Introduction
Colorectal cancer (CRC) is one of the three most
common types of cancer worldwide and is a major contributor to
cancer-associated mortality (1).
Effective screening to detect cancer is expected to decrease the
mortality rate of CRC (2). Novel
methods are currently under development for the detection of CRC,
including those based on the detection of microRNAs (miRNA/miR)
(3,4).
miRNAs are small (between 19 and 22 nucleotides)
non-coding RNA molecules that are encoded by eukaryotic genomic
DNA. miRNAs lead to translation repression or degradation of
specific target mRNAs by directly binding to their potential target
site in the 3′ untranslated region (5). Located in the spacer regions between
protein-coding genes or in the introns of protein-encoding genes,
miRNA coding sequences are transcribed as primary miRNAs in the
same manner as the mRNAs of the protein-coding genes (6). miRNAs are important mediators of
biological functions, and their dysregulation has been implicated
in a wide range of diseases, including malignancies, heart
diseases, inflammation and lung diseases (7–9).
As identified previously, miR-28 suppresses
viability, but activates metastasis in CRC cells (10), miR-381 increases radiosensitivity in
esophageal squamous cell carcinoma (11), and miRNA-27b targets vascular
endothelial growth factor C to inhibit tumor progression and
angiogenesis in CRC (12).
miR-106a (miRBase accession number MIMAT0000103),
located in Xq26.2, exhibits oncogenic activity in humans, and its
expression is often altered in carcinogenesis (13). It has been demonstrated to be highly
expressed in cancer tissues, including gastric carcinoma and
ovarian cancer, and may be a potential biomarker in the diagnosis
of certain malignant tumors (14–16).
In the present study, the expression levels of
miR-106a were determined in CRC tissues and plasma, compared with
the control group, and the potential association between the
expression of miR-106a and clinicopathological factors of all
patients was analyzed. In order to investigate the underlying
molecular mechanism for the effects of miR-106, apoptosis of CRC
cells transfected with miR-106a mimic and miR-106a inhibitor was
examined using flow cytometry.
Patients and methods
Patients
In total, 42 patients with CRC (23 male, 19 female;
median age, 63 years) and 42 healthy individuals (24 male, 18
female; median age, 60 years) were recruited from The First
Hospital of Hebei Medical University (Shijiazhuang, China) between
October 2013 and March 2014. No previous local or systemic
treatment had been administered to the 42 patients with CRC prior
to surgery. Written informed consent was obtained from all of the
patients, in accordance with the protocols of the Ethics Review
Board at Hebei Medical University (Shijiazhuang, China), which
approved the present study.
Samples
A total of 42 pairs of CRC and non-tumor adjacent
tissues were obtained from patients that underwent radical
resection between October 2013 and March 2014 at The First Hospital
of Hebei Medical University. The corresponding normal tissues were
obtained from a section of the resected specimen ≥5 cm from the
edge of the tumor. The samples were stored in liquid nitrogen or an
−80°C freezer until use.
Plasma samples were collected from 42 CRC patients
prior to surgical resection and 7 days following surgery
respectively. Furthermore, 42 plasma samples were obtained from 42
cases of age-matched healthy individuals as the control. Cell-free
plasma was extracted from all blood samples within 2 h of
collection using a two-step protocol at 2,140 × g for 10 min and
12,840 × g for 2 min. Plasma was transferred to fresh tubes and
stored in a −80°C freezer until use.
Cell culture
The human CRC cell line HCT116 was obtained from
Professor Xiaofeng Sun (Division of Oncology, Department of
Clinical and Experimental Medicine, Faculty of Health Sciences,
Linköping University, Linköping, Sweden). Cells were cultured in
McCoy's 5A medium (Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) containing 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) and 1% penicillin-streptomcyin (Invitrogen;
Thermo Fisher Scientific, Inc.) in a 37°C humidified incubator
containing 5% CO2.
Quantitative miRNA analysis
Total RNA was extracted from the frozen tissues
using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol, and resuspended in 60 µl
pre-heated (95°C) nuclease-free water.
Total RNA was extracted from 400 µl plasma samples
using the miRNeasy Serum/Plasma kit (Qiagen GmbH, Hilden, Germany),
according to the manufacturer's protocol, and eluted with 105 µl
pre-heated (95°C) double distilled water.
The concentration and purity of all RNA samples were
detected using a NanoDrop ND-2000 spectrophotometer (NanoDrop
Technologies; Thermo Fisher Scientific, Inc.). All RNA was stored
at −80°C until use following determination of the concentration RNA
using a spectrophotometer.
Reverse transcription was carried out using the One
Step PrimeScript® miRNA cDNA Synthesis kit (Takara Bio,
Inc., Otsu, Japan), according to the manufacturer's protocol.
Quantitative polymerase chain reaction (qPCR) was
performed using an All-in-One™ miRNA qPCR kit (QP101;
GeneCopoeia, Inc., Guangzhou, China) with an ABI 7500 Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. All-in-OneTM miRNA qPCR
Primers were used, including a hsa-miR-106a-5p primer pair
(HmiRQP0026) and a HsnRNA U6 primer pair (HmiRQP9001; GeneCopoeia,
Inc.). The PCR reaction mixture included 10 µl 2X
All-in-One™ qPCR mix, 2 µl All-in-One™ miRNA
qPCR Primer, 2 µl Universal Adaptor PCR Primer, 2 µl template cDNA
and 4 µl double distilled water, for a total volume of 20 µl. The
reaction mixture was incubated at 95°C for 10 min, followed by 40
cycles of 95°C for 15 sec and 60°C for 60 sec. All RT-qPCR
reactions were performed in triplicate.
