Introduction
Inverted papilloma (IP) is a benign tumor occurring
in the nasal cavity and paranasal sinuses (1–3). IPs
typically arise from the lateral wall of the nasal cavity and may
spread into the surrounding tissue, the ethmoid, the skull base and
the orbita (3). Although surgical
treatment has developed through the integration of skull base
surgery, navigation systems and endoscopic surgical techniques
(1), the management of this disease
has remained challenging as IPs tend to recur following incomplete
resection, and the malignant transformation to squamous cell
carcinoma (SCC) occurs in 5–15% of cases (2,3).
MicroRNAs (miRNA/miR) are small non-coding RNAs of
~19–22 nucleotides that regulate gene expression through
translational repression and mRNA cleavage (4). In total, >1,500 types of human miRNA
have been detected and registered, with a number of them reported
to be associated with carcinogenesis, differentiation, apoptosis
and metastasis (5–7). In the head and neck field, miRNAs are
aberrantly expressed in SCC tissue and are associated with
carcinogenesis and drug-resistance (5–7).
High-risk human papilloma virus (HPV) infection is
regarded as a potential etiological agent of IPs and their
malignant transformation (3,8). Carcinogenesis resulting from HPV
infection has previously been explained by the inhibition of tumor
suppressors, including the p53 and retinoblastoma protein (pRB)
signaling pathways (9). The E6 and E7
regions of high-risk HPV are transfected into the genes of
epithelial basal cells following infection (10). E6 binds to p53 and promotes the
ubiquitination and degradation of p53 itself. E7 also suppresses
the cell cycle regulators of p21 and p27, and binds directly to pRB
to promote aggressive cell growth (10). However, it is not always possible to
detect high-risk HPV in sinonasal SCCs arising from IPs; the
detection rate of high-risk HPV is ~37.8% (10). The rate of unapparent infection with
HPV remains unknown, but a considerable number of such cases are
speculated to exist (4). The majority
of IPs require a lengthy period for malignant transformation, but
the trigger underlying the malignant changes remains obscure
(10). SCC arising from IP is one of
the best models for studies, as certain tissue samples obtained via
surgical resection may contain normal epithelium, dysplasia and SCC
(4). The present study aimed to
identify the underlying mechanisms and roles of miRNAs in the
malignant transformation of IPs.
Materials and methods
Samples and cell lines
A total of 36 individual nasal and paranasal tumor
specimens from 25 patients (19 males and 6 females) were obtained
during surgery at Hokkaido University Hospital (Sapporo, Japan)
between January 1993 and October 2012. All samples were obtained
prior to radiotherapy and chemotherapy. The median age of the
patients was 64.7 years (range, 39–93 years) at the time of
diagnosis. The details of the pretreatment clinical and
pathological characteristics are summarized in Table I. SCC25 cells were purchased from the
American Type Culture Collection (Manassas, VA, USA). Written
informed consent was obtained from all patients. The present study
was approved by the Institutional Ethics Review Board of Hokkaido
University Hospital.
 | Table I.Baseline characteristics of patients
with IP and SCC. |
Table I.
Baseline characteristics of patients
with IP and SCC.
