Introduction
Ovarian cancer is the most lethal type of female
reproductive system cancer. In the United States of America, 14,180
patients were estimated to succumb to ovarian cancer and 21,290 new
patients were estimated to be diagnosed in 2015 (1). A histological study indicated that
90–95% of ovarian cancer cases were of epithelial ovarian carcinoma
(EOC) (2). The International
Federation of Gynecology and Obstetrics (FIGO) staging system
categorizes ovarian cancer into four stages (I–IV) from the least
to the most advanced (3). The more
advanced the ovarian cancer, the lower the five-year survival rate
of patients. The five-year survival rate at FIGO stage I is 90–95%,
stage II is 70–80%, stage III is 20–50% and Stage IV is only 1–5%
(4). However, patients with
early-stage EOC often do not exhibit noticeable symptoms and it is
therefore difficult to diagnose. Thus, the majority of patients
with EOC are diagnosed at an advanced stage.
Cancer antigen 125 (CA125), also termed MUC16, is a
plasma membrane glycoprotein on ovarian epithelial cells and the
most common type of biomarker currently used for EOC diagnosis
(5). A CA125 level ≥35 U/ml is a
marker of ovarian cancer (6). Ozols
(7) reported that the sensitivity of
the CA125 blood test is 50% with respect to the early stages and
80% with respect to the advanced stages of EOC. Therefore, the
current diagnosis assay presents an issue for patients, as it
cannot efficiently inform optimally-timed cancer treatment. An
improved assay that is sensitive to early-stage EOC has yet to be
established. The present study aimed to explore certain alternative
markers, with potentially improved sensitivity to early and
advanced malignancy.
Serum free circulating DNA (fcDNA) is a type of
sample that may be accessed without surgical biopsy. Numerous
studies have identified a correlation between fcDNA and cancer,
including ovarian, uterine, colorectal, breast, lung and prostate
cancer, cervical and malignant gastrointestinal tumors, glioma,
hepatocellular carcinoma, metastatic melanoma, leukemia and
lymphoma (8–13). The characteristics of fcDNA may also
make this type of DNA a promising tool for the diagnosis of
early-stage EOC.
In addition, in human somatic cells, methylation
primarily occurs at the cytosine of the 5′-C-phosphate-G-3′ (CpG)
site (14,15). The regulatory regions of genes are
rich in unmethylated CpGs, which form CpG islands (16). A total of three tumor suppressor
genes, Runt-related transcription factor 3 (RUNX3), tissue factor
pathway inhibitor 2 (TFPI2) and opioid binding protein/cell
adhesion molecule-like (OPCML), were considered as candidate
biomarkers in the present study. RUNX3 is a transcription factor,
which has methylated CpG islands in various types of cancer with
percentages ranging from 73 to 2.5% (17). A previous study reported that 53.1%
patients with primary ovarian cancer exhibited methylated RUNX3
(18). The expression of TFPI2, a
serine proteinase inhibitor, is inversely correlated with the
degree of tumor malignancy (19).
OPCML is a plasma membrane protein functioning as an opioid
receptor. Previous studies revealed that the expression of these
genes was suppressed by DNA methylation of their regulatory regions
in ovarian cancer (19–21).
Herman et al (22) developed a type of polymerase chain
reaction (PCR), methylation-specific PCR (MSP), to identify the
methylation status of CpG islands. This method requires two pairs
of primers, the M pair and the U pair. The M pair corresponds to
the modified and methylated sequence, and the U pair corresponds to
the modified and unmethylated sequence. Cytosine of CpG in the U
pair converts to thymine. A primer design program for MSP,
MethPrimer, is available at present (23). The present study investigated
potential new biomarkers on fcDNA to improve the sensitivity,
specificity and accuracy of early stage EOC screening.
Materials and methods
Patients and sample collection
Patients that were admitted to the Affiliated
Hospital of Guiyang Medical College (GMCAH; Guiyang, China) between
June 2011 and December 2014, comprising of 80 healthy donors, 43
donors with benign ovarian tumors and 71 donors with ovarian
epithelial carcinoma, provided informed consent to take part in the
present study. The healthy donor samples were collected from the
Blood Transfusion Station of GMCAH. Their median age was 46 years
(range 18–61 years). The median age of the patients with benign
tumors was 34 years (range 18–52 years) and of the patients with
malignant cancer was 54 years (range 28–67 years). The blood and
tissue samples of the patients with ovarian cancer were collected
and processed in the Department of Medical Laboratory of GMCAH, and
stored at −80°C prior to use. Based on the FIGO staging system, out
of the 71 patients with ovarian cancer, 39 had stage I or II, and
32 patients had stage III or IV disease. The present study regarded
stages I and II as early-phase cancer, and stages III and IV as
advanced-phase cancer. A total of 3 ml blood was collected from
each patient and the serum was isolated by 3,500 × g centrifugation
at 4°C for 5 min. In addition, 100 mg tissue sample from each
patient was collected during surgery and then soaked in 4°C
RNAlater Stabilization Solution (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) overnight prior to storage at
−80°C until required.
Cell culture
The epithelial ovarian cancer cell line HO8910 was
acquired from the Cell Bank of the Type Culture Collection of the
Chinese Academy of Sciences (Shanghai, China). This cell line was
cultured in RPMI-1640 media (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (Hangzhou Sijiqing
Biological Engineering Materials Co., Ltd., Hangzhou, China), 100
U/ml penicillin and 100 µg/ml streptomycin (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany). Cell cultures were maintained in an
atmosphere with 5% CO2 at 37°C.
