Introduction
As one of the tumors with the highest incidence in
China, the related research results show that (1) at present, the incidence of leukemia is
increasing year by year (2).
Currently, there is a high incidence of leukemia, especially acute
leukemia, which is a cancer that progresses rapidly (3). The current mechanism and cause of the
pathogenesis of leukemia is not clear, which is the reason why
there are no specific drugs available for leukemia (4). In recent years, with the development of
molecular biology techniques, increasing research shows that
abnormal expression of some human genes (e.g., HLA and other immune
regulation related genes) are correlated to the incidence of
leukemia (5). Results of this study
show that the HLA gene located on human chromosome 6 is related to
many diseases of the human body (6).
For example, in children with acute lymphoblastic leukemia
(7), HLA-DQA*101 and *014 in children
increased significantly compared to normal. At the same time, a
study on the correlation between the chronic myelogenous leukemia
(CML) and HLA showed that (8) there
was a correlation between the incidence of CML and the abnormal
increase and the decline of HLA antigen frequency (9). The abnormal changes of HLA-B8, -B14,
-DR3 and other antigen frequencies greatly promotes the occurrence
and progression of CMNL (10).
Currently, the research on the HLA gene polymorphism is mainly
focused on HLA-1. There are few studies on the polymorphism of
HLA-II gene, especially the association between HLA-II gene
polymorphism and leukemia. In this study, we investigated the
correlation between HLA class II gene polymorphism and leukemia for
the first time in order to reveal the correlation between HLA class
II gene polymorphism and acute leukemia that may exist in order to
provide certain theoretical and experimental basis for leukemia
diagnosis and treatment.
Materials and methods
Sample selection
For this study, we selected 53 patients with
leukemia from February 2014 to September 2015 at Zhongnan Hospital
of Wuhan University as the research subjects. This cohort included
26 males and 27 females with an average age of 24.3±18.2 years. We
also selected 46 healthy persons as the control group, including 23
males and 23 females, with an average age of 22.6±16.7 years. This
study was approved by the Ethics Committee of Wuhan University.
Signed written informed consents were obtained from all
participants before the study. Inclusion criteria: Patients must be
diagnosed with leukemia based on cell morphology and immune cell
typing. Exclusion criteria: Patients with other related diseases
such as other types of cancer and patients suffering from other
immune deficiency diseases such as AIDS.
Main instruments and reagents
The main instruments used were PCR, refrigerated
centrifuge and gel imaging instrument (all from Bio-Rad
Laboratories, Inc., Hercules, CA, USA). The main reagents were DNA
extraction and enzyme-linked immunosorbent assay (ELISA) kits (both
from Qiagen, Hilden, Germany), and HLA goat anti-mouse first
antibody (cat. no. ab6789; dilution, 1:1,250; Abcam, Cambridge, MA,
USA) and PCR reagents (Takara Bio, Dalian, China), SSO-HLA gene
type kit (Dynal, Wirral, UK).
Method
Genome extraction
We extracted 5ml of peripheral blood from both
patients in the observation and control group, centrifuged at 2,750
× g for 10 min, and added EDTA to the supernatant and stored at
−80°C. Using Qiagen's DNA extraction kit extract, and in accordance
with the instructions, we extracted cell genome from the patients'
blood. Genomic DNA concentration was measured using micro-UV points
spectrophotometric gauge (Biosharp, Hefei, China). The DNA purity
was A260/280=1.8–2.0, then the extraction of genomic DNA was used
in the following experiments.
Genotyping
In this study, different genotypes were classified
using polymerase chain reaction with sequence specific primers
(PCR-SSP) kit (KeyGen, Nanjing, China). The primers were
synthesized by the Shanghai Biological Engineering Co. Ltd.
(Shanghai, China). Primer sequences are shown in Table I. HLA-II genotypes with high
polymorphism were obtained by different primers amplification, and
then the PCR product was heated to 95°C in the PCR instrument. The
single stranded DNA was obtained by unzipping double-stranded DNA,
and then adding it to the nylon membrane labeled with a specific
nucleotide fragment. The single stranded DNA and specific
nucleotide fragments fused for double stranded DNA through
complementary pairing between base groups. Then, streptomycin
labeled HRP was added, and reacted at room temperature for 2 min.
