Introduction
Human proviral integration site for Moloney murine
leukemia virus (Pim) is a proto-oncogene that was initially
recognized 20 years ago (1); further
studies have revealed that Pim encodes serine/threonine
kinase and functions as a signaling regulator through cellular
substrate phosphorylation (2,3). Pim participates in multiple biological
functions, including numerous cellular signaling pathways, and
regulates cell cycle progression, cell proliferation and
differentiation, and inhibits apoptosis. Recently, the association
between androgen receptor (AR) and the proto-oncogene,
serine/threonine kinase, Pim-1, has been shown to be strong,
and studies have indicated that Pim-1 is expressed in
androgen-dependent prostate cancer (ADPC) and castration-resistant
prostate cancer (CRPC) (4). However,
the response of Pim-1 to androgen deprivation therapy (ADT),
particularly the dynamic changes in animal models, has not
previously been established. In the current study, reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
enzyme-linked immunosorbent assay (ELISA) and immunohistochemistry
(IHC) were performed to detect the difference in expression levels
of Pim-1 in mice models, in which prostate cancer was simulated to
develop into CRPC during ADT. In addition, the present study
discusses the known molecular mechanisms of Pim-1 kinase in
prostate tumorigenesis and progression, providing the opportunities
for targeting Pim-1 as an alternative therapeutic method for
prostate cancer, particularly in CRPC cases.
Materials and methods
Lab preparations
A total of 32 male BALB/c nude mice were purchased
from the Resources Research and Development Center of the Institute
of Laboratory Animal, Chinese Academy of Medical Sciences (Beijing
HFK Bioscience Co., Ltd., Beijing China; animal certification no.
SCXK2009-0004). All procedures involving mice were approved by the
University Committee on Use and Care of Animals at the Tianjin
Medical University, China, and conform to all regulatory standards.
LNCaP cells were purchased from the American Type Culture
Collection (Manassas, VA, USA). Chloroform, isopropanol, RNase-free
ddH2O and TRIzol were purchased from Tiangen Biotech
Co., Ltd. (Beijing, China). cDNA synthesis and the qPCR kit were
purchased from Fermentas (Thermo Fisher Scientific, Inc.,
Pittsburgh, PA, USA). The following primers were purchased from
Shanghai Sangon Biotech, Co., Ltd. (Shanghai, China): Upstream,
5′-GCCTCAACTCCTCCCATAGATAC-3′ and downstream,
5′-GCGGCATTCAGCAGAACTCAT-3′ for Pim-1 (product length, 147 bp);
upstream, 5′-TGACGTGGACATCCGCAAAG-3′ and downstream,
5′-CTGGAAGGTGGACAGCGAGG-3′ for β-actin (product length, 205
bp).
Animals
Thirty-two male BALB/c nude mice (age, 6 weeks;
weight, 16–18 g) were randomly divided into four groups: ADPC, ADT,
androgen-independent prostate cancer (AIPC) and control groups,
with 8 mice per group. Mice were housed at 25°C under a 12-h
light/dark cycle and were fed and watered on schedule.
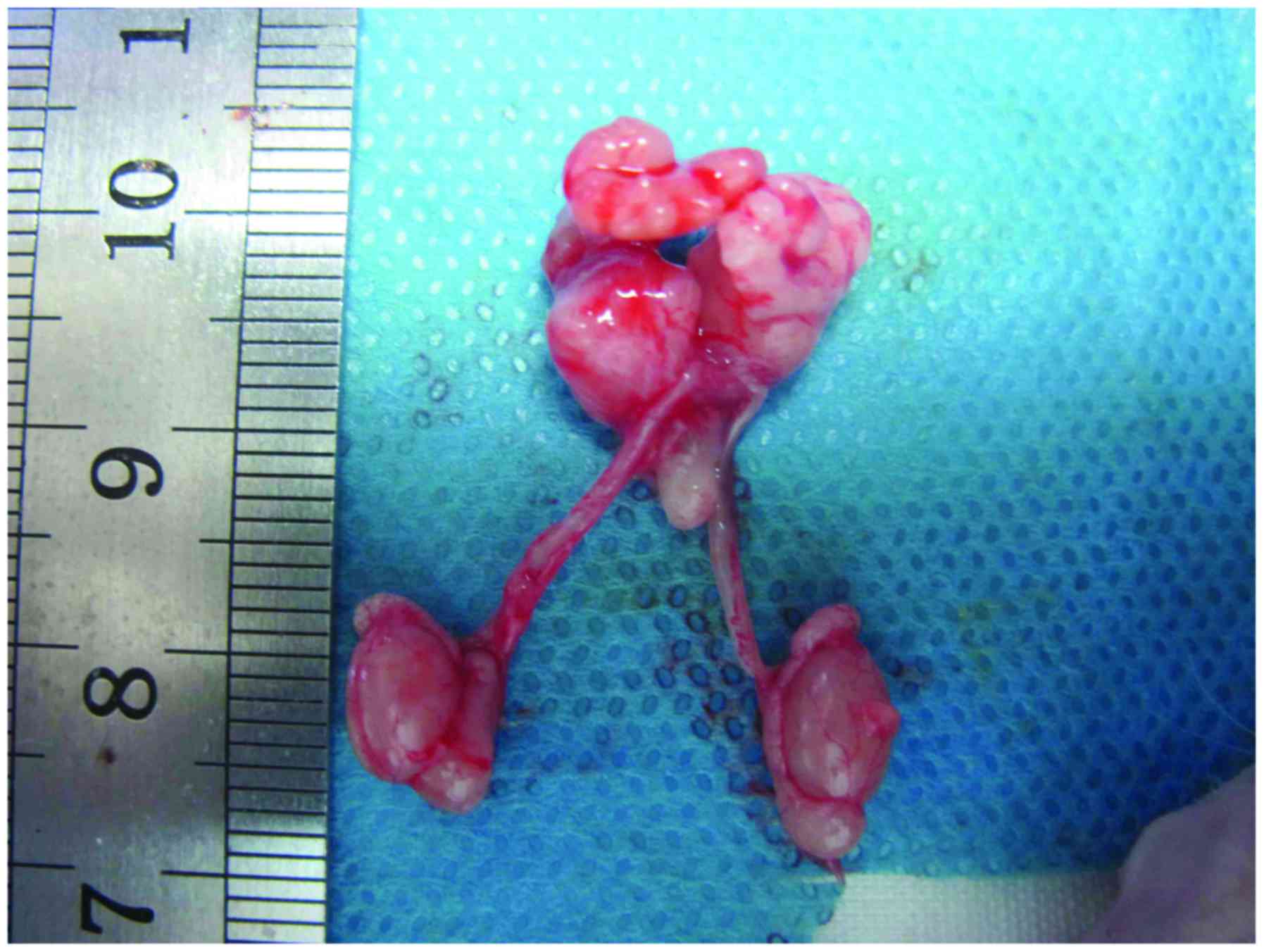
Xenograft tumor model in nude
mice
LNCaP cells (1×106 cells/mouse) were
suspended in 0.1 ml serum-free RPMI-1640 (Gibco; Thermo Fisher
Scientific, Inc.) and implanted subcutaneously into the flank of
each mouse. The mice were maintained in ventilated cages and fed
normal food and water for ~4 weeks after tumor inoculation to
obtain a subcutaneous tumor. The tumor volume was determined by
caliper measurements and divided into three equivalent segments to
implant into the anterior capsule of the nude mice prostate in the
ADPC, AIPC and ADT groups. Meanwhile, the tumor tissue was