DNA isolation and quantification
fcDNA from sera collected from the aforementioned
194 healthy donors and patients with ovarian epithelia carcinoma or
benign ovarian tumors was extracted using the QIAamp DNA blood mini
kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's
protocol. Briefly, 200 µl patient serum was mixed with 200 µl
Buffer AL plus 20 µl QIAGEN Protease from the kit described above
and incubated at 56°C for 10 min. The mixture was applied to the
QIAamp Mini spin column subsequent to the addition of 200 µl 100%
ethanol. Centrifugation at 6,000 × g at room temperature for 1 min
was performed to remove the majority of impurities and bind the DNA
to the column. The column was washed withAW1 and AW2 buffers.
Finally, the DNA was collected in buffer AE. Genomic DNA from 3
randomly-chosen patient tissues from each category and HO8910 cell
cultures was purified with a QIAamp DNA mini kit (Qiagen GmbH)
according to the manufacturer's protocol. To calculate the DNA
concentration, absorbance at a wavelength of 260 nm was measured
with a SmartSpec Plus spectrophotometer (Bio-Rad Laboratories,
Inc.) and compared with a standard curve. Only DNA with an
A260/A280 ratio of 1.7–2.0 was used in the present study.
Methylation status analysis of tumor
suppressor genes
The cytosine residues in target DNA were converted
into uracil and the product was purified with an EpiTect Bisulfite
kit (Qiagen GmbH). In total, 1 µg fcDNA was mixed with 85 µl
Bisulfite mix in a 200-µl PCR tube. The mixture was incubated at
60°C for 25 min, 1 h 25 min and 2 h 55 min with 5 min denaturation
at 95°C between each incubation in a C1000 Touch Thermal Cycler
(Bio-Rad Laboratories, Inc.). Once the reaction was completed, the
DNA was recovered by EpiTect spin column.
Methylation of DNA was detected using
nested PCR
The PCR primers used in this study were listed in
Table I. The PCR reactions included
0.2 µM forward and reverse primers, 0.1 µg template, 12.5 µl Taq 2X
Master mix (New England BioLabs, Inc., Ipswich, MA, USA) and
nuclease-free water. The total volume was 25 µl. PCR mix without
template DNA was used as a negative control. The external round PCR
reaction conditions were: i) Initialization at 95°C for 5 min; ii)
denaturation at 95°C for 30 s, annealing at 50°C for 1 min,
elongation at 72°C for 45 s, total 25 cycles; iii) final elongation
at 72°C for 10 min. The internal round PCR reaction conditions were
similar to the external round PCR, except that the total cycle
number was 35.
 | Table I.Primer sets used in the present
study. |
Table I.
Primer sets used in the present
study.
| Protein | Sequence |
|---|
| OPCML |
|
| M
system | 5′-ACC
GCT AAA
CGT AAC
GTC
CG-3′ |
| U
system | 5′-ACC
ACT AAA
CAT AAC
ATC
CA-3′ |
| Nested
F | 5′-AAA
AGT TTT AAG GYG GGG AT-3′ |
| Nested
R | 5′-TCA
AAA CTA CTC CCA AAA AA-3′ |
| RUNX3 |
|
| M
system |
5′-TCG GGA CGT ATA ATA TTT
TCG-3′ |
| U
system |
5′-TTG GGA TGT ATA ATA TTT
TTG
AG-3′ |
| Nested
F | 5′-GGT
TAG GGG TTT TTT AAT TTT AAT TYG-3′ |
| Nested
R | 5′-CAC
CRC RAA TAA AAT ACR AAC-3′ |
| TFPI2 |
|
| M
system | 5′-TGG
CGA AGT TGT
TAT TAG TC-3′ |
| U
system | 5′-T TGG
TGA AGT TGT
TAT TAG TT-3′ |
| Nested
F | 5′-ATT
TTT TGT AGA AAG TGA GAT G-3′ |
| Nested
R | 5′-AAT
ACA CAC AAA ACT ACC AC-3′ |
DNA gel quantification analysis
The nested MSP products were separated by gel
electrophoresis. Images of the DNA bands observed were captured
using the Gel DocXR+ Imaging System (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). JPEG-formatted images were used for
quantification analysis by ImageJ software [National Institutes of
Health (NIH), Bethesda, MD, USA]. Based on expected PCR product
size, DNA bands in images were selected and the Analyze>Set
Measurements command in ImageJ was used to record area, mean gray
value and integrated density. An integrated density <9,000 was
regarded as a negative result.
CA125 assay
A CA125 II assay kit was purchased from Ortho
Clinical Diagnostics, Inc., Raritan, NJ, USA. All assays were
performed according to the protocol of the manufacturer. The
principle of this assay was to evaluate CA125 antigen expressionin
samples using streptavidin-coated wells and biotinylated M11 mouse
anti-CA125 antibody, which was then incubated with horseradish
peroxidase (HRP)-labeled mouse anti-CA125 antibody to form a
sandwich. HRP catalyzed the signal reagent to generate
luminescence, which was monitored using a VITROS 5600 Integrated
System (Ortho Clinical Diagnostics, Inc.).
Statistical analysis
Microsoft office Excel 2013 (Microsoft Corporation,
Redmond, WA, USA) and GraphPad Prism version 6 (GraphPad Software,
Inc., La Jolla, CA, USA) were used for data calculation, and table
and chart construction. Data obtained from small sample sizes were
presented as the mean ± standard deviation. Data obtained from
large sample sizes were presented as a box and whisker plot, which
was analyzed by one-way analysis of the variance (ANOVA) to
statistically determine the significance of differences between
groups. P<0.05 was considered to indicate a statistically
significant difference. Cohen's κ coefficient was used to compare
the degree of agreement between observed and expected results. The
following κ values were considered to indicate the following:
<0.20, poor agreement; 0.20–0.40, fair agreement; 0.40–0.60,
moderate agreement; 0.60–0.80, good agreement; and 0.80–1.00, very
good agreement.