U6 small nuclear RNA was used as a housekeeping gene
to normalize miRNA expression, and the relative expression of
miR-106a in CRC cells was determined using the comparative
threshold cycle (Cq) method (2−ΔΔCq) (17).
Cell Counting kit-8 (CCK-8)
assays
HCT116 cells were seeded into 96-well plates and
then transfected with miR-106a mimic (miR10000103-1-5), miR-106a
mimic control (miR01101-1-5), miR-106a inhibitor (miR20000103-1-5)
or miR-106a inhibitor control (miR02101-1-5; all RiboBio,
Guangzhou, China) using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). At 24 h after culture, the cells were
harvested and absorbance values at 450 were determined using CCK-8
reagent (Dojindo Molecular Technologies, Inc., Kumamoto, Japan)
according to the manufacturer's protocol. Tests were performed in
triplicate.
Apoptosis assay
Fluorescein isothiocyanate (FITC)-conjugated Annexin
V and propidium iodide (PI) staining were performed by means of the
Annexin V-FITC Apoptosis Detection kit (Neobioscience, Shenzhen,
China). Following transfection with miR-106a mimic or inhibitor, or
negative control, cells were cultured for 48 h, resuspended in 1X
binding buffer, washed twice with PBS, and treated with Annexin
V-FITC and PI. Following 10 min of incubation at room temperature
in the dark, cells were submitted to flow cytometry with a BD
FACSCalibur flow cytometer calibrated with CaliBRITE beads, and the
results were analyzed using CellQuest software (all BD Biosciences,
San Jose, CA, USA). Tests were performed in quadruplicate.
Early apoptotic cells were located in the
lower-right quadrant, late apoptotic or necrotic cells were located
in the upper-right quadrant, normal cells were located in the
lower-left quadrant and mechanically damaged cells were located in
the upper-left quadrant of the flow cytometric dot plots (18).
Statistical analysis
Each experiment was conducted at least three times.
All values are presented as the median and 25th and 75th