implanted in subcutaneous tissue to assess growth changes of
orthotopic prostate cancer indirectly. In the ADPC group, the
prostate tumor was removed 8 weeks after implantation. In the ADT
group, the mice were castrated by surgery 8 weeks after
implantation and the prostate tumor was removed 3 days later,
following castration. In the AIPC group, the mice were castrated 15
days after tumor implantation and subsequently the prostate tumor
was removed 8 weeks later (Fig. 1).
The tumor volume was calculated based on weekly caliper measurement
using the following formula: Volume = (width2 x length)
/ 2. All tumors from the three individual groups were assessed by
pathologists at the Tianjin Institute of Urology, China. RT-PCR was
conducted to evaluate the mRNA expression levels of Pim-1 and IHC
was performed to analyze its protein expression.
RT-qPCR
Fresh prostate tumor tissue samples were immediately
immersed into the RNA later solution (Qiagen, Inc., Valencia, CA,
USA) for 24 h at 4°C and total mRNA was extracted using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA) according to the manufacturer's instructions. A Multisource
Total RNA Miniprep kit (Axygen; Corning Life Sciences, Shanghai,
China) was used in the AIPC, ADPC and ADT subgroups. The
concentration of RNA was measured using an ultraviolet
spectrophotometer (Biophotometer; Eppendorf, Hamburg, Germany).
cDNA was synthesized using the High-Capacity cDNA Synthesis kit
(Applied Biosystems; Thermo Fisher Scientific, Inc.). PCR was
performed using a one-step RT-PCR system (Beijing Transgen Biotech
Co., Ltd., Beijing, China). PCR was run in a 12.5 µl reaction
volume. The PCR specific primer pairs (Takara Biotechnology Co.,
Ltd., Dalian, China) were 5′-GCCTCAACTCCTCCCATAGATAC-3′ (forward)
and 5′-GCGGCATTCAGCAGAACTCAT-3′ (reverse). The PCR conditions were
as follows: Preprocessing of uracil-DNA glycosylase at 50°C for 2
min; pre-denaturation at 95°C for 10 min; 40 cycles of denaturation
at 95°C for 15 sec, annealing at 60°C for 30 sec and extension at
72°C for 30 sec; and full extension at 72°C for 10 min to complete
the amplification.
Analysis of RT-qPCR
The PCR products were separated on 1% agarose gels
containing ethidium bromide using the TC-96/G/H(b)A electrophoresis
apparatus (Beijing Liuyi Biotechnology Co., Ltd., Beijing, China).
The gels were photographed and analyzed using a Tanon-1600 gel
imaging analysis system (Shanghai Tanon Science & Technology
Co., Ltd., Shanghai, China) by measurement of the gray value of the
electrophoresis strip. The ratio between the target gene and
β-actin was used to calculate the relative quantitation. The
amplification products and the relative expression of RT-PCR were
analyzed using the ΔΔCq method (5).
ELISA of mice plasma prostate-specific
antigen (PSA)
Mouse plasma was obtained from the caudal vein every
2 weeks for measurement of PSA levels using a Quantikine human PSA
immunoassay kit (R&D Systems, Inc., Minneapolis, MN, USA)
according to the manufacturer's instructions. Concentration of
Pim-1 protein was measured using a commercially quantitative ELISA
kit (BioCheck USA, Scarborough, ME, USA), according to the
manufacturer's instruction. A total of three independent
experiments, each in triplicate, were assayed and the median Pim-1
protein concentration from each duplicate was used for statistical
analysis.
IHC
Tumor tissue was embedded in paraffin and sliced
into 3-µm thick sections for IHC. Sections were dewaxed in xylene,
hydrated through graded alcohols and rinsed in deionized water.