Results
Evaluating effectiveness of RUNX3,
TFPI2 and OPCML MSP primer sets to HO8910 cell line genomic
DNA
To verify if the chosen primers were able to
evaluate known methylation of tumor suppressor genes in patients
with EOC, genomic DNA was extracted from HO8910 cell culture.
HO8910 is an established human epithelial ovarian carcinoma cell
line. Bisulfite-modified genomic DNA from the HO8910 cells and
three sets of primers were used in the MSP. The three sets of
primers corresponded to non-coding regions of the tumor suppressor
genes OPCML, TFPI2 and RUNX3. Fig. 1A
demonstrates the quantitative results of separate MSP experiments
using each set of primers. Each reaction was repeated three times.
These results were analyzed by the NIH Java-based image processing
software, ImageJ. A 190-bp PCR product band was identified when
OPCML M pair primers were applied. The average integrated density
was ~130,000. No band was identified using the U pair primers.
Using the TFPI2 and RUNX3 M primer pairs, 140- and 80-bp bands were
amplified. However, same size PCR products were also present when U
primer pairs were used.
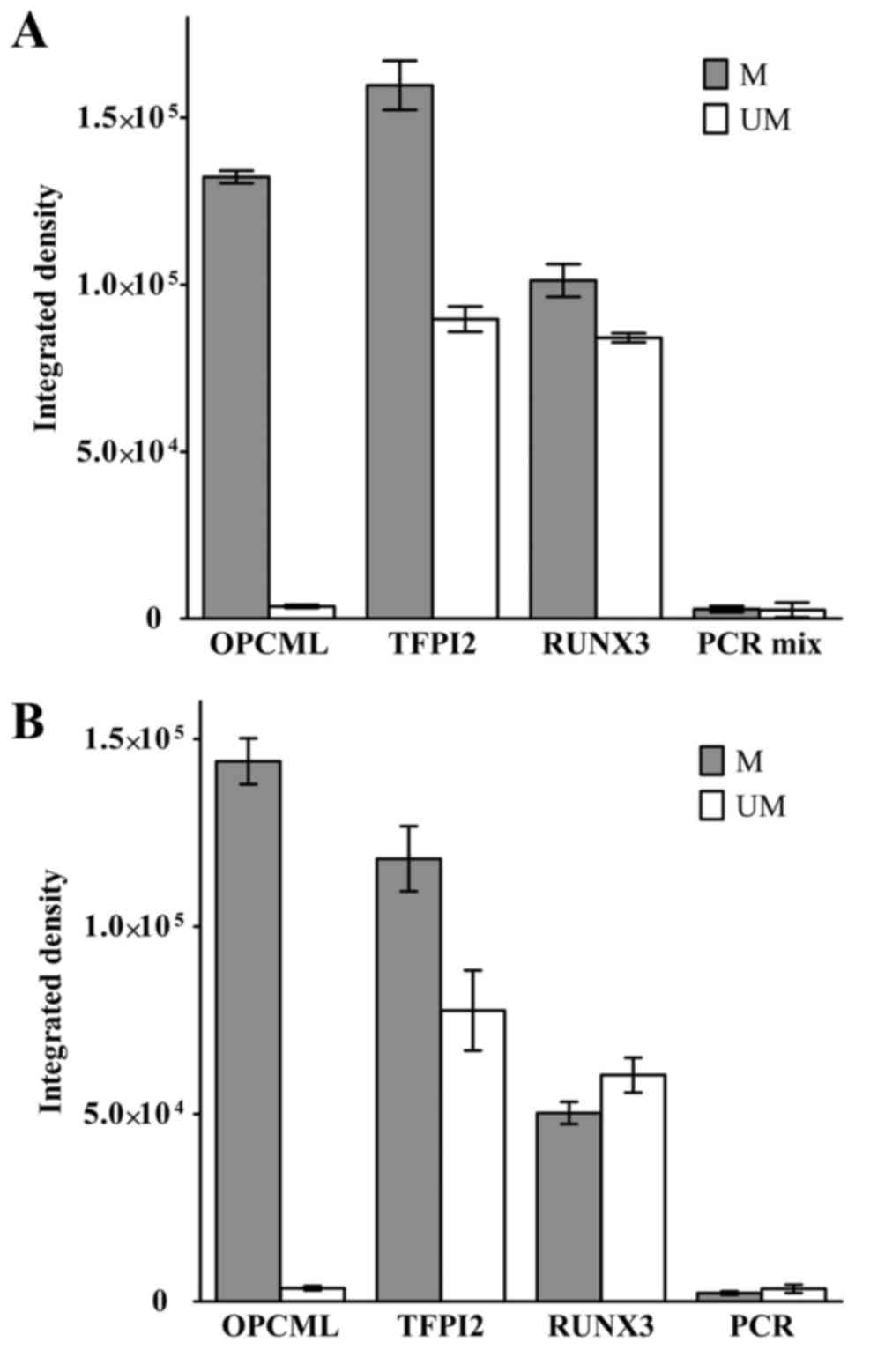 | Figure 1.MSP primer set detects OPCML
methylation status in epithelial ovarian cancer HO8910 cell line.
(A) Three sets of nested MSP primers, corresponding to OPCML, TFPI2
and RUNX3, and separate reactions were set up to test each single
set of primers. (B) Three sets of primers were mixed in one MSP
reaction. Integrated density represented relative PCR product
amount. PCR mix was the negative control, without DNA template.
Error bars represent standard deviation of the mean. OPCML, opioid
binding protein/cell adhesion molecule like; TFPI2, tissue factor
pathway inhibitor 2; RUNX3, runt-related transcription factor 3;
PCR, polymerase chain reaction; MSP, methylation-specific PCR; M,
methylated; UM, unmethylated. |
For manipulation convenience, the present study also
tested a PCR system with all three sets of primers in one reaction.