Nylon membrane after hybridization was then treated with color in
the gel imaging instrument, and the HLA-II poly-morphism was
identified by the hybridization bands.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Primer | Sequences |
|---|
| HLA-A-F |
GTGATCGTAGTGCTAGCTAGC |
| HLA-A-R |
CGTCGTAGCTGATCGTAGCTAG |
| HLA-B-F |
CTGCTAGTCGATCGGCATATACG |
| HLA-B-R |
CTGATCGTAGCTAGCCTAGATCG |
ELISA
Total protein extracted from different genotypic
samples and control samples were studied. The expression of HLA-II
protein in different samples was determined according to the
specification of ELISA kit (Qiagen). In the study, protein samples
for the ELISA standard curve were diluted by the elution buffer at
a ratio of 1:50. After diluted solution of different concentrations
were obtained and according to the specification of the operation,
the standard curve was produced. In samples of the observation and
the control group, PBS (pH 7.2), through the sterilization, was
diluted with a scale of 1:200. A 100 µl of solution was taken and
added into 96-well plates. Fifty microliters of test solution was
added to each well, and after incubation for 2 h at room
temperature, the TMB color substrate was added. The absorbance at
495 nm was measured and then the protein expression and
concentration of HLA-II in each sample were calculated according to
the standard curve.
Western blotting experiment
The total protein extraction from each subject was
the experimental material. Approximately 150 mg of the frozen
(−80°C) sample was rapidly ground in an EP tube in a mortar
containing liquid nitrogen. Then, we added 300 µg protein
extraction and 10 µl protease inhibitors, and then incubated in ice
water for 30 min. We centrifuged the sample at 10,000 × g for 150
min, collected the supernatant and analyzed total protein
extraction. Secondly, the expression of protein was detected by
western blotting, a mix of 10 µl supernatant was obtained and the
sample buffer for conventional SDS-PAGE and electrophoresis were
prepared. The membrane was incubated at room temperature for 2 h
with the HLA-II protein antibody (1:250, 4°C overnight). Then, it
was washed with TBST 5 times, each time for 10 min, and incubated
with the horseradish peroxidase-labeled secondary antibody (cat.
no. ab6789; dilution, 1:1,250; Abcam; 60 rpm incubated with shaker
for 1 h at room temperature). The membrane was washed 3 times, each
time for 10 min, and then the color was shown by diaminobenzidine,
and the image was obtained by the Fluorchem 9900 imaging system
(Alpha Innotech, San Leandro, CA, USA). We determined the integral
optical density of color belt of the protein, and the relative
content of HLA-II protein was calculated.
Statistical analysis
SPSS 20.0 software (IBM SPSS, Armonk, NY, USA) was
used for data statistical analysis. The experimental data were
expressed by mean ± standard error. The single factor analysis
method was used to analyze the data between different groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
HLA-A gene polymorphism and genotype
frequency detection in the observation group and the control
group
Based on the HLA-A alleles and gene frequency
detection in the observation and the control group, a total of 12
loci were found. Results are shown in Table II. Compared to the control group, 53
cases of leukemia patients had higher gene frequency of HLA-A04;
differences were significant (P<0.05). The frequency of the
HLA-A09 gene was lower, and the frequency of the gene had a
significant difference from the control group (P<0.05). A06,
A12, A08 loci gene frequencies were slightly higher than those of
the control group, but results were not significantly different
(P>0.05). A02 and A05 were slightly lower than those of the
control group, however, there was no significant difference between
the two proteins (P>0.05).
 | Table II.HLA-A allele and gene frequency
statistics of the observation group and the control group. |
Table II.
HLA-A allele and gene frequency
statistics of the observation group and the control group.
|
| Observation group
(53) | Control group
(46) |
|
|---|
|
|
|
|
|
|---|
| HLA-A | No. | Frequency (%) | No. | Frequency (%) | χ2 |
|---|
| A01 | 2 | 3.8 | 1 | 2.2 | 6.04 |
| A02 | 2 | 3.8 | 3 | 6.5 | 0.12 |
| A03 | 3 | 5.7 | 1 | 2.2 | 0.18 |
| A04 | 26 | 49 | 3 | 6.5 | 0.24 |
| A05 | 3 | 5.7 | 4 | 8.7 | 0.64 |
| A06 | 3 | 5.7 | 1 | 2.2 | 0.116 |
| A07 | 1 | 1.8 | 1 | 2.2 | 0.19 |
| A08 | 3 | 5.7 | 2 | 4.3 | 0.13 |
| A09 | 2 | 3.8 | 26 | 57 | 4.07 |
| A10 | 2 | 3.8 | 1 | 2.2 | 0.26 |
| A11 | 3 | 5.7 | 1 | 2.2 | 0.32 |
| A12 | 3 | 5.7 | 2 | 4.3 | 0.48 |
HLA-B gene polymorphism and genotype
frequency detection in the observation group and the control
group
Based on the HLA-A alleles and gene frequency
detection in the observation and the control group, a total of 14
loci was found. Results are shown in Table III. Compared to the control group,
in 53 cases of leukemia patients, the HLA-B08 gene frequency was
lower; results were found to be significantly different from the
control group (P<0.05). B03, B05 and B11 loci gene frequencies
were slightly higher than those of the control group, but there was
no significant difference (P>0.05). B01 and B07 were slightly
lower than those of the control group; there was no significant
difference between the two proteins (P>0.05).
