=0.0007) and Gx
(P=0.0004) and squamous histological type (P<0.00001) cervical
tissue samples, compared with non-cancerous tissue (Table II). TET1 transcript levels were
significantly higher in CC tissue samples from patients aged 45–60
(P=0.0002) and patients aged >60 years (P=0.003), compared with
controls (Table II). Furthermore,
lower TET2 transcript levels were observed in CC tissue samples
characterized as stage II (P=0.043), and lower TET3 transcript
levels were detected in samples characterized as stage III
(P=0.010), with a grade of differentiation of G3 (P=0.025) and a
squamous histological type (P=0.047), compared with samples from
the non-cancerous control group (Tables
III and IV). However, there were
no significant differences between TET1, TET2 and TET3 transcript
levels for I vs. II, III or IV, II vs. III or IV and III vs. IV
FIGO stage (data not shown). Additionally, there were no
significant differences between these transcript levels for G1 vs.
G2, G3 or Gx, G2 vs. G3 or Gx and G3 vs. Gx for grade of
differentiation, as well as for the histological type (data not
shown).
 | Table II.Statistical analysis of TET1
transcript levels in cervical cancer and non-cancerous tissues from
patients stratified by age, FIGO stage, grade of differentiation
and histological type of cancer. |
Table II.
Statistical analysis of TET1
transcript levels in cervical cancer and non-cancerous tissues from
patients stratified by age, FIGO stage, grade of differentiation
and histological type of cancer.
|
| No. of cases | Transcript | P-value |
|---|
|
|
|
|
|
|---|
| Variables | Patients (%) | Controls (%) | Cancerous tissue,
median (range) | Non-cancerous tissue,
median (range) | Patients vs.
controlse | Patients vs.
patientsf |
|---|
| Total no. of
cases | 80 | 41 | 2.563 | 3.262 |
0.0000001c | – |
|
|
|
| (1.740–4.740) | (2.212–3.212) |
|
|
| Agea |
|
|
|
|
|
|
|
>45 | 11 (13.75) | 13 (31.70) | – | – |
0.157000b | – |
|
45–60 | 31(38.75) | 23 (56.10) | 2.522 | 3.378 |
0.000200c | – |
|
|
|
| (1.740–4.740) | (2.441–3.441) |
|
|
|
>60 | 38 (47.50) | 5 (12.20) | 2.561 | 3.523 |
0.003000c | – |
|
|
|
| (2.033–3.033) | (2.997–3.997) |
|
|
| FIGO stage |
|
|
|
|
|
|
| I | 4 (5.00) | – | 2.385 |
|
0.016000c |
0.380176d |
|
|
|
| (2.272–3.272) |
|
|
|
| II | 26 (32.50) | – | 2.507 |
|
<0.000100c |
|
|
|
|
| (1.999–3.999) |
|
|
|
|
III | 43 (53.75) | – | 2.578 |
|
0.000070c |
|
|
|
|
| (1.740–4.740) |
|
|
|
| IV | 7 (8.75) | – | 2.827 |
|
0.075000c |
|
|
|
|
| (2.096–3.096) |
|
|
|
| Grade of
differentiation |
|
|
|
|
|
|
| G1 | 5 (6.25) | – | 2.412 | 3.262 |
0.026000c |
0.464650d |
|
|
|
| (2.033–3.033) | (2.212–3.212) |
|
|
| G2 | 36 (45.00) | – | 2.582 |
|
0.000060c |
|
|
|
|
| (2.074–3.074) |
|
|
|
| G3 | 11 (13.75) | – | 2.418 |
|
0.000700c |
|
|
|
|
| (1.999–3.999) |
|
|
|
|
Gx | 28 (35.00) | – | 2.577 |
|
0.000400c |
|
|
|
|
| (1.740–4.740) |
|
|
|
| Histological
type |
|
|
|
|
|
|
|
Squamous | 78 (97.50) | – | 2.564 |
|
<0.000010c |
|
|
|
| (1.740–4.740) |
|
|
|
|
Adenocarcinoma | 2 (2.50) | – | 3.394 |
|
0.840000c |
0.295441c |
|
|
|
| (2.514–4.514) |
|
|
|
 | Table III.Statistical analysis of TET2
transcript levels in cervical cancer and non-cancerous tissues from
patients including age, FIGO stage, grade of differentiation and
histological type of cancer. |
Table III.