| A, Cases for microRNA
analysis |
|---|
|
|---|
| Characteristics | Patients, n |
|---|
| Total | 8 |
| Sex |
|
| Male | 7 |
|
Female | 1 |
| Age, years |
|
| Range
(median) | 38–77 (54) |
|
| B, Cases for
immunohistochemistry |
|
| Total | 25 |
| Sex |
|
| Male | 19 |
|
Female | 6 |
| Age, years |
|
| Range,
median | 32–84 (63) |
| Histology |
|
| IP
non-dysplastic | 6 |
| IP
coexisting with SCC | 12 |
| SCC | 7 |
RNA isolation and miRNA TaqMan™ low
density array (TLDA) assay
Total RNA was isolated from the formalin-fixed
tissue using a Recover All Total Nucleic Acid Isolation kit
(Ambion; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
according to the manufacturer's protocol. Formalin-fixed tumor
tissues were cut into 4-um thick sections, deparaffinized in
xylene, rehydrated through graded ethanol, and stained with
hematoxylin and eosin. These sections were examined for
histopathological accuracy under a light microscope. Unstained
sections for RNA isolation were cut into 8-um thick sections and
macro-dissected to avoid contamination with other tissues. The
quality and quantity of the RNA was determined using an Agilent RNA
6000 Nano LabChip kit and an Agilent 2100 Bioanalyzer (Agilent
Technologies, Inc., Santa Clara, CA, USA). Total RNA was used to
run the ABI Megaplex™ protocol without pre-amplification
(Applied Biosystems; Thermo Fisher Scientific, Inc.). A reaction
was run using an ABI miRNA Reverse Transcription (RT) kit (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and
Megaplex™ RT Human Pool A primers (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The cDNA from each sample was then
transferred to a new tube and diluted with water and
TaqMan™ Universal polymerase chain reaction (PCR) Master
Mix (2X) without uracil-DNA glycosylase (Applied Biosystems; Thermo
Fisher Scientific, Inc.). Each sample was then loaded onto its own
TLDA card. TLDAs were queued into the ABI7900HT Real Time PCR
machine (Applied Biosystems; Thermo Fisher Scientific, Inc.) and
run according to the Applied Biosystems default TLDA protocol. Data
were normalized based on the endogenous control on the TLDA card
and analyzed using DataAssist™ v3.0 software (Applied
Biosystems; Thermo Fisher Scientific, Inc).
Transfection of miRNA
The miRNA mimics/inhibitor and negative controls
were purchased from GE Healthcare Dharmacon, Inc. (Lafayette, CO,
USA) and introduced into SCC25 cells using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) at
37°C according to the manufacturer's protocol. Following 6 h, the
transfection medium was aspirated and Dulbecco's modified Eagle's
medium (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) with 10%
fetal calf serum (Gibco; Thermo Fisher Scientific, Inc.) was added.
Cells were lysed 48 h following transfection as described below.
Transfection was confirmed using miRIDIAN miRNA Mimic Transfection
Control with Dy547 (GE Healthcare Dharmacon, Inc.).
Immunoblotting
Cells were lysed with radioimmunoprecipitation assay
lysis buffer (1 mM NaVO3, 1 mM dithioreitol, 1 mM
phenylmethylsulfonyl fluoride (Sigma-Aldrich; Merck KGaA),
phosphatase inhibitor cocktail and protease inhibitor cocktail mini
tablet (Roche Diagnostics, Indianapolis, IN, USA) on ice. The
samples were centrifuged at 10,000 × g for 4 min and the
supernatant was collected. Protein concentration was quantified
with a standard Bradford absorbance assay according to the
manufacturer's protocol (Bio-Rad, Hercules, CA, USA). Protein from
each sample was fractionated on a 4–15% SDS-PAGE gel (Bio-Rad). The
proteins were then transferred to nitrocellulose membranes and
blocked with skim milk at room temperature for 1 h. Then, membranes
were incubated for 12 h at 4°C with the appropriate primary
antibodies phosphatase and tensin homolog (PTEN; cat no. 9559;
dilution, 1:1,000), p21 (cat no. 2947; dilution, 1:1,000) (both
Cell Signaling Technology, Inc., Danvers, MA, USA) and β-actin (cat
no. ab8226; Abcam, Cambridge, UK; dilution, 1:1,000); followed by
incubation with secondary anti-rabbit or anti-mouse HRP-linked
antibodies (cat nos., 7074 and 7076, respectively; Cell Signaling
Technology, Inc., Danvers, MA, USA) at the manufactures' indicated
dilution ratio. Protein bands were visualized using ECL Western
Blotting Detection reagent (GE Healthcare Life Sciences, Chalfont,
UK) Signal intensities were determined using Image Gauge software
(version 4.1; Fujifilm, Tokyo, Japan). Each gel was normalized to
β-actin.