 | Table III.HLA-B allele and gene frequency
statistics of the observation group and the control group. |
Table III.
HLA-B allele and gene frequency
statistics of the observation group and the control group.
|
| Observation group
(53) | Control group
(46) |
|
|---|
|
|
|
|
|
|---|
| HLA-B | No. | Frequency (%) | No. | Frequency (%) | χ2 |
|---|
| B01 | 1 | 1.9 | 2 | 4.3 | 0.14 |
| B02 | 2 | 3.8 | 2 | 4.3 | 0.116 |
| B03 | 6 | 11.3 | 3 | 6.5 | 0.32 |
| B04 | 3 | 5.7 | 2 | 4.3 | 0.21 |
| B05 | 8 | 15.1 | 4 | 8.7 | 0.204 |
| B06 | 6 | 11.3 | 5 | 10.9 | 0.165 |
| B07 | 2 | 3.8 | 4 | 8.7 | 0.186 |
| B08 | 7 | 13.2 | 13 | 28.3 | 6.05 |
| B09 | 5 | 9.4 | 3 | 6.5 | 0.214 |
| B10 | 3 | 5.7 | 2 | 4.3 | 0.24 |
| B11 | 4 | 7.5 | 3 | 6.5 | 0.253 |
| B12 | 2 | 3.8 | 1 | 2.2 | 0.32 |
| B13 | 2 | 3.8 | 1 | 2.2 | 0.36 |
| B14 | 2 | 3.8 | 1 | 2.2 | 0.42 |
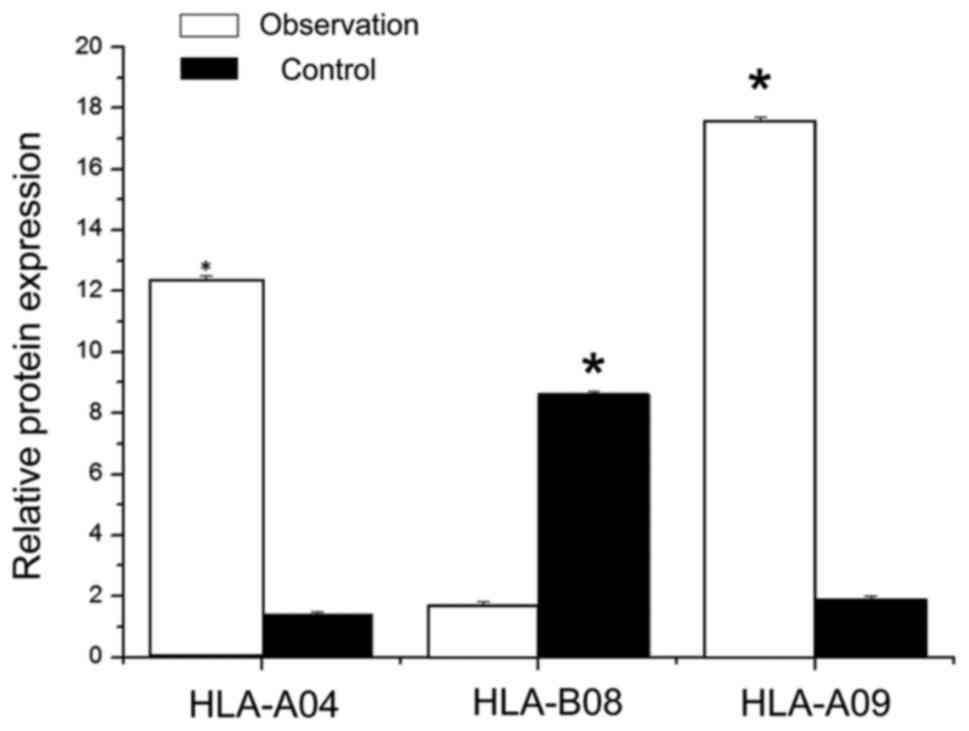
ELISA to detect the expression of
HLA-II protein in different genotypes
The total protein extraction of patients in the
control group as well as the observation group were analyzed. The
expression of HLA-II protein in different genotypes was determined
by ELISA method, and the results are shown in Fig. 1. Compared to the control group, the
HLA-A04 genotype protein content (12.3±0.2 µg/l) in the observation
group was significantly higher than HLA-A04 protein content in the
control group (1.4–0.1 µg/l); there were no significant differences
between the two groups (P<0.05). In the observation group, the
HLA protein content (1.7±0.13 µg/l) was significantly lower than
HLA-B08 protein content in the control group (8.6±0.12); there were
significant differences between the two (P<0.05). The content of
HLA-09 protein in the observation group (17.6±0.11 µg/l) was
significantly higher than HLA-09 protein of the control group
(1.9±0.12 µg/l), and there was a significant difference between the
two groups (P<0.05).