Statistical analysis of TET2
transcript levels in cervical cancer and non-cancerous tissues from
patients including age, FIGO stage, grade of differentiation and
histological type of cancer.
|
| No. of cases | Transcript | P-value |
|---|
|
|
|
|
|
|---|
|
|
| Variables | Patients (%) | Controls (%) | Cancerous tissue,
median (range) | Non-cancerous
tissue, median (range) | Patients vs.
controlse | Patients vs.
patientsf |
|---|
| Total no. of
cases | 80 | 41 | 3.075 | 3.185 |
0.084040c | – |
|
|
|
| (2.246–4.246) | (1.256–3.256) |
|
|
| Agea |
|
|
|
|
|
|
| >45 | 11 (13.75) | 13 (31.70) | – | – |
0.763000b | – |
|
45–60 | 31 | 23 | 3.075 | 3.185 |
0.239000c | – |
|
| (38.75) |
(56.10) | (2.246–4.246) | (1.256–3.256) |
|
>60 | 38 (47.50) | 5 (12.20) | 3.099 | 3.243 |
0.116000c | – |
|
|
|
| (2.362–4.362) | (3.122–3.122) |
| FIGO stage |
|
|
|
|
|
|
| I | 4 (5.00) | – | 3.077 |
|
0.735000c |
0.804017d |
|
|
|
| (2.887–3.887) |
|
| II | 26 (32.50) | – | 3.015 |
|
0.043000c |
|
|
|
|
| (2.489–4.489) |
|
|
|
|
III | 43 (53.75) | – | 3.100 |
|
0.256000c |
|
|
|
|
| (2.246–4.246) |
|
|
|
| IV | 7 (8.75) | – | 3.086 |
|
0.539000c |
|
|
|
|
| (2.625–3.625) |
|
|
|
| Grade of
differentiation |
|
|
|
|
|
|
| G1 | 5 (6.25) | – | 2.899 | 3.185 |
0.230040c |
0.809431d |
|
|
|
| (2.647–4.647) | (1.256–3.256) |
|
|
| G2 | 36 (45.00) | – | 3.095 |
|
0.264000c |
|
|
|
|
| (2.362–4.362) |
|
|
|
| G3 | 11 (13.75) | – | 3.029 |
|
0.093000c |
|
|
|
|
| (2.489–3.489) |
|
|
|
|
Gx | 28 (35.00) | – | 3.099 |
|
0.243000c |
|
|
|
|
| (2.246–3.246) |
|
|
|
| Histological
type |
|
|
|
|
|
|
|
Squamous | 78 (97.50) | – | 3.075 |
|
0.085000c |
|
|
|
|
| (2.246–4.246) |
|
|
|
|
Adenocarcinoma | 2 (2.50) | – | 3.089 |
|
|
0.902289c |
|
|
|
| (2.833–3.833) |
|
0.708000c |
|
 | Table IV.Statistical analysis of TET3
transcript levels in cervical cancer and non-cancerous tissues
according to age, FIGO stage, grade of differentiation and
histological type of cancer. |
Table IV.
Statistical analysis of TET3
transcript levels in cervical cancer and non-cancerous tissues
according to age, FIGO stage, grade of differentiation and
histological type of cancer.
|
| No. of cases | Transcript | P-value |
|---|
|
|
|
|
|
|---|
| Variables | Patients (%) | Controls (%) | Cancerous tissue,
median (range) | Non-cancerous
tissue, median (range) | Patients vs.
controlse | Patients vs.