Antigen retrieval was performed by boiling the slides in a cooker
for 10 min in a citrate buffer (pH 6.0; Wuhan Boster Biological
Engineering Co., Ltd., Wuhan, China). Subsequent to a 10-min
treatment with 3% H2O2, tissue sections were
blocked with 5% normal goat serum (Genomapping Technology Co., Ltd.
Tianjin, China) in Tris-buffered saline (pH 8.0; Beijing Zhongshan
Golden Bridge Biotechnology Co., Ltd., Tianjin, China) for 1 h at
room temperature, incubated with rabbit polyclonal antibody AR
(dilution, 1:100; cat. no. ab133273; Abcam, Cambridge, MA, USA) and
rabbit monoclonal antibody Pim-1 (dilution, 1:2,000; cat. no.
ab75776; Abcam) at 4°C overnight, and incubated with horseradish
peroxidase-conjugated secondary antibodies (dilution, 1:20,000; cat
no. ab136636; Abcam) for 30 min at room temperature. After the
application of the diaminobenzidine kit (dilution, 1:1; cat. no.
SP-9000-D; Beijing Zhongshan Jinqiao Biotech Co., Ltd., Beijing,
China), tissue sections were stained with hematoxylin, dehydrated
and mounted. The slides were scanned with a Hamamatsu NanoZoomer
scanner (Nikon ECLIPSE90ir; Nikon Corporation, Tokyo, Japan).
Positive cells were defined as cell cytoplasm or nuclei that were
immunostained dark yellow or brown. Microscopic imaging was used to
quantify the positive immunoreactivity, which was recorded by a
microscope equipped with a digital camera. Integrated optical
density (IOD) was calculated to analyze semiquantitative expression
of AR and Pim-1 using Image-Pro Plus 6.0 software (Media
Cybernetics, Inc., Rockville, MD, USA). The immunoreactivity
reaction and staining intensity of the prostate tumor cells were
compared by calculating the mean optical density (MOD) from 15
different microscopic fields.
Statistical analysis
Statistical analyses were performed using SPSS
software (version 14.0; SPSS, Inc., Chicago, IL, USA). Data are
expressed as the mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference. The
differences between multiple groups were determined by one-way
analysis of variance followed by χ2 analysis.
Results
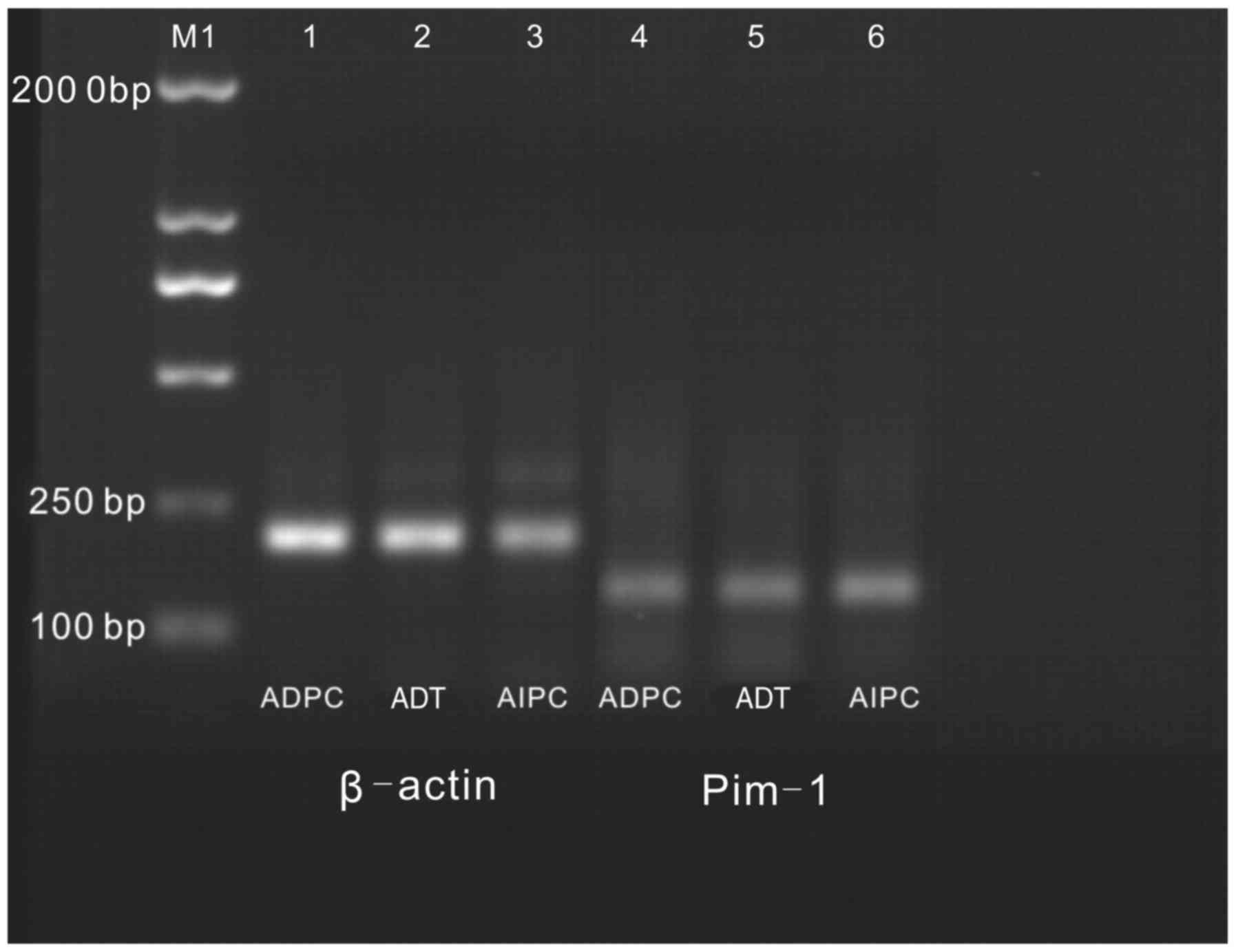
RT-PCR electrophoretogram
The lengths of the amplified products of β-actin and
Pim-1 were 205 and 147 bp, respectively. An agarose gel
electrophoretogram revealed that the relative mRNA expression
levels of Pim-1 in the ADPC, AIPC and ADT groups were 0.59±0.01,
1.14±0.015 and 0.62±0.026, respectively (Fig. 2). A statistically significant
difference was observed for the ADPC and AIPC groups, as compared
with the ADT group (P<0.05).