Immunohistochemistry
Paraffin-embedded tumor specimens from the primary
sites were available from all 25 patients involved. These specimens
were cut into 4-um thick sections, deparaffinized in xylene,
rehydrated through graded ethanol and then placed in 0.1% hydrogen
peroxide to quench any endogenous peroxidase activity at room
temperature for 10 min. Following pretreatment in a 0.01 M citrate
buffer (pH 6.0) for three cycles of 5 min at 750 W in a microwave,
these sections were treated with 10% normal rabbit serum
(Histofine® SAB-PO M kit; Nichirei Biosciences, Inc.,
Tokyo, Japan) at room temperature for 10 min to prevent any
nonspecific binding of the antibody. The slides were then incubated
with a specific monoclonal antibody against human PTEN or p21 (Cell
Signaling Technology) in a humid chamber at 4°C overnight. The
sections were then incubated with a biotin-labeled rabbit
anti-mouse secondary antibody according to the manufacturer's
protocol (Histofine SAB-PO M kit) for 30 min at room temperature,
followed by incubation with a streptavidin-biotin horseradish
peroxidase complex (Histofine SAB-PO M kit). The reaction products
were observed by immersing the slides in a freshly prepared
diaminobenzidine solution for 10 min and counterstaining with
hematoxylin at room temperature prior to dehydration and mounting.
Stained slides were scored based on the staining intensity and the
percentage of positive cells. Staining intensity was scored using a
4-point system as previously described (11) (0+, negative; 1+, weak; 2+, moderate;
3+, strong). The positive cell percentage was scored using a
5-point system (0, 0%; 1+, 1–10%; 2+, 11–50%; 3+, 51–80%; 4+,
>80%). PTEN expression was estimated by cytoplasmic staining,
and p21 expression was estimated by nuclear staining. The
expression of the two proteins was scored semi-quantitatively using
the immunoreactive score (IRS), which was calculated as follows:
IRS=staining intensity score × score of percentage of positive
cells. Positivity for PTEN and p21 was defined as an IRS score ≥3,
according to the Nagata score (10).
In all cases, the procedure was performed by two pathologists, who
were blinded to the clinical outcomes. Unpaired t-tests were
performed using GraphPad Prism software (version 5.0; GraphPad
Software, Inc., La Jolla, CA, USA), and P<0.05 was considered to
indicate a statistically significant difference.
Biological targets of miRNAs were predicted by
searching for the presence of sites that match the seed region of
each miRNA using the TargetScan 6.2 database (http://www.targetscan.org).
Results
Differential miRNA expression between
IP and SCC cells
Using a total of 10 samples (IP=5 and SCC=5), the
role of miRNAs in the carcinogenesis of IP was examined. All five
IP samples exhibited transformation to SCCs and all five SCC
samples had arisen from IPs. A total of two IP and SCC samples were
obtained from the same case (Table
IA). miRNA expression analyses were performed using
RT-PCR-based arrays to examine 754 unique miRNAs. Among the
differentially-expressed miRNAs, the 10 that exhibited the greatest
differential expression between IP and SCC are presented in
Table II. miR-296-3p exhibited a
23-fold decrease in IP, compared with in SCC. miR-296-3p also
demonstrated a 76-fold difference in expression between the IP and
SCC from the same cases. A list of 66 target genes was obtained
using TargetScan 6.2, including 66 conserved sites and 26 poorly
conserved sites that are putatively targeted by miR-296-3p. PTEN,
which was identified as a tumor suppressor and involved in the p53
and RB pathways, was included in the list of miR-296-3p target
genes.
 | Table II.Top 10 miRNAs that exhibited
differential expression between SCC and IP. |
Table II.
Top 10 miRNAs that exhibited
differential expression between SCC and IP.