percentiles or the mean ± standard deviation. Differences were
tested for significance using a Student's t-test, unpaired t-test,
Mann-Whitney U test or a two-way analysis of variance followed by
Tukey's post hoc test in SPSS software (version 19.0; IBM SPSS,
Armonk, NY, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Effects of miR-106a on CRC cell
viability
Our previous study (19) identified that miR-106a expression was
increased in the plasma of patients with CRC compared with healthy
controls. Therefore, it was hypothesized that miR-106a expression
may be associated with the progress of CRC. In the present study,
the effects of miR-106a on cell viability and apoptosis in CRC
cells were analyzed. First, to measure the effect of miR-106a on
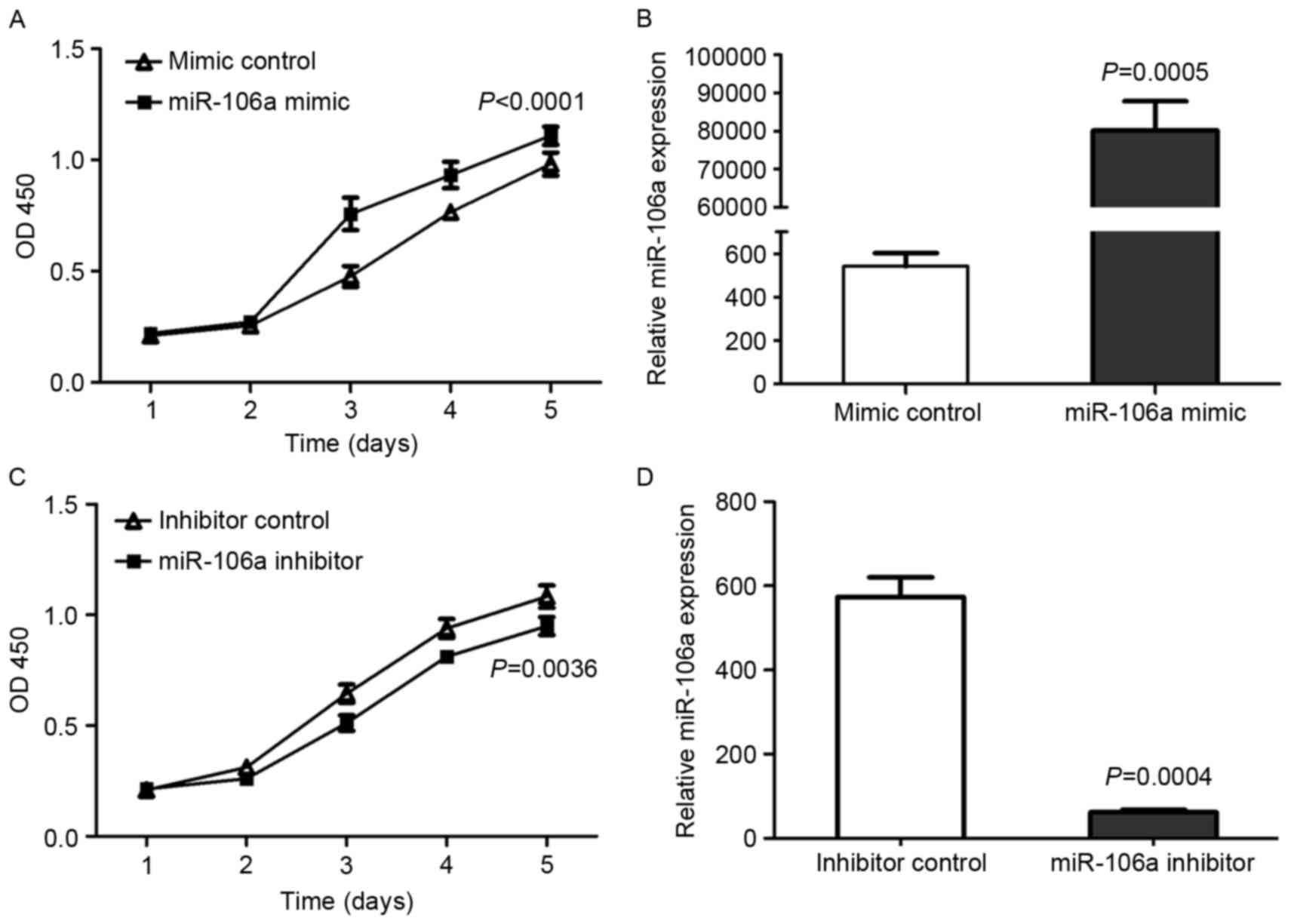
cell viability, a CCK-8 assay was carried out. HCT116 cells were
transfected with miR-106a mimic or mimic control, miR-106a
inhibitor or inhibitor control. Cell viability curves and, at 24 h
after transfection, miR-106a expression were examined (Fig. 1).
The expression of miR-106a and cell viability were
significantly upregulated by the miR-106a mimic comparison with
mimic control in HCT116 cells (P<0.01; Fig. 1A and B, respectively). Conversely,
following transfection with miR-106a inhibitor, the viability of
HCT116 cells was also significantly decreased compared with
inhibitor control-transfected cells (P=0.0036; Fig. 1C) and the expression of miR-106a was
significantly decreased (P=0.0004; Fig.
1D). These results indicated that the miR-106a expression level
was positively associated with CRC cell viability.
Effects of miR-106a on CRC cell
apoptosis
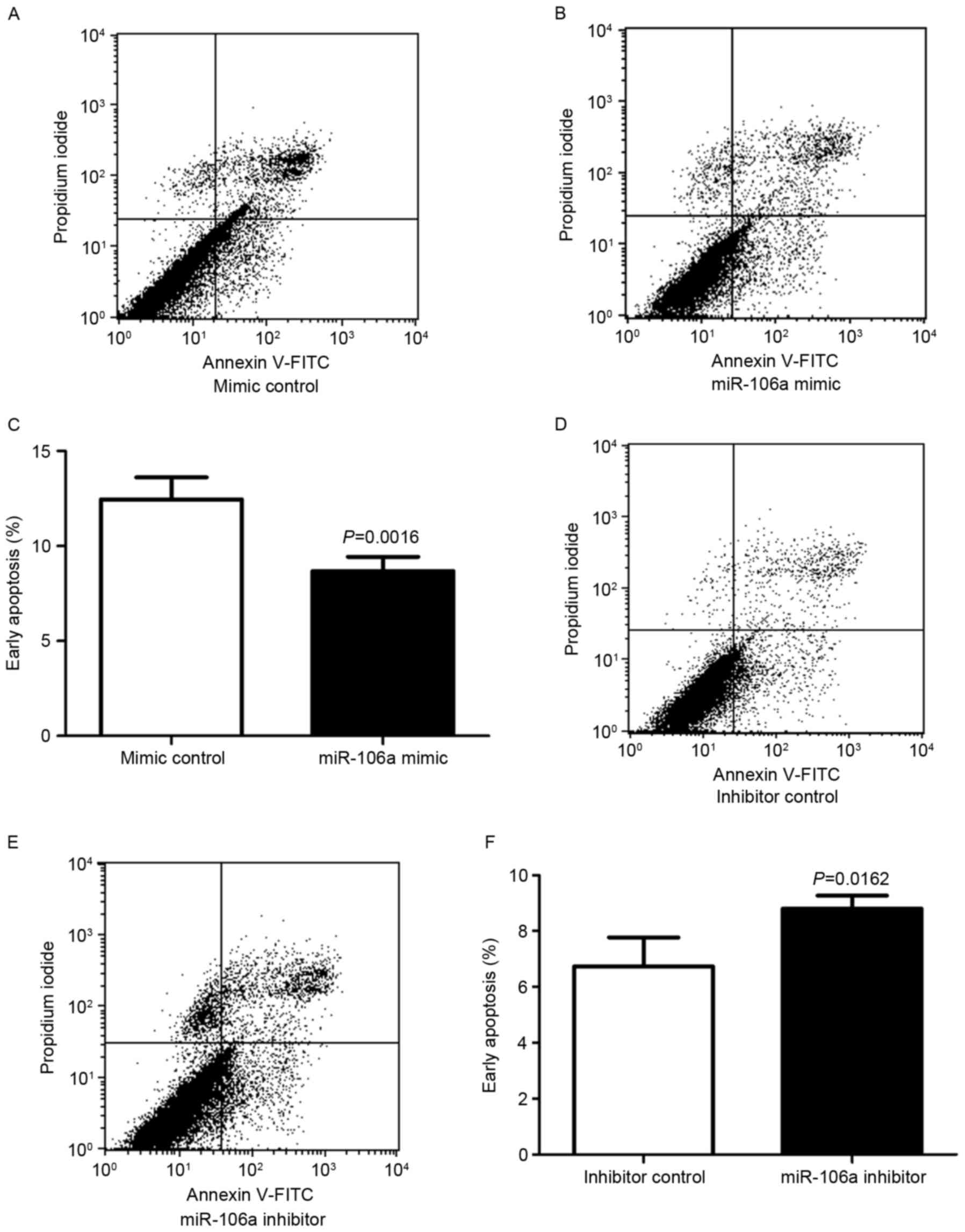
The function of miR-106a on cell apoptosis was
determined using flow cytometry. HCT116 cells were transfected with
miR-106a mimic or mimic control, miR-106a inhibitor or inhibitor
control. At 48 h after transfection, the apoptosis rates of HCT116
cells were determined (Fig. 2).