Amplification of RT-PCR
Only one Pim-1 amplification peak appeared during
the entire amplification process, without non-specific peaks,
non-specific amplification products or primer dimers. All
amplification curves showed as typical S-shaped curves. The
amplicon of the target gene, Pim-1, showed a rapid increase from
the 20–25th cycles and the amplicon for the internal control gene,
β-actin, appeared in the 15–20th cycles. The expression level of
Pim-1 mRNA was significantly different between the ADT and ADPC
groups (P<0.05), and that of AIPC was higher than the ADPC group
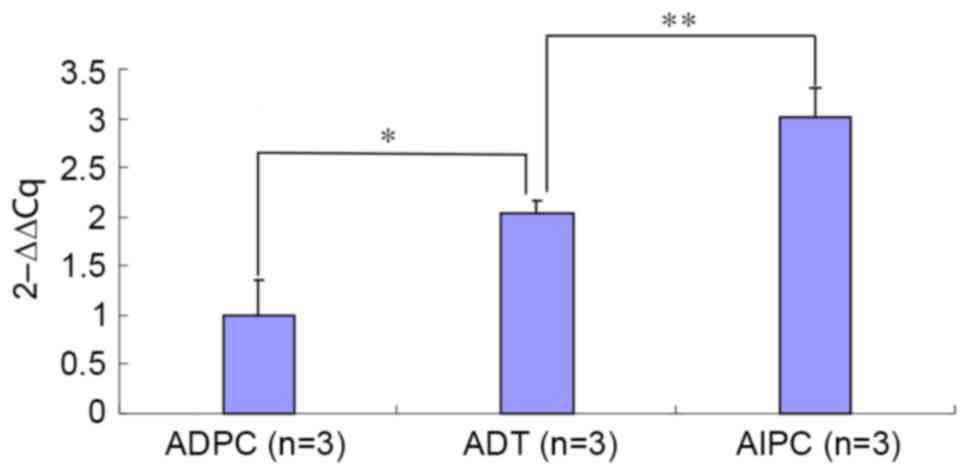
(Fig. 3; P<0.01). The ΔCq and ΔΔCq
values of the Pim-1 gene in the ADPC, ADT and AIPC groups are shown
in Table I. Compared with the ΔCq
value of Pim-1 in the ADT group, a significant difference was found
in the ADPC group and AIPC group (P<0.05). Analyzed using the
relative quantification method, the Pim-1 amplification product was
increased by 2.05 and 3.01 times in the ADT and AIPC groups,
respectively, as compared with the ADPC group (Fig. 4).
 | Table I.Cq value analysis of Pim-1 in the
three groups. |
Table I.
Cq value analysis of Pim-1 in the
three groups.
| Group | Pim-1 Cq value | β-actin Cq value | ΔCq value | ΔΔCq value |
2−ΔΔCq |
|---|
| ADPC | 22.76±0.11 | 16.61±0.27 |
6.15±0.34a | 0.00±0.10 | 1.00 |
| ADT | 22.69±0.38 | 17.58±0.20 | 5.11±0.21 | −1.04±0.12 | 2.05 |
| AIPC | 22.51±0.45 | 17.95±0.46 |
4.56±0.23a | −1.59±0.30 | 3.01 |
PSA concentration of mouse blood
serum
The PSA concentration of the blank control group
mice was 0 ng/ml; by contrast, that of the ADPC, ADT and AIPC
groups were 0.48±0.025, 0.17±0.032 and 0.87±0.023 µg/l,
respectively. The concentrations of the ADT and AIPC groups were
significantly reduced and increased, respectively, as compared with
the ADPC group (P<0.01).