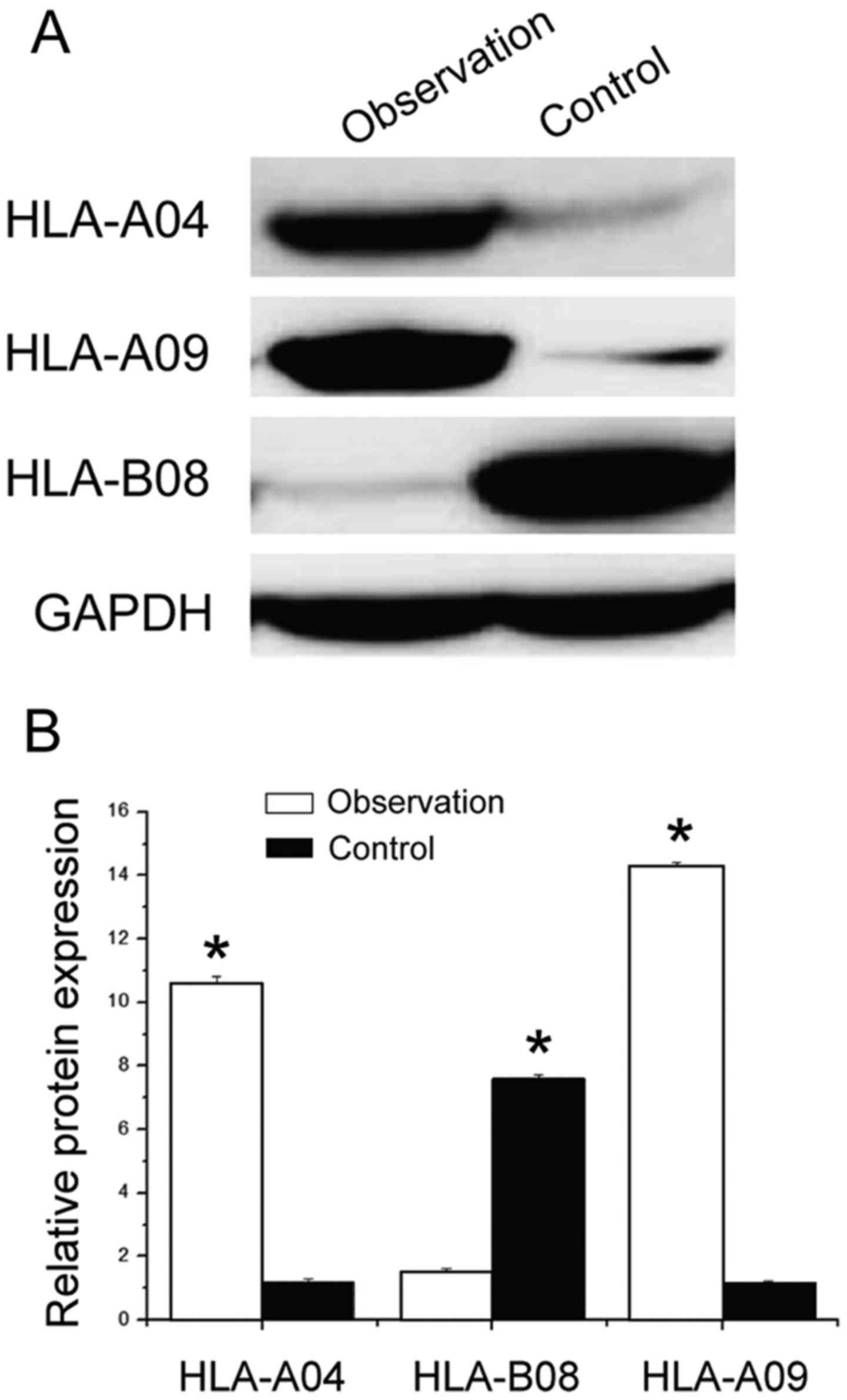
Western blotting to detect the
expression of HLA-II protein in different genotypes
The total protein extracted from patients in the
control group as well as the observation group was investigated.