The results are presented in Fig. 1B,
and were similar to those for separate reactions. These data
suggested that the primer set for OPCML was appropriate for
additional investigation, and not the primer sets for TFPI2 and
RUNX3.
Validating OPCML MSP primer set in
patient samples
To examine if the MSP primer set for OPCML was
effective in real patient samples, plasma and tissue from patients
were tested. Sampling involved 3 randomly chosen patients in each
category (Fig. 2). In this
experiment, patients with EOC were classified as early or advanced
EOC based on their clinical signs and symptoms. fcDNA from plasma
or genomic DNA from cancer tissue were templates in the MSP. The
integrated density of PCR products corresponding to fcDNA from
early and advanced patients with EOC was ~63,000 and 52,000,
respectively. The integrated density of MSP products amplified
through genomic DNA from patients with early and advanced EOC was
32,000 and 35,000, respectively. Although there was a relatively
wide range of standard deviation, the scores were still
significantly higher compared with those from reactions utilizing
fcDNA of healthy donors, patients with benign tumors or the
background integrated density scores (~3,000).
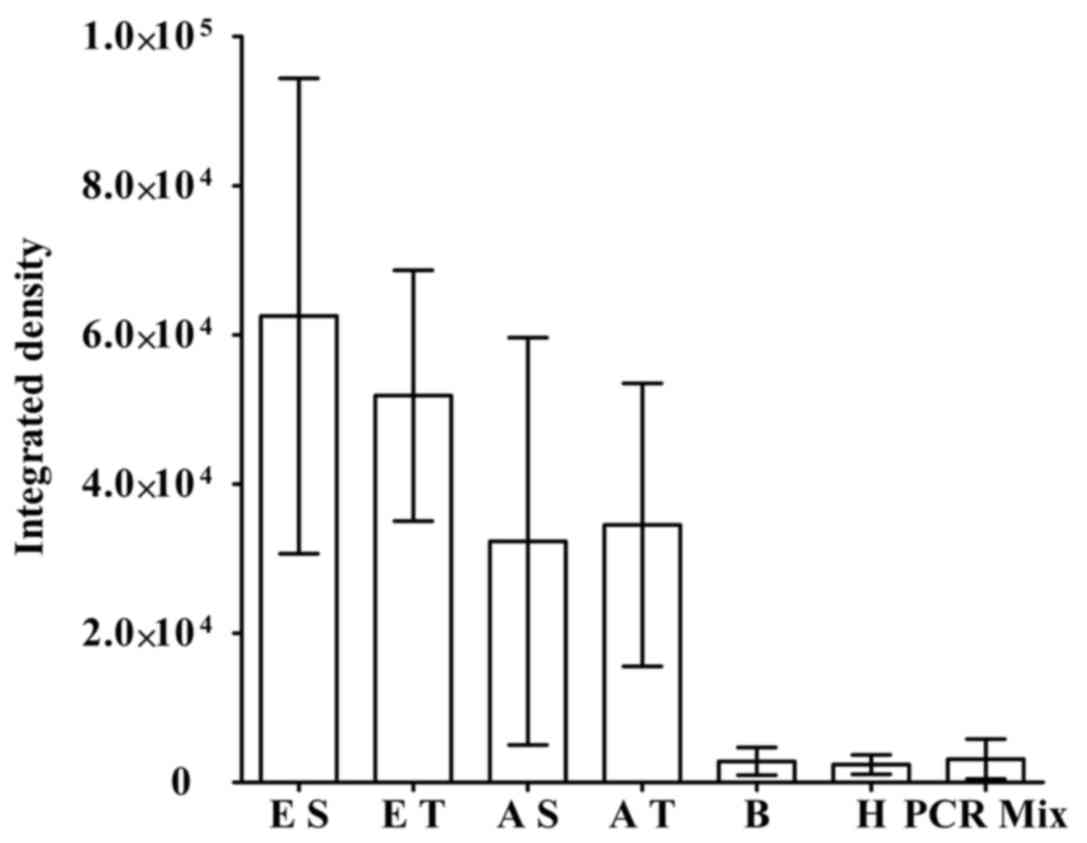 | Figure 2.OPCML MSP primer set may also be valid
for samples from patients with early- and advanced-stage EOC.