| Upregulated in
SCC | Downregulated in
SCC |
|---|
|
|
|---|
| Name | Fold-change | Name | Fold-change |
|---|
| hsa-miR-206 | 37.74 | hsa-miR-892b | 183.83 |
| hsa-miR-675 | 28.9 | hsa-miR-874 | 144.4 |
| hsa-miR-105 | 25.38 | hsa-miR-371-3p | 121.14 |
| hsa-miR-296-3p | 23.92 | hsa-miR-16–2 | 116.27 |
| hsa-miR-577 | 21.88 | hsa-miR-129 | 85.26 |
| hsa-miR-574-3p | 12.94 | hsa-miR-518f | 70.78 |
| hsa-miR-93 | 9.09 | hsa-miR-431 | 59.55 |
| hsa-miR-936 | 8.87 | hsa-miR-593 | 49.27 |
| hsa-miR-922 | 8.75 |
hsa-miR-1225-3p | 45.06 |
| hsa-miR-616 | 8.68 | hsa-miR-92b | 43.71 |
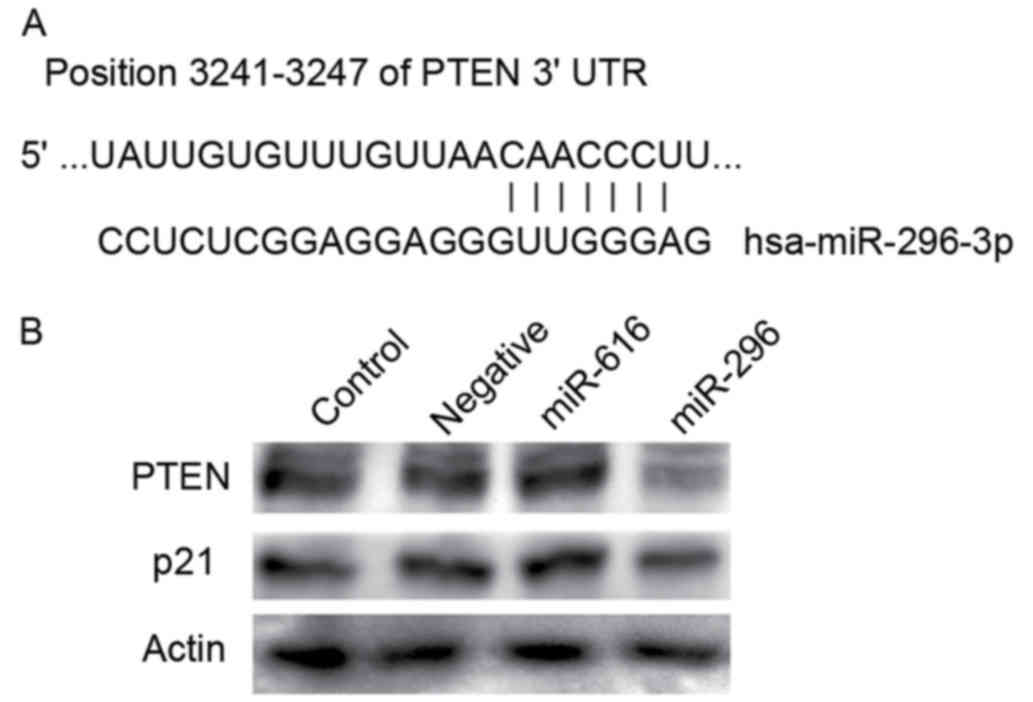
PTEN is a direct target of miR-296-3p
in head and neck SCC (HNSCC) cells
Target prediction using TargetScan 6.2 revealed that
miR-296-3p potentially targets PTEN, as seven nucleotides of the 5′
seed region were complementary to the PTEN 3′UTR (Fig. 1A). It was established that miR-296-3p
negatively regulates PTEN by adding a miR-296-3p mimic into SCC25
cells. The HNSCC cell line, SCC25, was transfected with a
miR-296-3p mimic, miR-616 mimic, and a negative control. miR-616
mimic, which was upregulated in SCC cells but did not target PTEN
or p21 in silico, was used as the negative control.
Following transfection with the miR-296-3p, PTEN expression was
markedly decreased, compared with that for the negative control
(Fig. 1B). p21, a cyclin-dependent
kinase inhibitor, is directly regulated by p53 and is involved in
the carcinogenesis of HNSCC. p21 was also reported to be a target
of miR-296-5p; therefore, p21 expression was investigated in HNSCC
cells transfected with miRNA-296-3p. No significant difference was
observed between the p21 expression following transfection with the
miRNA-296-3p mimic and with the control (Fig. 1B).