The expression of the HLA-II protein in different genotypes was
determined by western blotting; the results are shown in Fig. 2. The content of the HLA-04 protein in
the observation group was significantly higher than that in the
control group; there were significant differences between the two
(P<0.05). In the observation group, the content of HLA protein
in HLA-A09 was significantly lower than that in the control group,
and there was a significant difference between the two (P<0.05).
Whereas, the content of HLA protein in the HLA-B08 genotype in the
observation group was significantly higher than that in the control
group, and there were significant differences between the two
(P<0.05). These results are in agreement with the ELISA
results.
Discussion
In recent years, research has shown that (11) the HLA gene is located on the human
sixth chromosome. It is related to a complex gene regulation and
control system in the human body (12). Since the 1990s, research on (13) HLA alleles in the human body (14) and HLA related diseases (15,16) in
chronic myeloid leukemia has shown that the frequencies of HLA-A32,
B27, B45 and B44 were significantly higher than those in the
control group (17). The HLA-A32 gene
frequency was lower compared to the control group, and with further
study of the HLA-II genotype, it was found that HLA-II genotype in
the human body is not only involved in an external immune response
of the body, but also has an important role in the promotion of the
maturation of lymphocytes and other immune cells in vivo
(18).
As a type of genetic disease that is very harmful,
the diagnosis and treatment of leukemia has been paid increasing
attention, but because of the complexity of leukemia (19), its incidence is often affected by the
external environment and the genetic factors. Therefore, there is
no cure for leukemia. Early diagnosis and treatment of leukemia has
become the key to the treatment and prevention of leukemia
(20). In this study, we found
associations between different polymorphisms of the HLA-II gene
polymorphism and leukemia. Particularly, the polymorphisms of
HLA-A04, A09 and B08 are associated with the occurrence and
suppression of leukemia to a large extent; HLA-A04 and HLA-A09 can
promote the occurrence and progression of leukemia, and HLA-B08, to
a certain extent, can inhibit the occurrence and progression of
leukemia through the detection of different genotypes of HLA
protein activity. The HLA activity of HLA-A04 and HLA-A09
significantly increased, which showed that the above mutations
promote the deterioration of leukemia by promotion of the activity
of HLA. However, due to the experimental conditions and the time
limit, this study did not analyze the spatial structure of protein
of different genotypes from the aspects of protein structure, or
reveal the potential relevance between different genotypes and
protein spatial structure.
References
|
1
|
McErlean C, Gonzalez AA, Cunningham R,
Meenagh A, Shovlin T and Middleton D: Differential RNA expression
of KIR alleles. Immunogenetics. 62:431–440. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bao X, Hou L, Sun A, Chen M, Chen Z and He
J: An allelic typing method for 2DS4 variant used in study of
haplotypes of killer cell immunoglobulin-like receptor gene. Int J
Lab Hematol. 32(6p2): 625–632. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhuang YL, Zhu CF, Zhang Y, Song YH, Wang
DJ, Nie XM, Liu Y and Ren GJ: Association of KIR2DS4 and its
variant KIR1D with syphilis in a Chinese Han population. Int J
Immunogenet. 39:114–118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Salim PH, Jobim M, Jobim LF and Xavier RM:
Autoimmune rheumatic diseases and their association with killer
immunoglobulin-like receptor genes. Rev Bras Reumatol. 51:351–356,
362-364. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mosaad YM, Mansour M, Al-Muzairai I,
Al-Otabi T, Abdul-Moneam M, Al-Attiyah R and Shahin M: Association
between human leukocyte antigens (HLA-A, -B, and -DR) and end-stage