Nested MSP reactions examined OPCML methylation status in various
types of patients with EOC. Relative PCR product amount was
compared with respect to integrated density. Values indicate the
mean ± standard deviation. OPCML, opioid binding protein/cell
adhesion molecule like; EOC, epithelial ovarian carcinoma; PCR,
polymerase chain reaction; MSP, methylation-specific PCR; E,
patients with early-stage EOC; A, patients with advanced-stage EOC;
fcDNA, free circulating DNA; S, fcDNA as template; T, tissue
genomic DNA as template; B, fcDNA of patients with benign ovarian
tumors as template; H, fcDNA of healthy donors as template; PCR
mix, PCR reaction without templates. |
Methylation status of OPCML in
patients with EOC screened by MSP
To determine the efficacy of MSP compared with the
CA125 test with respect to the diagnosis of epithelial ovarian
carcinoma, plasma samples and fcDNA from 80 healthy donors, 43
patients with benign ovarian tumors and 71 ovarian epithelial
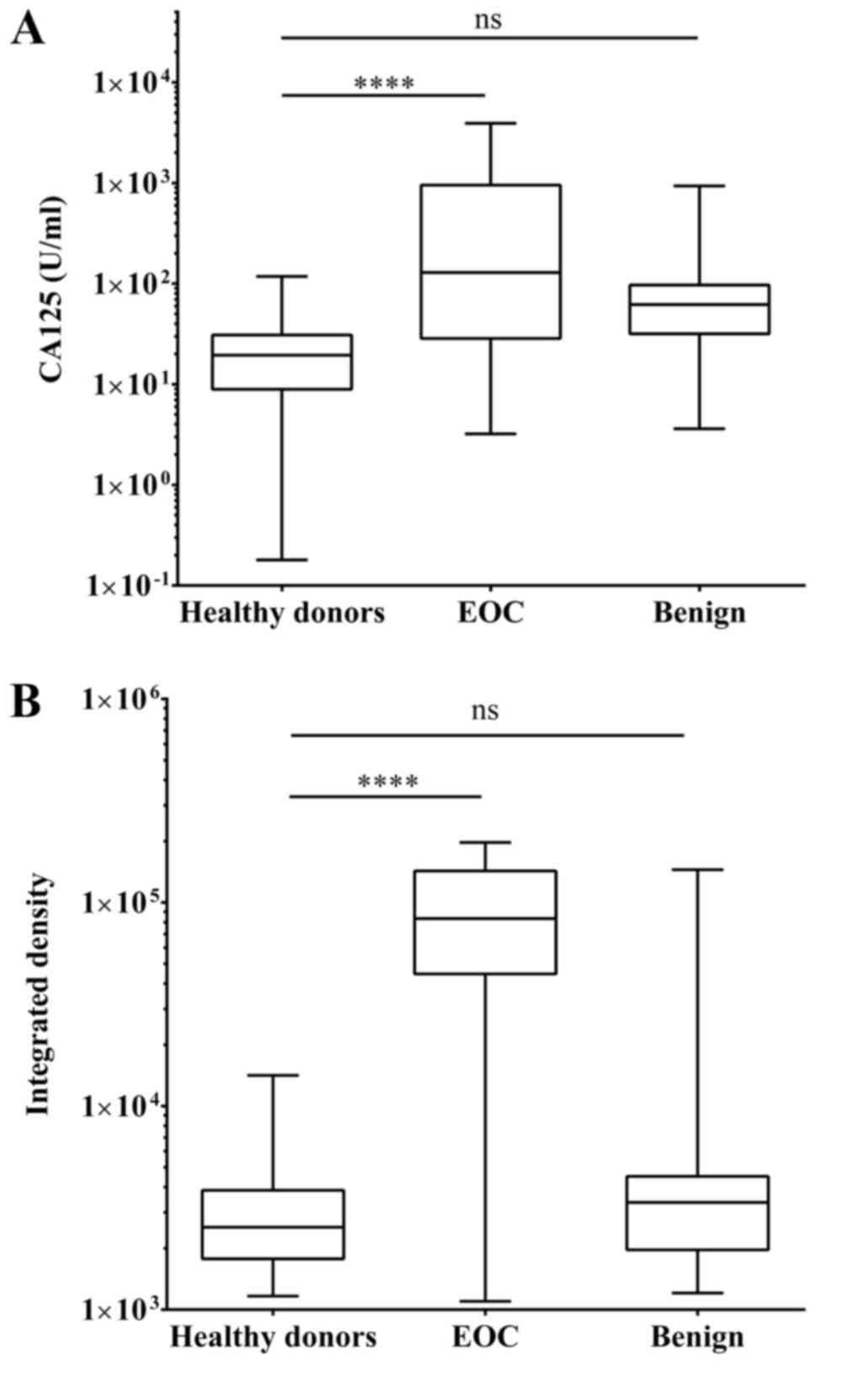
carcinoma donors were assessed by the two methods. The box and
whisker plot in Fig. 3A illustrates
that the median value of CA125 in healthy donors was 19.45 U/ml.
Comparing the aforementioned value with 128.93 U/ml CA125 in
patients with EOC, the P-value subsequent to one-way ANOVA was
<0.0001. Additionally, the median CA125 level in patients with
benign ovarian tumors was 62.13 U/ml, which was not statistically
significant compared with the healthy donors. One-way ANOVA for the
MSP results revealed that there was a statistically significant
difference between healthy donors and patients with EOC
(P<0.0001), but not between healthy donors and patients with
benign ovarian tumors (Fig. 3B).
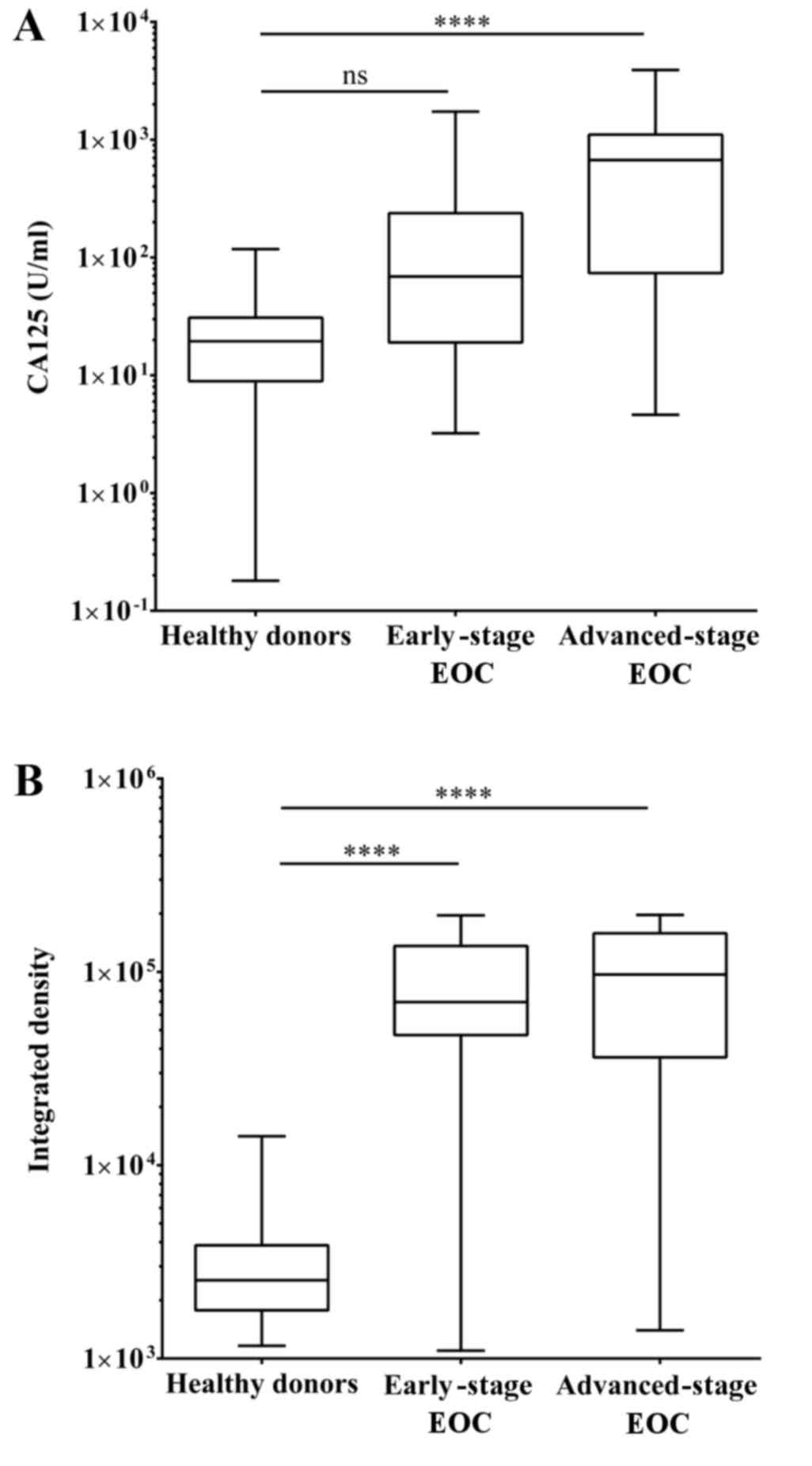
Significance of OPCML methylation
status in early-stage EOC reflected by MSP
The conventional CA125 test is not efficient to
detect early-stage EOC. To evaluate the detection sensitivity of
early-stage EOC using MSP and CA125 tests, patients with EOC in the
cohort of the present study were sorted into 39 early- and 32
advanced-stage patients based on the FIGO stages. Patients
exhibiting stages I and II were early-stage, while stages III and
IV were considered advanced-stage EOC. The median serum CA125 level
in patients with advanced-stage EOC reached up to 672.02, however
the median level of patients with early-stage EOC was 147 (Fig. 4). One-way ANOVA indicated that the
P-value of CA125 level with respect to the difference between
healthy donors and patients with advanced-stage EOC was <0.0001,
but no statistical significance was observed between healthy donors
and patients with early-stage EOC. The median integrated densities
of PCR products from healthy donors and patients with early- and
advanced-stage EOC were 2540.54, 69795.43 and 96921.52,
respectively. The P-values between healthy donors and patients with
early- or advanced-stage EOC were all <0.0001.