patientsf |
|---|
| Total no. of
cases | 80 | 41 | 2.720 | 2.803 |
0.068636b | – |
|
|
|
| (1.985–4.985) | (2.443–4.443) |
|
|
| Agea |
|
|
|
|
|
|
|
>45 | 11 (13.75) | 13 (31.70) | 2.782 | 2.294 |
0.242000b | – |
|
|
|
| (2.341–3.341) | (2.443–4.443) |
|
|
|
45–60 | 31 (38.75) | 23 (56.10) | 2.675 | 2.761 |
0.077000b | – |
|
|
|
| (2.055–3.055) | (2.446–4.446) |
|
|
|
>60 | 38 (47.50) | 5 (12.20) | 2.742 | 2.725 |
0.955000b | – |
|
|
|
| (1.986–4.986) | (2.616–3.616) |
|
|
| FIGO stage |
|
|
|
|
|
|
| I | 4 (5.00) | – | 3.061 |
|
0.984000b |
0.222237c |
|
|
|
| (2.617–3.617) |
|
|
| II | 26 (32.50) | – | 2.912 |
|
|
|
|
|
|
| (2.268–4.268) |
|
0.676000b |
|
|
III | 43 (53.75) | – | 2.621 |
|
|
|
|
|
|
| (1.986–4.986) |
|
0.010000b |
|
| IV | 7 (8.75) | – | 2.904 |
|
|
|
|
|
|
| (2.368–3.368) |
|
0.726000b |
|
| Grade of
differentiation |
|
|
|
|
|
|
| G1 | 5 (6.25) | – | 2.396 | 2.803 |
0.138000b |
0.498179c |
|
|
|
| (2.331–3.331) | (2.443–4.443) |
|
|
| G2 | 36 (45.00) | – | 2.846 |
|
0.453000b |
|
|
|
|
| (1.994–4.994) |
|
|
|
| G3 | 11 (13.75) | – | 2.680 |
|
0.025000b |
|
|
|
|
| (2.341–3.341) |
|
|
|
|
Gx | 28 (35.00) | – | 2.756 |
|
0.158000b |
|
|
|
|
| (1.986–3.986) |
|
|
|
| Histological
type |
|
|
|
|
|
|
|
Squamous | 78 (97.50) | – | 2.703 |
|
0.047000b |
0.109049b |
|
|
|
| (1.986–4.986) |
|
|
|
|
Adenocarcinoma | 2 (2.50) | – | 3.542 |
|
0.215000b |
|
|
|
|
| (3.466–3.466) |
|
|
|
Discussion
The involvement of TET1, TET2 and TET3 proteins in
active demethylation at CpG islands in DNA has been previously
documented (14,16,30). A
previous study reported that the three mouse Tet proteins (Tet1,
Tet2 and Tet3) may catalyze a similar reaction (12). TET1 is crucial for mouse embryonic
stem (ES) cell maintenance via the regulation of methylation and
the expression of the gene Nanog, which encodes a
transcription factor essential for self-renewal of undifferentiated
ES cells (12). A loss of TET
proteins in ES cells has been demonstrated to be involved in the
maintenance of DNA methylation patterns at several other DNA
methylation regions (31).
The role of TET proteins in malignant transformation
has been reported in animal models (12,32).
Removal of TET function induces the development of aggressive
myeloid leukemia in a mouse model (32). TET1 expression is responsible for DNA
methylation of tissue inhibitors of metalloproteinase proteins 2
and 3 (TIMP2, TIMP3) in prostate and breast cancer (19). Reduced levels of TET1, TET2 and TET3
have been associated with decreased 5-hmC levels in human breast,
liver, lung, pancreatic and prostate cancer compared with the
surrounding non-cancerous tissue (20). Decreased TET1 expression levels
correspond to reduced 5-hmC levels in breast, prostate and
hepatocellular cancer compared with normal tissue (19,22). Du
et al (27) demonstrated that
the loss of 5-hmC in tumors is correlated with the downregulation
of TET1 expression.
Rawłuszko-Wieczorek et al (13) observed reduced levels of TET1, TET2
and TET3 mRNA in in colorectal cancer tissue compared with
non-cancerous tissue. The decreased TET1, TET2 and TET3 mRNA levels
were associated with various groups, including age, gender, cancer
localization, histological grade, tumor node and metastasis
classification. Furthermore, this study also demonstrated that
patients with elevated TET2 transcript levels have more favorable
overall survival (13). Another
previous study demonstrated significantly lower levels of TET1
transcripts and protein in gastric cancer, which was correlated
with gender, age and certain clinicopathological features including
tumor localization, depth of invasion, lymph node metastasis,
histological grade and histological type (17). Du et al (27) used liquid chromatography mass
spectrometry/mass spectrometry to demonstrate very low levels of
5-fC and 5-caC and decreased levels of 5-hmC in gastric cancer
tissue compared with adjacent non-cancerous tissue. In addition,
the authors revealed that the reduction of 5-hmC in gastric cancer
was primarily associated with decreased TET1 expression (27). Using immunochemistry analysis, Müller
et al (21) demonstrated that
the exclusion of TET1 from nuclei was associated with a loss of
5-hmC in the genomic DNA of gliomas. The depletion of TET1 in
prostate and breast cancer tissues has also been observed. TET1
deficiency promotes tumor growth, cell invasion and cancer
metastasis in prostate xenograft mouse models (19). Furthermore, TET1 reduction corresponds
to a poor survival outcome in patients with breast cancer (19). Decreased TET1 levels are responsible
for maintaining the methylation of TIMP2 or TIMP3, which correlates
with advanced node status in clinical samples (19).