Following transfection with miR-106a mimic or mimic
control, cell status was determined using flow cytometry, with
early apoptotic cells located in the lower right quadrant of the
plot (Fig. 2A and B). HCT116 cells
transfected with miR-106a mimic exhibited a lower rate of early
apoptosis compared with the control group. The mean ± standard
deviation early apoptosis rates were significantly decreased for
the miR-106a mimic compared with the mimic control (8.68±0.75 vs.
12.45±1.16%, respectively; P=0.0016; n=4; Fig. 2C).
Flow cytometry was also employed to analyze cells
subsequent to transfection with an miR-106a inhibitor or an
inhibitor control (Fig. 2D and E).
HCT116 cells transfected with miR-106a inhibitor exhibited a higher
rate of early apoptosis compared with the control group. The mean ±
standard deviation early apoptosis rates were significantly
increased for the miR-106a inhibitor compared with the inhibitor
control (8.803±0.27 vs. 6.733±0.60%, respectively; P=0.0162; n=4;
Fig. 2F).
These results indicated that the miR-106a mimic
decreased early apoptosis in HCT116 cells, and that the miR-106a
inhibitor increased early apoptosis in HCT116 cells, suggesting
that miR-106a promoted cell viability by inhibiting early apoptosis
in CRC cells.
Expression of miR-106a is upregulated
in human CRC tissues and plasma
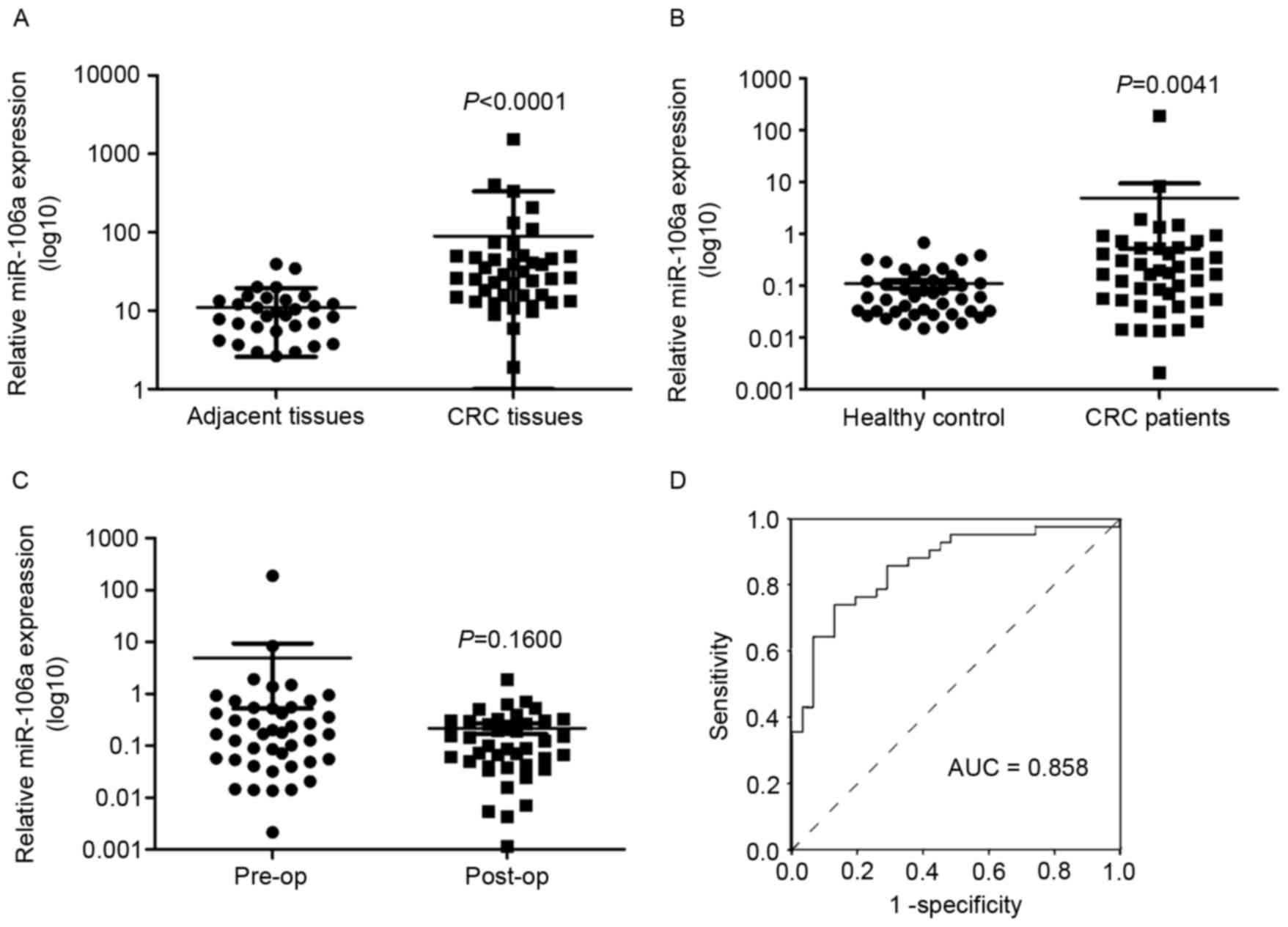
In previous studies (19), miR-106a expression was upregulated in
the serum of patients with CRC. In the present study, the
expression of miR-106a in human CRC tissues and plasma was
determined. It was identified that the expression level of miR-106a
was significantly upregulated in tumor tissues compared with
adjacent tissues [median (25th percentile, 75th percentile, 26.2882
(14.5358, 49.3492) vs. 8.7187 (5.4891, 13.6726); P<0.0001;
Fig. 3A]. The association between
miR-106a expression in cancer tissues and clinicopathological
characteristics of patients with CRC was also analyzed (Table I).
 | Table I.Association between miR-106a
expression in cancer tissues and clinicopathological
characteristics of patients with colorectal cancer. |
Table I.
Association between miR-106a
expression in cancer tissues and clinicopathological