Diagnostic performance of MSP
method
To assess the diagnostic performance of the MSP test
in terms of early, advanced and overall EOC cases, the sensitivity,
specificity and accuracy of the CA125 test and the OPCML MSP test
were analyzed (Table II). A CA125
level <35 U/ml or integrated density <9,000 was defined as
negative results in the present study. The sensitivity of the CA125
test with respect to patients with early, advanced and overall EOC
was 53.85, 81.25 and 67.61%, respectively. The sensitivity of the
fcDNA MSP test, which was 87.18, 93.75 and 90.14%, respectively,
was higher than that of CA125 test for each group of patients. The
results of Cohen's κ coefficient for the CA125 test, whereby κ
represents the degree of agreement for categorizing subjects into
groups, were 0.14, 0.308 and 0.291 in the aforementioned order,
therefore the degrees of agreement are poor, fair and fair.
However, κ of the fcDNA MSP were 0.757, 0.784 and 0.813 and the
degrees of agreement were good, good, and very good,
respectively.
 | Table II.Performance of fcDNA MSP for
diagnosing patients with early-stage EOC. |
Table II.
Performance of fcDNA MSP for
diagnosing patients with early-stage EOC.
|
| EOC | Early-stage EOC | Advanced-stage
EOC |
|---|
|
|
|
|
|
|---|
| Variables | CA125 test | fcDNA MSP | CA125 test | fcDNA MSP | CA125 test | fcDNA MSP |
|---|
| Sensitivity, % | 67.61 | 90.14 | 53.85 | 87.18 | 81.25 | 93.75 |
|
| (48/71) | (64/71) | (21/39) | (34/39) | (26/32) | (30/32) |
| Specificity, % | 63.41 | 91.87 | 63.41 | 91.87 | 63.41 | 91.87 |
|
| (78/123) | (113/123) | (18/39) | (5/39) | (78/123) | (113/123) |
| Accuracy, % | 64.95 | 91.24 | 61.11 | 90.74 | 67.10 | 92.26 |
|
| (126/194) | (177/194) | (99/162) | (147/162) | (104/155) | (143/155) |
| PPV, % | 51.61 | 86.49 | 31.82 | 77.27 | 36.62 | 75.00 |
|
| (48/93) | (64/74) | (21/66) | (34/44) | (26/71) | (30/40) |
| NPV, % | 63.40 | 63.40 | 75.93 | 75.93 | 79.35 | 79.35 |
|
| (123/194) | (123/194) | (123/162) | (123/162) | (123/155) | (123/155) |
| κ | 0.291 | 0.813 | 0.14 | 0.757 | 0.308 | 0.784 |
| Strength of
agreement | Fair | Very good | Poor | Good | Fair | Good |
Discussion
Epithelial ovarian carcinoma is the most common type
of ovarian cancers, accounting for 90% of cases. Due to few or no
symptoms in the majority of early cases, 85% patients were revealed
to be diagnosed at an advanced stage (24). However, an issue with respect to
diagnosis and medical intervention is that the five-year survival
rate of advanced-stage patients with EOC may be <50%, whereas
the five-year survival rate of patients with early-stage EOC may be
>95% (4). At present, serum CA125
level is a common marker used to assist clinical diagnosis. The
inability of this measurement to screen for early-stage EOC makes
it unsatisfactory to aid resolving this challenge. The aim of the
present study was to reveal another biomarker to improve the
diagnosis of early-stage EOC.