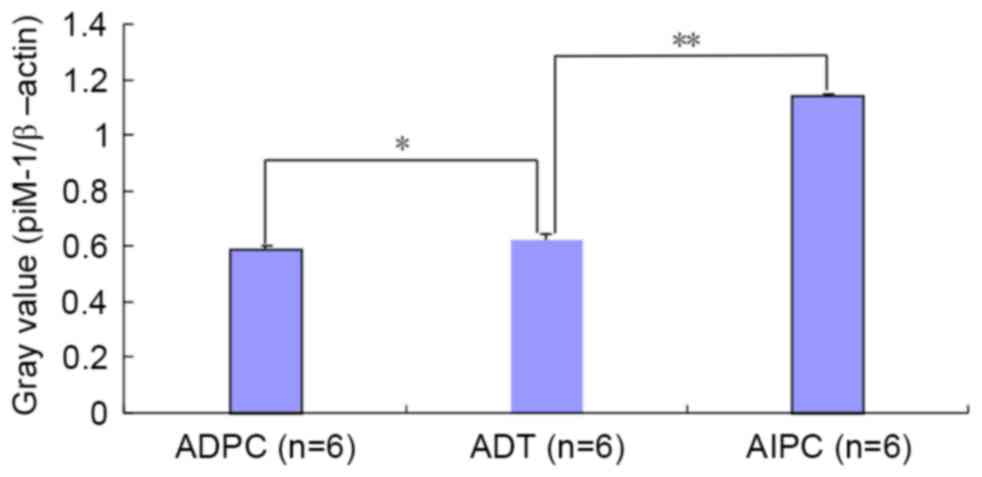
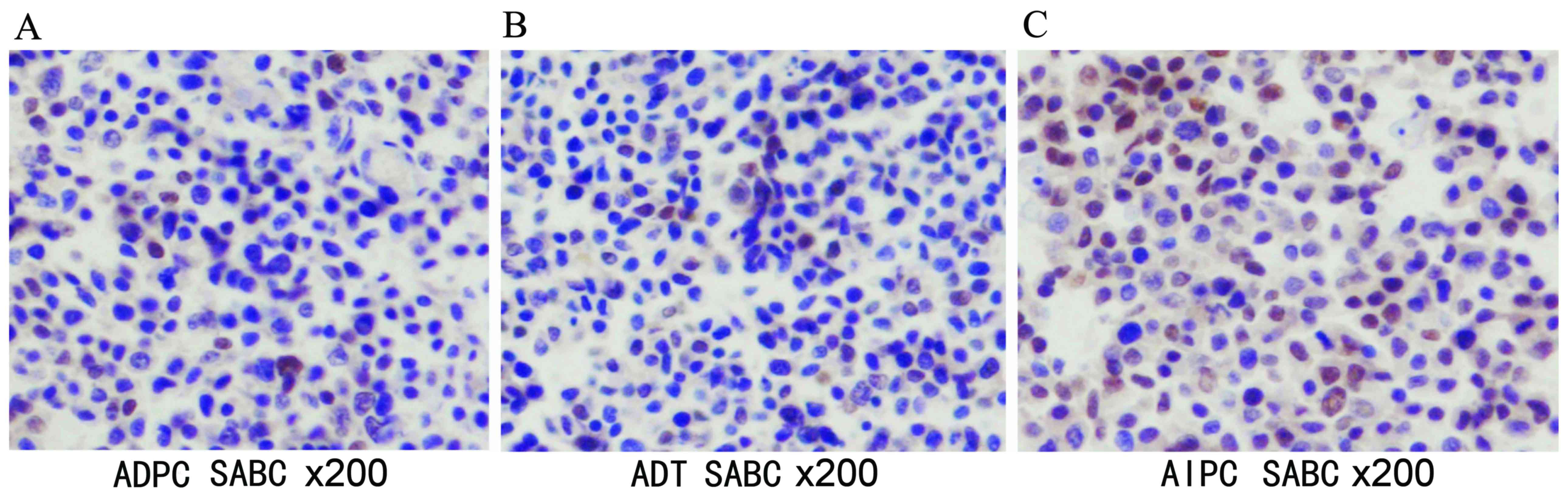
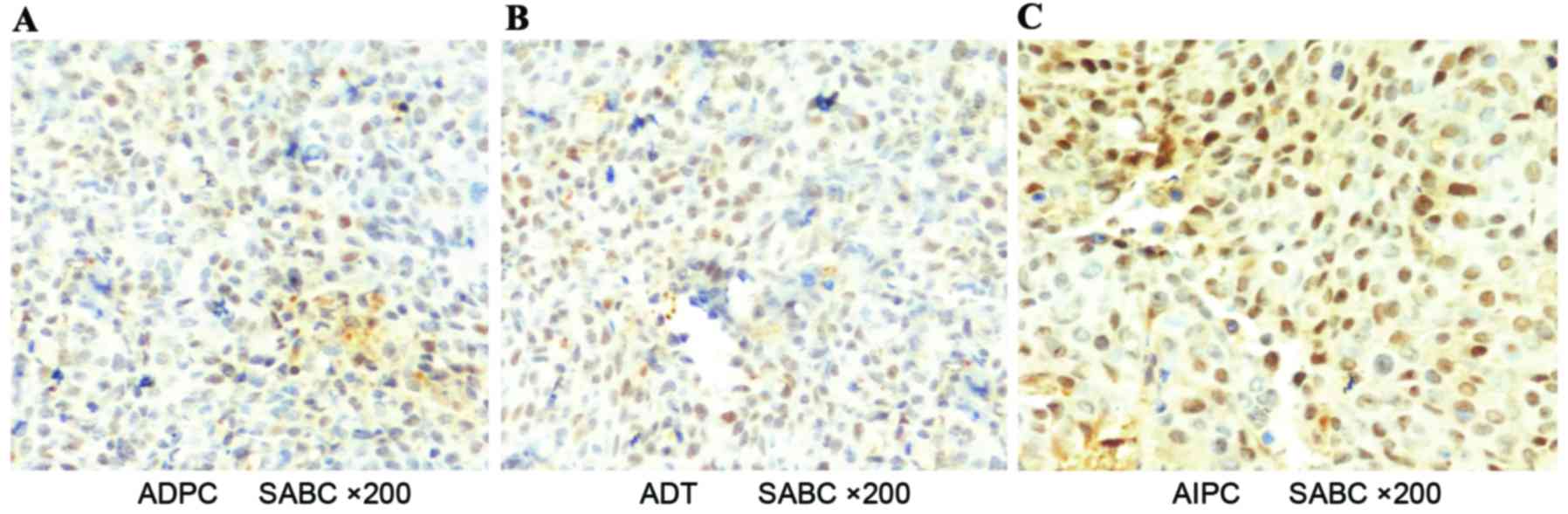
Results of IHC
Histologically, AR was predominantly present in the
cell nucleus (Fig. 5) and Pim-1 was
present in the cytoplasm (Fig. 6).