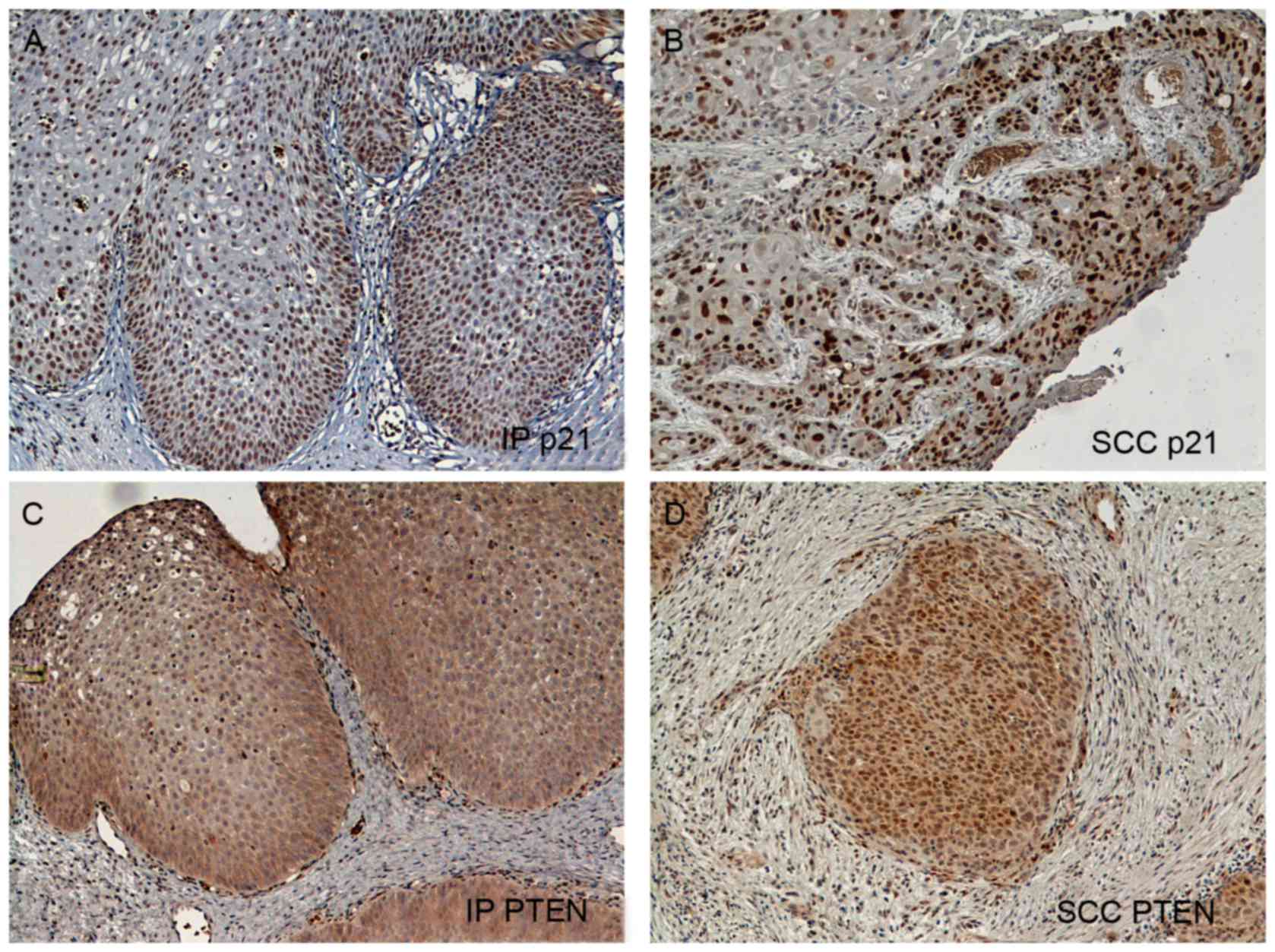
Expression of PTEN and p21 in IP and
SCC, and its association with clinicopathological features
Immunohistochemical staining for PTEN and p21 was
performed for 17 IP and 19 SCC tissue samples. A total of 11 IPs
coexisted with severe dysplasia or SCC, while the remaining six
samples were non-dysplastic IPs. A total of 12 SCC samples were
differentiated from IPs and the other 7 SCC samples did not coexist
with IPs. One IP that coexisted with SCC samples was too small and
badly damaged and, therefore, it was excluded from the present
study. Representative images of staining for PTEN and p21 are
presented in Fig. 2. PTEN expression
was observed primarily in the cytoplasm of the tumor cells
(Fig. 2C and D). Positive PTEN
staining was observed in 13/17 IPs (76.4%), a rate significantly
increased compared with that observed in SCCs (4/19, 21.0%;
P=0.0005; unpaired t-test with Welch's correlation; Table III). Positive p21 staining was
observed in 11/17 IPs (64.7%), which was also a significantly
increased rate compared with that in SCCs (5/19, 26.3%; P=0.0086;
Table III). No significant
differences were observed in PTEN or p21 expression between
non-dysplastic and dysplastic IPs, or between SCCs with IPs and
SCCs without IPs. Double positive staining was observed in 12 of
all the tissue samples, and double negative staining was observed
in 15 tissue samples.