In conclusion, to the best of our knowledge the
current study is the first to demonstrate a significant reduction
in the levels of TET1 transcripts in cancerous tissues compared
with non-cancerous samples. In addition, TET1, TET2 and TET3
transcript levels were revealed to be reduced in patients with
primary CC stratified according to their clinicopathological data,
in comparison with non-cancerous tissues. The present study did not
evaluate TET protein in conjunction with 5-hmC levels. Therefore,
further studies are required to evaluate the potential correlation
between 5-hmC levels and TET expression in CC tissues, and their
associations with clinical characteristics.
Acknowledgements
The authors would like to thank the patients
enrolled in the study for their participation. In addition, the
authors would like to acknowledge Hanna Drzewiecka and
BartoszSłowikowski (both Poznań University of Medical Sciences,
Poznań, Poland) or their invaluable assistance. This study was
supported by Poznań University of Medical Sciences, Poland (grant
no. 502-01-01124182-07474).
References
|
1
|
Deręgowska J: Women with breast cancer in
the network of social support-a quantitative context. Nowiny
Lekarskie. 81:203–213. 2012.
|
|
2
|
Wang X, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and
tumor-suppressive MicroRNAs in cervical cancer is required for
cancer cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Estimating the world cancer burden: Globocan 2000. Int J Cancer.
94:153–156. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ibeanu OA: Molecular pathogenesis of
cervical cancer. Cancer Biol Ther. 11:295–306. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Asih TS, Lenhart S, Wise S, Aryati L,
Adi-Kusumo F, Hardianti Ms and Forde J: The dynamics of Hpv
infection and cervical cancer cells. Bull Math Biol. 78:4–20. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jiménez-Wences H, Peralta-Zaragoza O and
Fernández-Tilapa G: Human papilloma virus, DNA methylation and
microRNA expression in cervical cancer (Review). Oncol Rep.
31:2467–2476. 2014.PubMed/NCBI
|
|
7
|
Faridi R, Zahra A, Khan K and Idrees M:
Oncogenic potential of Human Papillomavirus (HPV) and its relation
with cervical cancer. Virol J. 8:2692011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fang J, Zhang H and Jin S: Epigenetics and
cervical cancer: From pathogenesis to therapy. Tumour Biol.
35:5083–5093. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang X, Zhang L, Tian C, Yang L and Wang
Z: Genetic variants and risk of cervical cancer: Epidemiological
evidence, meta-analysis and research review. BJOG. 121:664–674.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li D, Guo B, Wu H, Tan L and Lu Q: TET
family of dioxygenases: crucial roles and underlying mechanisms.
Cytogenet Genome Res. 146:171–180. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Abdel-Wahab O, Mullally A, Hedvat C,
Garcia-Manero G, Patel J, Wadleigh M, Malinge S, Yao J, Kilpivaara
O, Bhat R, et al: Genetic characterization of TET1, TET2 and TET3
alterations in myeloid malignancies. Blood. 114:144–147. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ito S, D'Alessio AC, Taranova OV, Hong K,
Sowers LC and Zhang Y: Role of tet proteins in 5mC to 5hmC
conversion, ES-cell self-renewal and inner cell mass specification.
Nature. 466:1129–1133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rawłuszko-Wieczorek AA, Siera A, Horbacka
K, Horst N, Krokowicz P and Jagodziński PP: Clinical significance
of DNA methylation mRNA levels of TET family members in colorectal
cancer. J Cancer Res Clin Oncol. 141:1379–1392. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rawłuszko-Wieczorek AA, Siera A and
Jagodziński PP: TET proteins in cancer: Current ‘state of the art’.