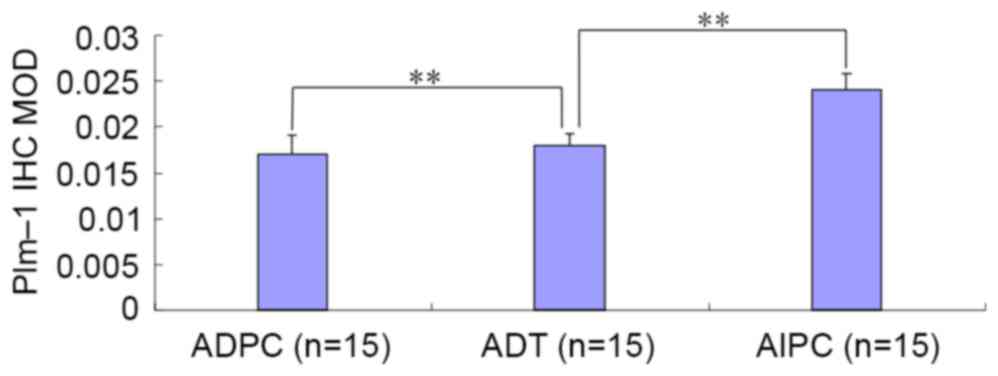
Image-Pro Plus software was used to analyze the IHC results
(Table II); a statistically
significant difference was observed or the ADPC and AIPC groups, as
compared with the ADT group with regard to the MOD ratio (Fig. 7).
 | Table II.Immunohistochemical-stained MOD of
Pim-1 and AR expression in the three groups. |
Table II.
Immunohistochemical-stained MOD of
Pim-1 and AR expression in the three groups.
|
|
| MOD |
|---|
|
|
|
|
|---|
| Group | n | Pim-1 | AR |
|---|
| ADPC | 15 |
0.017±0.0021a |
0.032±0.009a |
| ADT | 15 | 0.018±0.0013 | 0.019±0.006 |
| AIPC | 15 |
0.024±0.0019a |
0.040±0.011a |
Discussion
Prostate cancer remains the most common type of
cancer in males in the USA and the western world. It was estimated
that in the USA in 2016, prostate cancer would account for 21% of
all newly diagnosed cancer cases representing 841,390 men. In 2016,
26,120 men in the USA succumbed to the disease (6). Prostate cancer mortality rates have been
declining, which was attributed to PSA screening and androgen
ablation treatment. Although most prostate cancer grow this
dependent on the presence of androgens and initially responds well
to ADT, the majority of cases progress to CRPC, which is currently
incurable. An improved understanding of the underlying mechanisms
of castration resistance is critical to reduce associated morbidity
and mortality rates. There have been numerous studies attempting to
clarify the mechanisms for the development of CRPC, including
ligand-independent AR activation (7,8), AR
mutations (9) or abnormal
amplification of AR (10).
Pim is an oncogene that is closely associated
with the proliferation and differentiation of cancer cells
(11). They are primarily regulated
at the transcription level without a specifically regulatory
domain. Pim controls cellular proliferation, differentiation
or apoptosis functions. Additionally, it regulates tumorigenesis
via various signaling pathways. Pim-1 is regulated by
cytokines, growth factors or hormones, and it encodes a
serine/threonine kinase, which is implicated in tumor malignant
transformation and progression (12).
For example, the overexpression of Pim-1 and Pim-2 are implicated
in prostate cancer progression (13).
To date, three members of oncogenic serine/threonine kinases genes,
including the highly homologous Pim-1, Pim-2 and Pim-3, have been
reported in the PIM family, and possess overlapping structures and
functions. Among them, Pim-1 has been widely investigated
and its oncogenic nature has been confirmed (14). The crystal structure reveals that
Pim-1 is a constitutively active kinase. Notably, it was reported
that Pim-1 is predominantly located in the cytoplasm and nucleus,
although it may also be found at the membrane. The Pim-1
gene is a proto-oncogene that encodes two ubiquitous protein kinase
isoforms, including Pim-1L (44 kDa) and PIM-1S (33 kDa), with high
degrees of sequence and structural similarities. In humans,
Pim-1 is often expressed in normal and transformed cells,
and its overexpression has been documented in various tumors
(15). The expression level of
Pim-1 is higher in prostate cancer than in human benign
prostatic hypertrophy (16). In
vitro, the overexpression of exogenous Pim-1 increases prostate
cancer cell proliferation and progression (17). In vivo, it has the potential to
serve as a diagnostic and prognostic biomarker (18). According to clinical findings,
Pim-1 expression is associated with a poor prognosis and
hormone insensitivity to ADT (11).
There are two Pim-1 kinase isoforms, namely Pim-1S
and Pim-1L, which modulate AR stability and transcriptional
activity (19); these isoforms
modulate AR activity by phosphorylation of AR to promote prostate
cancer cell growth (20). In a
previous study, the decreased AR expression coincided with
increased Pim-1 expression, indicating that Pim-1 may
contribute to regulation of AR turnover (21). AR is critical in prostate cancer
development to CRPC, therefore, Pim-1 may serve as a potential
target for the treatment of hormone-refractory prostate cancer
(22). However, the molecular
mechanisms of Pim kinases in specific signaling pathways for
prostate cancer cell proliferation and progression are complicated
and not well established (11). Pim
kinases may be associated with the regulation of gene transcription
through interactions with c-Myc and AKT to enhance tumorigenesis
and inactivate cell cycle inhibitors by phosphorylating and
downregulating p27Kip1 at the transcriptional and
post-transcriptional levels (23,24).