characteristics of patients with colorectal cancer.
| Variables | Patients (n=42) | (25th and 75th
percentile) miR-106a expression, median | P-value |
|---|
| Age, years |
|
| 0.5800 |
| ≤63 | 21 | 26.643 (13.741,
72.249) |
|
|
>63 | 21 | 25.934 (14.554,
40.273) |
|
| Sex |
|
| 0.6133 |
| Male | 23 | 26.643 (16.049,
50.062) |
|
|
Female | 19 | 25.934 (12.790,
47.944) |
| Dukes |
|
| 0.6311 |
| A, B | 19 | 26.643 (16.049,
74.260) |
|
| C, D | 23 | 25.934 (13.316,
47.944) |
|
| Pathological
type |
|
| 0.0423a |
|
Adenocarcinoma | 36 | 30.493 (15.941,
49.824) |
|
| Mucinous
carcinoma | 6 | 11.939 (4.928,
32.244) |
|
| Depth of
invasion |
|
| 0.0431a |
| T1,
T2 | 9 | 37.528 (27.910,
286.856) |
|
| T3,
T4 | 33 | 24.340 (13.223,
48.528) |
|
| Location |
|
| 0.2610 |
|
Colon | 21 | 18.367 (12.665,
42.628) |
|
|
Rectum | 21 | 39.325 (22.801,
80.544) |
|
| Lymph node
metastasis |
|
| 0.4375 |
|
Absent | 23 | 26.643 (15.791,
70.237) |
|
|
Present | 19 | 25.934 (13.316,
46.261) |
|
| Distant
metastasis |
|
| 0.8700 |
|
Absent | 33 | 26.643 (14.129,
50.144) |
|
|
Present | 9 | 23.090 (14.460,
48.162) |
|
|
Differentiation |
|
| 0.0382a |
|
Poor | 8 | 12.539 (5.423,
33.613) |
|
|
Moderate-well | 34 | 35.731 (20.439,
60.149) |
|
miR-106a expression in plasma of patients with CRC
or healthy controls, and pre- or post-surgery in patients with CRC,
was also investigated. The expression level of miR-106a in plasma
was significantly higher in patients with CRC compared with in the
healthy control group, consistent with our previous study (19) (P=0.0041; Fig. 3B). The healthy control group contained
42 age-matched healthy individuals with patients with CRC (data not
shown). miR-106a expression in plasma post-surgery was decreased
compared with in plasma pre-surgery, but no statistically
significant difference was identified (0.16±0.18 vs. 8.80±40.50;
P=0.1600; Fig. 3C). No significant
association was identified between miR-106a expression in plasma
and the clinicopathological parameters of the patients with CRC
(data not shown). The sensitivity and specificity of miR-106a were
analyzed using a receiver operating characteristic curve in CRC
diagnosis (Fig. 3D).