Crit Rev Oncol Hematol. 96:425–436. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saavedra KP, Brebi PM and Roa JC:
Epigenetic alterations in preneoplastic and neoplastic lesions of
the cervix. Clin Epigenetics. 4:132012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Głowacki S and Błasiak J: Role of
5-hydroxymethylcytosine and TET proteins in epigenetic regulation
of gene expression. Postepy Biochem. 59:64–69. 2013.(In Polish).
PubMed/NCBI
|
|
17
|
Frycz BA, Murawa D, Borejsza-Wysocki M,
Marciniak R, Murawa P, Drews M, Kołodziejczak A, Tomela K and
Jagodziński PP: Decreased expression of ten-eleven translocation 1
protein is associated with some clinicopathological features in
gastric cancer. Biomed Pharmacother. 68:209–212. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kudo Y, Tateishi K, Yamamoto K, Yamamoto
S, Asaoka Y, Ijichi H, Nagae G, Yoshida H, Aburatani H and Koike K:
Loss of 5-hydroxymethylcytosine is accompanied with malignant
cellular transformation. Cancer Sci. 103:670–676. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hsu CH, Peng KL, Kang ML, Chen YR, Yang
YC, Tsai CH, Chu CS, Jeng YM, Chen YT, Lin FM, et al: TET1
suppresses cancer invasion by activating the tissue inhibitors of
metalloproteinases. Cell Rep. 2:568–579. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu
J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al: Tumor development is
associated with decrease of TET gene expression and
5-methylcytosine hydroxylation. Oncogene. 32:663–669. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Müller T, Gessi M, Waha A, Isselstein LJ,
Luxen D, Freihoff D, Freihoff J, Becker A, Simon M, Hammes J, et
al: Nuclear exclusion of TET1 is associated with loss of
5-hydroxymethylcytosine in IDH1 wild-type gliomas. Am J Pathol.
181:675–683. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu C, Liu L, Chen X, Shen J, Shan J, Xu
Y, Yang Z, Wu L, Xia F, Bie P, et al: Decrease of
5-hydroxymethylcytosine is associated with progression of
hepatocellular carcinoma through downregulation of TET1. PLoS One.
8:e628282013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Afanas'ev I: Mechanisms of superoxide
signaling in epigenetic processes: Relation to aging and cancer.
Aging Dis. 6:216–227. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fan M, He X and Xu X: Restored expression
levels of TET1 decrease the proliferation and migration of renal
carcinoma cells. Mol Med Rep. 12:4837–4842. 2015.PubMed/NCBI
|
|
25
|
Tahiliani M, Koh KP, Shen Y, Pastor WA,
Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L and
Rao A: Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in
mammalian DNA by MLL partner TET1. Science. 324:930–935. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guz J, Jurgowiak M and Oliński R:
Oxidation and deamination of nucleobases as an epigenetic tool.
Postepy Hig Med Dosw (Online). 66:275–286. 2012.(In Polish).
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Du C, Kurabe N, Matsushima Y, Suzuki M,
Kahyo T, Ohnishi I, Tanioka F, Tajima S, Goto M, Yamada H, et al:
Robust quantitative assessments of cytosine modifications and
changes in the expressions of related enzymes in gastric cancer.
Gastric Cancer. 18:516–525. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Böcker W: WHO classification of breast
tumors and tumors of the female genital organs: Pathology and
genetics. Verh Dtsch Ges Pathol. 86:116–119. 2002.(In German).
PubMed/NCBI
|
|
29
|
Chomczynski P and Sacchi N: Single-step
method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction. Anal Biochem.
162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q,
Ding J, Jia Y, Chen Z, Li L, et al: Tet-mediated formation of
5-carboxylcytosine and its excision by TDG in mammalian DNA.
Science. 333:1303–1307. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu L, Mao SQ, Ray C, Zhang Y, Bell FT, Ng
SF, Xu GL and Li X: Differential regulation of genomic imprinting
by TET proteins in embryonic stem cells. Stem Cell Res. 15:435–443.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
An J, González-Avalos E, Chawla A, Jeong
M, López-Moyado IF, Li W, Goodell MA, Chavez L, Ko M and Rao A:
Acute loss of TET function results in aggressive myeloid cancer in
mice. Nat Commun. 6:100712015. View Article : Google Scholar : PubMed/NCBI
|