Furthermore, Pim may be involved in the regulation of cell
proliferation and viability (14).
Due to the above-mentioned findings, Pim-1 is
considered to be a potential tumor target for prostate cancer
therapy. The development of specific Pim inhibitors is imperative
for the treatment of CRPC patients. Notably, various Pim family
small-molecule inhibitors targeting these kinases have been
identified, and the majority of Pim kinase inhibitors are specific
for Pim-1. These inhibitors display anticancer activity in prostate
cancer cell lines, including those that are sensitive and resistant
to chemotherapy (11). Despite the
fact that the Pim-1 kinase inhibitors that have been investigated
exhibit potential anticancer effects, there are a few small
molecule inhibitors showing an inhibitory effect (25). In addition, other studies revealed
that a Pim-1 specific monoclonal antibody markedly inhibited the
growth of the human prostate cancer cell line, DU145 in a mouse
model and thus may be considered another potential treatment
modality for CRPC (26). Furthermore,
Pim-1 contributes to the regulation of DNA repair when CRPC is
treated with paclitaxel. Cytotoxic drugs such as docetaxel activate
Pim-1 kinase in DU145 cells (27).
The ability of DNA repair significantly decreases in the absence of
Pim-1, leading to severe DNA damage and apoptosis (22).
In the current study, subcutaneous planting and
in situ embedding tumor methods were used to generate ADPC
mouse models. Subsequently, surgical castration was performed to
simulate ADT of human prostate cancer. PSA, AR and Pim-1 expression
levels were analyzed using RT-qPCR, ELISA and IHC in three
subgroups. The expression of AR and PSA revealed varying degrees of
reduction in the ADT treatment subgroup; by contrast, the
expression levels of AR and PSA were increased significantly in the
AIPC subgroup, which was similar to the trend of ADT in human
prostate cancer. These findings demonstrated that the in
situ prostate cancer mouse model may successfully simulate an
ADT model of humans. In the present mouse model, the expression
levels of genes and proteins were significantly different in the
ADPC and AIPC groups (P<0.05); Pim-1 was highly expressed and
implicated in the AIPC model during ADT, which implied that the
Pim-1 level was not influenced during ADT. It appeared that Pim-1
exerted a regulatory role in cell proliferation and tumor
differentiation. Additionally, Pim-1may affect the ADT effect on
the treatment of prostate cancer. The exact interaction mechanism
of Pim-1 and AR remains unclear; the current findings revealed that
the expression level of AR was decreased, while PIM-1 expression
was elevated in the ADT subgroup. Notably, AR and Pim-1 were highly
expressed in the AIPC subgroup. These findings revealed that there
were high expression levels of Pim-1 during the ADT treatment
period, indicating that Pim-1 is important in the progression and
metastasis of prostate cancer. This requires further confirmation
by subsequent experiments, such as using shRNA interference or
Pim-1 inhibitors to decrease the expression of Pim-1 and
systematically evaluate the mechanism of Pim-1 interaction with AR
during ADT.
There were several limitations of the current study.
First, we had to adjust the sample size from 8 to 6 for calculating
statistical differences due to the successful operation rate of
orthotopic implantation and to keep the same number of samples in
the four groups. However, a sample size of less than 5 may yield a
difference in subgroup analyses; a similar small sample especially
in the animal model was observed in another study (28). In addition, there was lack of a Pim-1
inhibitor to treat the mice in order to observe the change during
the ADT period. Furthermore, there was lack of Pim-1 data regarding
metastatic tumors and a lack of tumor growth curves.
In conclusion, the expression level of Pim-1 was
high during ADT, which may contribute to the progression or
metastasis of prostate cancer; therefore, further studies are
required to investigate the specific underlying mechanism of Pim-1
interaction with AR and ADT.
Acknowledgements
The current study was supported by the National
Natural Science Foundation for Young Scholars of China (grant no.
81302211) and the Tianjin Research Program of Application
Foundation and Advanced Technology (grant no. 14CYBJC29800).
References
|
1
|
Cuypers HT, Selten G, Quint W, Zijlstra M,
Maandag ER, Boelens W, van Wezenbeek P, Melief C and Berns A:
Murine leukemia virus-induced T-cell lymphomagenesis: Integration
of proviruses in a distinct chromosomal region. Cell. 37:141–150.
1984. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Song JH, Padi SK, Luevano LA, Minden MD,
DeAngelo DJ, Hardiman G, Ball LE, Warfel NA and Kraft AS: Insulin
receptor substrate 1 is a substrate of the Pim protein kinases.
Oncotarget. 7:20152–20165. 2016.PubMed/NCBI
|
|
3
|
Natarajan K, Xie Y, Burcu M, Linn DE, Qiu
Y and Baer MR: Pim-1 kinase phosphorylates and stabilizes 130 kDa
FLT3 and promotes aberrant STAT5 signaling in acute myeloid
leukemia with FLT3 internal tandem duplication. PLoS One.
8:e746532013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Holder SL and Abdulkadir SA: PIM1 kinase
as a target in prostate cancer: Roles in tumorigenesis, castration
resistance, and docetaxel resistance. Curr Cancer Drug Targets.