Herman et al (22) reported MSP as a novel assay to detect
hypermethylation in tumor suppressor genes, which are often
silenced in patients with cancer. In the present study three
promising markers, methylated OPCML, TFPI2 and RUNX3, were selected
in an attempt to improve the efficacy of early-stage EOC diagnosis
assays. The initial study with HO8910 genomic DNA as a template
suggested that the OPCML MSP primer set may be an appropriate
marker, but TFPI2 and RUNX3 MSP primer sets may introduce too many
false positive results.
Furthermore, test results from patient tissue and
fcDNA samples indicated that nested MSP with the OPCML primer set
clearly differentiated patients with advanced-stage EOC from
patients with benign ovarian tumors and healthy donors, and
patients with early-stage EOC from healthy and non-malignant
samples. To validate the methylated OPCML maker and the MSP method
in screening early-stage EOC, the sample size was expanded to 80
healthy donors, 43 patients with benign tumors, 39 patients with
early-stage EOC and 32 patients with advanced-stage EOC. CA125
level results were consistent with previously known clinical data
(7). A total of 35 U/ml is an
appropriate threshold value to distinguish between healthy donors
and patients with EOC. Notably, even though the differences between
patients with benign ovarian tumors and healthy donors were not
statistically significant, >75% of patients with benign tumors
exhibited CA125 levels >30 U/ml, leading to false positive
results. Similar to the CA125 assay, the MSP test for methylated
OPCML also distinguished healthy donors and patients with benign
tumors from patients with EOC. In addition, the novel approach made
this difference more marked.
In diagnostic performance analysis, compared with
the CA125 test, the MSP assay increased Cohen's κ coefficient from
0.291 to 0.813. A κ value is considered good or excellent when it
is >0.81. Further investigation confirmed that the CA125 test
failed to provide unambiguous evidence of early-stage EOC; however,
the test was revealed to highlight patients with advanced-stage
EOC. Notably, the κ value of the MSP assay in early-stage EOC,
0.757, suggested a good agreement with the expected test results,
contrasted with 0.14 in the CA125 test, which is extremely low.
Thus, the present study revealed that the hypermethylation of OPCML
is a promising biomarker to identify early-stage EOC. Additionally,
the biomarker exhibits an excellent degree of agreement when
testing patients with EOC at all stages.
In addition, the present study also demonstrated
that multiple biomarkers may be tested for in a single MSP
reaction, as long as the sizes of the amplified DNA fragments are
different. This system may reduce the time required and increase
the sensitivity and accuracy of clinical diagnosis. Therefore, this
MSP assay for EOC may be a novel supplement or replacement for the
traditional CA125 test.
To develop an effective diagnostic kit, the present
authors aimed to improve the screening technique to provide a
higher sensitivity and a κ value >0.81 for early-stage EOC. A
potential method is the investigation of additional biomarkers.
Sung et al (25) reported the
absence of p150 expression in ovarian cancer cells. This occurs due
to the methylation of the P2 promoter in a putative tumor
suppressor gene, spalt-like transcription factor 2. Other potential
candidates include p16 (26), cyclin
dependent kinase inhibitor 2B, cadherin 13 or ras association
domain family member 1 (27).
Multiple biomarkers may compensate for individual differences among
patients, which may account for some of the false negative cases in
single biomarker assays.
Investigating and designing MSP primers may be
another key aspect to improve the assay developed in the present
research. TFPI2 and RUNX3 MSP primers were not adopted for
screening patient cohorts since they introduced unacceptable false
positive results. One of the criteria for the design of MSP primers
is the inclusion of at least one CpG site at the 3′-end (22,23).
Unfortunately, the RUNX3 U pair of MSP primers does not exhibit a
CpG site at the 3′-end. In addition, adequate CpG sites are
necessary for good quality MSP primers (23). The U pair of the TFPI2 MSP primers
exhibits two CpG sites, which may be not be competent enough to
differentiate methylated DNA templates from unmethylated ones. As a
result, the two primers may be imperfect MSP primers. Therefore,
designing and testing high-quality MSP primers may be part of
prospective investigation to optimize the assay of the present
study. The present study demonstrates a preliminary effort to
identify a promising novel biomarker and develop an improved
screening method for EOC. Building upon the results of the present
study, a clinical screening test kit for EOC, particularly early
stage EOC, will be investigated.
Acknowledgements
The present study was supported by the Guangdong
Academician Workstation Development (grant no.
2012/397-2012B09050007), the Guangdong International Cooperation
(grant no. 2010B050100022), and the Guizhou Guiyang Maternal and
Child Health Care Hospital Academician Workstation (grant no.
20165603).
References
|
1
|
American Cancer Society, . Cancer Facts
& Figures 2015. American Cancer Society; Atlanta, GA: 2015
|
|
2
|
Ries LAG, Young JL, Keel GE, Eisner MP,
Lin YD and Horner MJ: SEER Survival Monograph: Cancer Survival
Among Adults. U.S. Seer Program, 1988–2001, Patient And Tumor
Characteristics. National Cancer Institute, Seer Program, NIH Pub.