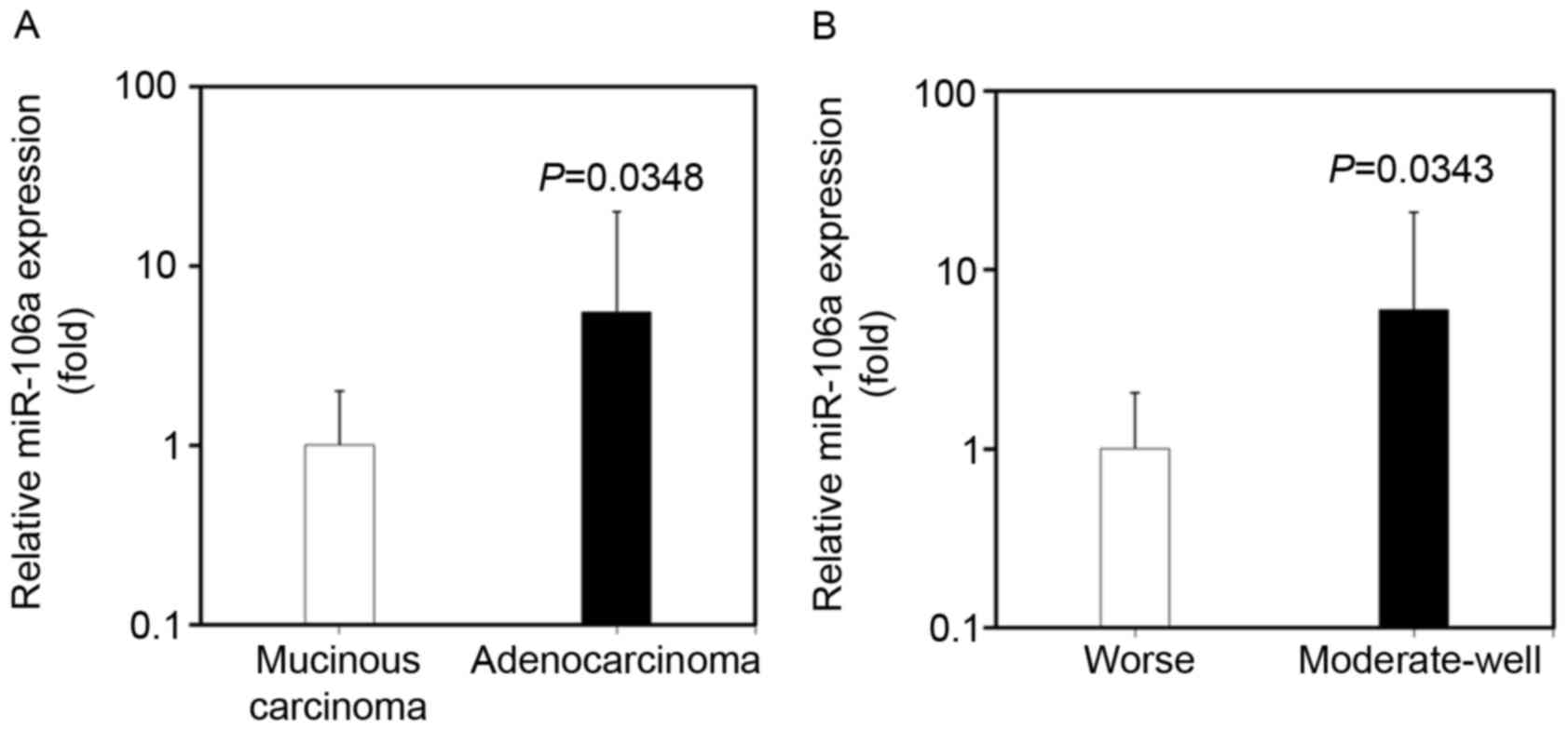
miR-106a expression in CRC tissues is
associated with differentiation and pathological type
The expression of miR-106a in CRC tissues was
identified to be associated with the differentiation and
pathological type of tumor, by analyzing the clinicopathological
characteristics of CRC patients. The expression level of miR-106a
in CRC tissues was significantly increased in patients with
adenocarcinoma compared with in patients with mucinous carcinoma
(100.96±262.20 vs. 18.14±18.22; P=0.0348; Fig. 4A). miR-106a expression in CRC tissues
was also increased in patients exhibiting moderate-well
differentiation compared with that in patients exhibiting poor
differentiation (108.87±272.79 vs. 18.12±19.20; P=0.0343; Fig. 4B).
The underlying molecular mechanism of increased
miR-106a expression in adenocarcinoma and moderate-well
differentiation CRC requires further investigation. No significant
association was identified between miR-106a expression and other
clinicopathological characteristics, such as sex, age, distant
metastasis, Dukes staging, tumor location and lymph node
metastasis.
Discussion
miR-106a was identified to serve an oncogenic role
in carcinogenesis (20,21). miR-106a enhanced the invasiveness of
human glioma stem cells, pancreatic cancer, gastric cancer and
glioblastoma (22–24).