14:105–114. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Siegel RL, Miller KD and Jemal A: Cancer
Statistics, 2017. CA Cancer J Clin. 66:7–30. 2017. View Article : Google Scholar
|
|
7
|
Mizokami A and Namiki M: Reconsideration
of progression to CRPC during androgen deprivation therapy. J
Steroid Biochem Mol Biol. 145:164–171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J,
Chen Z, Beroukhim R, Wang H, Lupien M, et al: Androgen receptor
regulates a distinct transcription program in androgen-independent
prostate cancer. Cell. 138:245–256. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Grasso CS, Wu YM, Robinson DR, Cao X,
Dhanasekaran SM, Khan AP, Quist MJ, Jing X, Lonigro RJ, Brenner JC,
et al: The mutational landscape of lethal castration-resistant
prostate cancer. Nature. 487:239–243. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Robinson D, Van Allen EM, Wu YM, Schultz
N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC,
Attard G, et al: Integrative clinical genomics of advanced prostate
cancer. Cell. 161:1215–1228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shah N, Pang B, Yeoh KG, Thorn S, Chen CS,
Lilly MB and Salto-Tellez M: Potential roles for the PIM1 kinase in
human cancer-a molecular and therapeutic appraisal. Eur J Cancer.
44:2144–2151. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rimon E, Sasson R, Dantes A, Land-Bracha A
and Amsterdam A: Gonadotropin-induced gene regulation in human
granulosa cells obtained from IVF patients: Modulation of genes
coding for growth factors and their receptors and genes involved in
cancer and other diseases. Int J Oncol. 24:1325–1338.
2004.PubMed/NCBI
|
|
13
|
Cibull TL, Jones TD, Li L, Eble JN,
Baldridge Ann L, Malott SR, Luo Y and Cheng L: Overexpression of
Pim-1 during progression of prostatic adenocarcinoma. J Clin
Pathol. 59:285–288. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Brasó-Maristany F, Filosto S, Catchpole S,
Marlow R, Quist J, Francesch-Domenech E, Plumb DA, Zakka L,
Gazinska P, Liccardi G, et al: PIM1 kinase regulates cell death,
tumor growth and chemotherapy response in triple-negative breast
cancer. Nat Med. 22:1303–1313. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu J, Zhang T, Wang T, You L and Zhao Y:
PIM kinases: An overview in tumors and recent advances in
pancreatic cancer. Future Onco. 10:865–876. 2014. View Article : Google Scholar
|
|
16
|
He HC, Bi XC, Zheng ZW, Dai QS, Han ZD,
Liang YX, Ye YK, Zeng GH, Zhu G and Zhong WD: Real-time
quantitative RT-PCR assessment of PIM-1and hK2 mRNA expression in
benign prostate hyperplasia and prostate cancer. Med Oncol.
26:303–308. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen WW, Chan DC, Donald C, Lilly MB and
Kraft AS: Pim family kinases enhance tumor growth of prostate
cancer cells. Mol Cancer Res. 3:443–451. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu Y, Zhang T, Tang H, Zhang S, Liu M, Ren
D and Niu Y: Overexpression of PIM-1 is a potential biomarker in
prostate carcinoma. J Surg Oncol. 92:326–330. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van der Poel HG, Zevenhoven J and Bergman
AM: Pim1 regulate androgen-dependent survival signaling in prostate
cancer cells. Urol Int. 84:212–220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ha S, Iqbal NJ, Mita P, Ruoff R, Gerald
WL, Lepor H, Taneja SS, Lee P, Melamed J, Garabedian MJ and Logan
SK: Phosphorylation of the androgen receptor by PIM1 in hormone
refractory prostate cancer. Oncogene. 32:3992–4000. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim O, Jiang T, Xie Y, Guo Z, Chen H and
Qiu Y: Synergism of cytoplasmic kinases in IL6-induced
ligand-independent activation of androgen receptor in prostate
cancer cells. Oncogene. 23:1838–1844. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hsu JL, Leong PK, Ho YF, Hsu LC, Lu PH,
Chen CS and Guh JH: Pim-1 knockdown potentiates paclitaxel-induced
apoptosis in human hormone-refractory prostate cancers through
inhibition of NHEJ DNA repair. Cancer Lett. 319:214–222. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J, Kim J, Roh M, Franco OE, Hayward
SW, Wills ML and Abdulkadir SA: Pim1 kinase synergizes with c-MYC
to induce advanced prostate carcinoma. Oncogene. 29:2477–2487.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Morishita D, Katayama R, Sekimizu K,
Tsuruo T and Fujita N: Pim kinases promote cell cycle progression
by phosphorylating and down-regulating p27Kip1 at the
transcriptional and posttranscriptional levels. Cancer Res.
68:5076–5085. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Holder S, Zemskova M, Zhang C, Tabrizizad
M, Bremer R, Neidigh JW and Lilly MB: Characterization of a potent
and selective small-molecule inhibitor of the PIM1 kinase. Mol
Cancer Ther. 6:163–172. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hu XF, Li J, Vandervalk S, Wang Z,
Magnuson NS and Xing PX: PIM-1-specific mAb suppresses human and
mouse tumor growth by decreasing PIM-1 levels, reducing Akt
phosphorylation, and activating apoptosis. J Clin Invest.
119:362–375. 2009.PubMed/NCBI
|
|
27
|
Zemskova M, Sahakian E, Bashkirova S and
Lilly M: The PIM1 kinase is a critical component of a survival
pathway activated by docetaxel and promotes survival of
docetaxel-treated prostate cancer cells. J Biol Chem.
283:20635–20644. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nakagawa S and Cuthill IC: Effect size,
confidence interval and statistical significance: A practical guide
for biologists. Biol Rev Camb Philos Soc. 82:591–605. 2007.
View Article : Google Scholar : PubMed/NCBI
|