renal disease in Kuwaiti patients awaiting transplantation. Ren
Fail. 36:1317–1321. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weitkamp LR, Moss AJ, Lewis RA, Hall WJ,
MacCluer JW, Schwartz PJ, Locati EH, Tzivoni D, Vincent GM,
Robinson JL, et al: Analysis of HLA and disease susceptibility:
chromosome 6 genes and sex influence long-QT phenotype. Am J Hum
Genet. 55:1230–1241. 1994.PubMed/NCBI
|
|
7
|
Ben Hamad M, Mahfoudh N, Marzouk S,
Kammoun A, Gaddour L, Hakim F, Fakhfakh F, Bahloul Z, Makni H and
Maalej A: Association study of human leukocyte antigen-DRB1 alleles
with rheumatoid arthritis in south Tunisian patients. Clin
Rheumatol. 31:937–942. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Karahan GE, Kekik C, Oguz FS, Onal AE,
Bakkaloğlu H, Calişkan YK, Yazici H, Turkmen A, Aydin AE, Sever MS,
et al: Association of HLA phenotypes of end-stage renal disease
patients preparing for first transplantation with anti-HLA antibody
status. Ren Fail. 32:380–383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jiyun Y, Guisen L, Li Z, Yi S, Jicheng L,
Fang L, Xiaoqi L, Shi M, Cheng J, Ying L, et al: The genetic
variants at the HLA-DRB1 gene are associated with primary IgA
nephropathy in Han Chinese. BMC Med Genet. 13:332012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xiao X, Liu L, Li WJ, Liu J and Chen DJ:
HLA-A, HLA-B, HLA-DRB1 polymorphisms and risk of cervical squamous
epithelial cell carcinoma: a population study in China. Asian Pac J
Cancer Prev. 14:4427–4433. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Amirzargar AA, Yalda A, Hajabolbaghi M,
Khosravi F, Jabbari H, Rezaei N, Niknam MH, Ansari B, Moradi B and
Nikbin B: The association of HLA-DRB, DQA1, DQB1 alleles and
haplotype frequency in Iranian patients with pulmonary
tuberculosis. Int J Tuberc Lung Dis. 8:1017–1021. 2004.PubMed/NCBI
|
|
12
|
Franco C, Yoo W, Franco D and Xu Z:
Predictors of end stage renal disease in African Americans with
lupus nephritis. Bull NYU Hosp Jt Dis. 68:251–256. 2010.PubMed/NCBI
|
|
13
|
El-Gezawy EM, Baset HA, Nasif KA, Osama A,
AbdelAzeem HG, Ali M and Khalil RY: Human leukocyte antigens as a
risk factor for the primary diseases leading to end stage renal
disease in Egyptian patients. Egypt J Immunol. 18:13–21.
2011.PubMed/NCBI
|
|
14
|
Korva M, Saksida A, Kunilo S, Jeras B
Vidan and Avsic-Zupanc T: HLA-associated hemorrhagic fever with
renal syndrome disease progression in slovenian patients. Clin
Vaccine Immunol. 18:1435–1440. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Catamo E, Zupin L, Crovella S, Celsi F and
Segat L: Non-classical MHC-I human leukocyte antigen (HLA-G) in
hepatotropic viral infections and in hepatocellular carcinoma. Hum
Immunol. 75:1225–1231. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang X, Jia J, Dong J, Yu F, Ma N, Li M,
Liu X, Liu W, Li T and Liu D: HLA-DQ polymorphisms with HBV
infection: different outcomes upon infection and prognosis to
lamivudine therapy. J Viral Hepat. 21:491–498. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pollicino T, Cacciola I, Saffioti F and
Raimondo G: Hepatitis B virus PreS/S gene variants: pathobiology
and clinical implications. J Hepatol. 61:408–417. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim YJ, Kim HY, Lee JH, Yu SJ, Yoon JH,
Lee HS, Kim CY, Cheong JY, Cho SW, Park NH, et al: A genome-wide
association study identified new variants associated with the risk
of chronic hepatitis B. Hum Mol Genet. 22:4233–4238. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang Q, Yin J, Zhang Y, Deng Y, Ji X, Du
Y, Pu R, Han Y, Zhao J, Han X, et al: HLA-DP polymorphisms affect
the outcomes of chronic hepatitis B virus infections, possibly
through interacting with viral mutations. J Virol. 87:12176–12186.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mansouri L, Messalmani M, Klai S, Bedoui
I, Derbali H, Gritli N, Mrissa R and Fekih-Mrissa N: Association of
HLA-DR/DQ polymorphism with Alzheimer's disease. Am J Med Sci.
349:334–337. 2015. View Article : Google Scholar : PubMed/NCBI
|