The results of the present study indicated that
overexpression of miR-106a by transfection with miR-106a mimic
increased the viability and suppressed early apoptosis of HCT116
cells. Knocking down the miR-106a expression by miR-106a inhibitor
identified that cell viability was inhibited and the number of
pro-apoptotic cells was upregulated. These results suggested that
miR-106a served an important role in the viability and apoptosis of
CRC cells.
miR-106a expression was determined in CRC tissues,
and the association between miR-106a expression and
clinicopathological data was analyzed. The expression level of
miR-106a in CRC tissues was significantly upregulated in CRC
tissues compared with adjacent tissues. More importantly, an
increased association was identified between miR-106a expression
and pathological type, differentiation and depth of invasion. The
expression levels of miR-106a were increased in adenocarcinoma
compared with mucinous carcinoma (P=0.0423). The expression of
miR-106a was identified to be positively associated with
differentiation. The well-differentiated CRC tissues exhibited an
increased expression level of miR-106a compared with poorly
differentiated CRC tissues (P=0.0382). However, miR-106a expression
was not identified to be associated with other clinicopathological
characteristics, including sex, age, distant metastasis, Dukes
staging, tumor location and lymph node metastasis (P>0.05).
These results are in contrast with a previous study identifying
that miR-106a exhibited an increased expression level in rectal
tumors (25).
A number of studies have focused on circulating
miRNAs, which have been reported as an effective and non-invasive
biomarker for detecting various cancers or other diseases (26–28). In
our previous study, the plasma level of miR-106a was significantly
increased in patients with CRC compared with healthy controls
(19). The expression of miR-106a was
also determined in the plasma of 40 patients with CRC and 40
healthy individuals, and the expression level of miR-106a was
significantly increased in patients with CRC compared with in the
healthy control group. In the present study, although the plasma
levels of miR-106a were analyzed in patients with CRC at different
clinical status (age, tumor size, tumor-node-metastasis stage or
metastasis), no significant difference was identified. No
significant alterations in the miR-106a expression were identified
following surgery compared with its preoperative level, which is in
contrast with previous results (19).
It is hypothesized that this may be due to the limited sample size.
Therefore, expanding the sample size in future research may provide
more insight into the underlying molecular mechanism.
It was hypothesized that miR-106a served an
important role in carcinogenesis of CRC. In the present study, the
focus was on analyzing the effect of miR-106a in CRC cell
apoptosis, viability and the alterations in miR-106a expression in
CRC tissues.
miR-106a may be a potential biomarker in the
diagnosis of CRC, particularly in the diagnosis of colorectal
adenocarcinoma. miR-106a serves important roles in the
differentiation of colorectal adenocarcinoma, CRC progression, CRC
cell viability and apoptosis. miR-106a may be associated with the
migration and invasion of cancer cells. Increased accuracy could be
obtained if a number of types of circulating miRNAs are combined to
provide a clinical diagnosis of CRC. Further research is required
to elucidate the role of miR-106a in tumor progression and its
complicated interactions with oncogenes and tumor suppressors.
Further study is required to elucidate the underlying molecular
mechanism of miR-106a in the development of CRC.
Acknowledgements
The authors are indebted to Professor Xiaofeng Sun
(Linköping University, Linköping, Sweden) for generously providing
the HCT116 cells. The present study was supported by the National
Natural Science Foundation of China (grant no. 81072034), the
Excellent Youth Foundation of Hebei Scientific Committee (grant no.
2013206252), the International Science and Technology Cooperation
Program of China (grant no. 2014DFA31150) and Hebei Province (grant
no. 12396105D).
Glossary
Abbreviations
Abbreviations:
|
miRNA
|
microRNA
|
|
miR-106a
|
miRNA-106a
|
|
CRC
|
colorectal cancer
|
|
CCK-8
|
Cell Counting kit-8
|
|
PI
|
propidium iodide
|
|
FITC
|
fluorescein isothiocyanate
|
References
|
1
|
Wasserman M, Baxter NN, Rosen B, Burnstein
M and Halverson AL: Systematic review of internet patient
information on colorectal cancer surgery. Dis Colon Rectum.
57:64–69. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yang W, Lee DY and Ben-David Y: The roles
of microRNAs in tumorigenesis and angiogenesis. Int J Physiol
Pathophysiol Pharmacol. 3:140–155. 2011.PubMed/NCBI
|
|
3
|
Hamaya Y, Yoshida K, Takai T, Ikuma M,
Hishida A and Kanalka S: Factors that contribute to faecal
cyclooxygenase-2 mRNA expression in subjects with colorectal
cancer. Br J Cancer. 102:916–921. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mo ZH, Wu XD, Li S, Fei BY and Zhang B:
Expression and clinical significance of microRNA-376a in colorectal
cancer. Asian Pac J Cancer Prev. 15:9523–9527. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fayyad-Kazan H, Bitar N, Najar M, Lewalle
P, Fawad-Kazan M, Badran R, Hamade E, Daher A, Hussein N, ElDirani
R, et al: Circulating miR-150 and miR-342 in plasma are novel
potential biomarkers for acute myeloid leukemia. J Transl Med.