No. 07-6215; Bethesda, MD: 2007
|
|
3
|
Prat J; FIGO Committee on Gynecologic
Oncology, : Staging classification for cancer of the ovary,
fallopian tube and peritoneum. Int J Gynaecol Obstet. 124:1–5.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Seiden MV: Gynecologic
malignanciesHarrison's Principles of Internal Medicine. Longo DL,
Kasper DL, Jameson JL, Fauci AS, Hauser SL and Loscalzo J: 1. 18th.
McGraw-Hill; New York, NY: pp. 810–816. 2012
|
|
5
|
Suh KS, Park SW, Castro A, Patel H, Blake
P, Liang M and Goy A: Ovarian cancer biomarkers for molecular
biosensors and translational medicine. Expert Rev Mol Diagn.
10:1069–1083. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gupta D and Lis CG: Role of CA125 in
predicting ovarian cancer survival-a review of the epidemiological
literature. J Ovarian Res. 2:132009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ozols RF: The current role of gemcitabine
in ovarian cancer. Semin Oncol. 28 2 Suppl 7:S18–S24. 2001.
View Article : Google Scholar
|
|
8
|
Leon SA, Shapiro B, Sklaroff DM and Yaros
MJ: Free DNA in the serum of cancer patients and the effect of
therapy. Cancer Res. 37:646–650. 1977.PubMed/NCBI
|
|
9
|
Flamini E, Mercatali L, Nanni O, Calistri
D, Nunziatini R, Zoli W, Rosetti P, Gardini N, Lattuneddu A,
Verdecchia GM and Amadori D: Free DNA and carcinoembryonic antigen
serum levels: An important combination for diagnosis of colorectal
cancer. Clin Cancer Res. 12:6985–6988. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Silva JM, Dominguez G, Garcia JM, Gonzalez
R, Villanueva MJ, Navarro F, Provencio M, San Martin S, España P
and Bonilla F: Presence of tumor DNA in plasma of breast cancer
patients: Clinicopathological correlations. Cancer Res.
59:3251–3256. 1999.PubMed/NCBI
|
|
11
|
Gordian E, Ramachandran K, Reis IM,
Manoharan M, Soloway MS and Singal R: Serum free circulating DNA is
a useful biomarker to distinguish benign versus malignant prostate
disease. Cancer Epidemiol Biomarkers Prev. 19:1984–1991. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kamat AA, Bischoff FZ, Dang D, Baldwin MF,
Han LY, Lin YG, Merritt WM, Landen CN Jr, Lu C, Gershenson DM, et
al: Circulating cell-free DNA: A novel biomarker for response to
therapy in ovarian carcinoma. Cancer Biol Ther. 5:1369–1374. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mazurek AM, Fiszer-Kierzkowska A,
Rutkowski T, Składowski K, Pierzyna M, Scieglińska D, Woźniak G,
Głowacki G, Kawczyński R and Małusecka E: Optimization of
circulating cell-free DNA recovery for KRAS mutation and HPV
detection in plasma. Cancer Biomark. 13:385–394. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ehrlich M, Gama-Sosa MA, Huang LH, Midgett
RM, Kuo KC, McCune RA and Gehrke C: Amount and distribution of
5-methylcytosine in human DNA from different types of tissues of
cells. Nucleic Acids Res. 10:2709–2721. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jabbari K and Bernardi G: Cytosine
methylation and CpG, TpG (CpA) and TpA frequencies. Gene.
333:143–149. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Deaton AM and Bird A: CpG islands and the
regulation of transcription. Genes Dev. 25:1010–1022. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim TY, Lee HJ, Hwang KS, Lee M, Kim JW,
Bang YJ and Kang GH: Methylation of RUNX3 in various types of human
cancers and premalignant stages of gastric carcinoma. Lab Invest.
84:479–484. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang S, Wei L, Zhang A, Zhang L and Yu H:
RUNX3 gene methylation in epithelial ovarian cancer tissues and
ovarian cancer cell lines. OMICS. 13:307–311. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sierko E, Wojtukiewicz MZ and Kisiel W:
The role of tissue factor pathway inhibitor-2 in cancer biology.
Semin Thromb Hemost. 33:653–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sellar GC, Watt KP, Rabiasz GJ, Stronach
EA, Li L, Miller EP, Massie CE, Miller J, Contreras-Moreira B,
Scott D, et al: OPCML at 11q25 is epigenetically inactivated and
has tumor-suppressor function in epithelial ovarian cancer. Nat
Genet. 34:337–343. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chuang LS and Ito Y: RUNX3 is
multifunctional in carcinogenesis of multiple solid tumors.
Oncogene. 29:2605–2615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: A novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA. 93:pp.
9821–9826. 1996; View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ledermann JA, Raja FA, Fotopoulou C,
Gonzalez-Martin A, Colombo N and Sessa C; ESMO Guidelines Working
Group, : Newly diagnosed and relapsed epithelial ovarian carcinoma:
ESMO clinical practice guidelines for diagnosis, treatment and
follow-up. Ann Oncol. 24 Suppl 6:vi24–vi32. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sung CK, Li D, Andrews E, Drapkin R and
Benjamin T: Promoter methylation of the SALL2 tumor suppressor gene
in ovarian cancers. Mol Oncol. 7:419–427. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Katsaros D, Cho W, Singal R, Fracchioli S,
De La Longrais IA Rigault, Arisio R, Massobrio M, Smith M, Zheng W,
Glass J and Yu H: Methylation of tumor suppressor gene p16 and
prognosis of epithelial ovarian cancer. Gynecol Oncol. 94:685–692.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ozdemir F, Altinisik J, Karateke A,
Coksuer H and Buyru N: Methylation of tumor suppressor genes in
ovarian cancer. Exp Ther Med. 4:1092–1096. 2012.PubMed/NCBI
|