11:312013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ye JJ and Cao J: MicroRNAs in colorectal
cancer as markers and targets-Recent advances. World J
Gastroenterol. 20:4288–4299. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oglesby IK, McElvaney NG and Greene CM:
MicroRNAs in inflammatory lung disease-master regulators or target
practice? Respir Res. 11:1482010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Holloway JW, Francis S Savarimuthu, Fong
KM and Yang IA: Genomics and the respiratory effects of air
pollution exposure. Respirology. 17:590–600. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Small EM and Olson EN: Pervasive roles of
microRNAs in cardiovascular biology. Nature. 469:336–342. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Almeida MI, Nicoloso MS, Zeng L, Ivan C,
Spizzo R, Gafa R, Xiao L, Zhang X, Vannini I, Fanini F, et al:
Strand-specific miR-28-5p and miR-28-3p have distinct effects in
colorectal cancer cells. Gastroenterology. 142:886–896. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhou S, Ye W, Ren J, Shao Q, Qi Y, Liang J
and Zhang M: MicroRNA-381 increases radiosensitivity in esophageal
squamous cell carcinoma. Am J Cancer Res. 5:267–277.
2014.PubMed/NCBI
|
|
12
|
Ye J, Wu X, Wu D, Wu P, Ni C, Zhang Z,
Chen Z, Qiu F, Xu J and Huang J: miRNA-27b targets vascular
endothelial growth factor C to inhibit tumor progression and
angiogenesis in colorectal cancer. PLoS One. 8:e606872013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tong AW and Nemunaitis J: Modulation of
miRNA activity in human cancer: A new paradigm for cancer gene
therapy? Cancer Gene Ther. 15:341–355. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xiao B, Guo J, Miao Y, Jiang Z, Huan R,
Zhang Y, Li D and Zhong J: Detection of miR-106a in gastric
carcinoma and its clinical significance. Clin Chim Acta.
400:97–102. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xie X, Liu HT, Mei J, Ding FB, Xiao HB, Hu
FQ, Hu R and Wang MS: miR-106a promotes growth and metastasis of
non-small cell lung cancer by targeting PTEN. Int J Clin Exp
Pathol. 8:3827–3834. 2015.PubMed/NCBI
|
|
16
|
Kim YW, Kim EY, Jeon D, Liu JL, Kim HS,
Choi JW and Ahn WS: Differential microRNA expression signatures and
cell type-specific association with Taxol resistance in ovarian
cancer cells. Drug Des Devel Ther. 8:293–314. 2014.PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu D, Zhang S, Du W, Zhang J, Fan Z, Hao
H, Liu Y, Zhao X, Qin T and Zhu H: Expression of intracellular
interferon-alpha confers antiviral properties in transfected bovine
fetal fibroblasts and does not affect the full development of SCNT
embryos. PLoS One. 9:e944442014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang L, Meng L, Fan Z, Liu B, Pei Y and
Zhao Z: Expression of plasma miR-106a in colorectal cancer and its
clinical significance. Nan Fang Yi Ke Da Xue Xue Bao. 34:354–357.
2014.(In Chinese). PubMed/NCBI
|
|
20
|
Wang Z, Liu M, Zhu H, Zhang W, He S, Hu C,
Quan L, Bai J and Xu N: miR-106a is frequently upregulated in
gastric cancer and inhibits the extrinsic apoptotic pathway by
targeting FAS. Mol Carcinog. 52:634–646. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yuan R, Zhi Q, Zhao H, Han Y, Gao L, Wang
B, Kou Z, Guo Z, He S, Xue X and Hu H: Upregulated expression of
miR-106a by DNA hypomethylation plays an oncogenic role in
hepatocellular carcinoma. Tumour Biol. 36:3093–3100. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li P, Xu Q, Zhang D, Li X, Han L, Lei J,
Duan W, Ma Q, Wu Z and Wang Z: Upregulated miR-106a plays an
oncogenic role in pancreatic cancer. FEBS Lett. 588:705–712. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Z, Wang B, Shi Y, Xu C, Xiao HL, Ma
LN, Xu SL, Yang L, Wang QL, Dang WQ, et al: Oncogenic miR-20a and
miR-106a enhance the invasiveness of human glioma stem cells by
directly targeting TIMP-2. Oncogene. 34:1407–1419. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu M, Zhang N and He S: Similarly
up-regulated microRNA-106a in matched formalin-fixed
paraffin-embedded and fresh frozen samples and the dynamic changes
during gastric carcinogenesis and development. Pathol Res Pract.
210:909–915. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schee K, Boye K, Abrahamsen TW, Fodstad Ø
and Flatmark K: Clinical relevance of microRNA miR-21, miR-31,
miR-92a, miR-101, miR-106a and miR-145 in colorectal cancer. BMC
Cancer. 12:5052012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Criscitiello C, Sotiriou C and Ignatiadis
M: Circulating tumor cells and emerging blood biomarkers in breast
cancer. Curr Opin Oncol. 22:552–558. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cheng H, Zhang L, Cogdell DE, Zheng H,
Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR and Zhang W:
Circulating plasma MiR-141 is a novel biomarker for metastatic
colon cancer and predicts poor prognosis. PLoS One. 6:e177452011.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Qin X, Xu H, Gong W and Deng W: The tumor
cytosol miRNAs, fluid miRNAs, and exosome miRNAs in lung cancer.
Front Oncol. 4:3572015. View Article : Google Scholar : PubMed/NCBI
|