 | Table III.The expression status of PTEN and
p21. |
Table III.
The expression status of PTEN and
p21.
| Cancer type | Samples, n | PTEN-positive, n
(%) | PTEN-negative, n
(%) | P-value | p21-positive, n
(%) | p21-negative, n
(%) | P-value |
|---|
| Total | 36 |
|
|
|
|
|
|
| IP
non-dysplastic | 6 | 5 | 1 |
| 3 | 3 |
|
| IP with SCC | 11 | 8 | 3 |
| 8 | 3 |
|
|
|
|
|
| n.s. |
|
| n.s. |
| SCC arise from
IP | 12 | 2 | 10 |
| 3 | 9 |
|
| SCC | 7 | 2 | 5 |
| 2 | 5 |
|
|
|
|
| n.s. |
|
| n.s. |
| All IP | 17 | 13 (76.5) | 4 (23.5) |
| 11 (64.7) | 6 (35.3) |
|
| All SCC | 19 | 4 (21.1) | 15 (79.9) |
| 5 (26.3) | 14 (73.7) |
|
|
|
|
|
| 0.0005 |
|
| 0.0086 |
Discussion
In the present study, it was revealed that the
expression of numerous miRNAs was altered during the transition
between sinonasal IP and SCC. In view of the multistep
carcinogenetic process, a large number of miRNAs are considered to
be associated with the malignant transformation through the
regulation of oncogenes (12). In the
present study, number of miRNAs were detected that demonstrated
differential expression between IP and SCC, but the majority were
not expected to regulate the expression of oncogenes in in
silico analysis. Regulation of PTEN by miRNA 296–3p may be one
of the critical processes associated with multiple molecular
alterations for transformation to invasive SCC.
PTEN is a well-known tumor suppressor gene that is
associated with human carcinogenesis (13). PTEN transcription is activated by p53
and negatively regulates cellular survival by inhibiting the
phosphoinositide 3-kinase (PI3K)/protein kinase B (Akt) signaling
pathway (13). The PI3K/Akt signaling
pathway performs important roles in the carcinogenesis of HNSCC
(14). However, a PTEN deficiency of
≤30% is observed in HNSCC (15).
Recent genome-wide sequence analysis of HNSCC has revealed a large
number of gene alterations, including the molecules involved in the
PI3K/Akt signaling pathway (16,17). PTEN
mutations were detected in only 5–10% of cases, and
phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α
mutations in 8–10% of cases (16,17).
Therefore, activation of the PI3K/Akt pathway due to PTEN deletion
alone is insufficient for the induction of invasive HNSCC. Bian
et al (18) promoted HNSCC via
the deletion of transforming growth factor-β1 and PTEN. Squarize
et al (19) demonstrated that
mice harboring a PTEN deficiency developed multiple oral SCC
lesions on exposure to a tobacco surrogate, whereas the control
mice did not.
p21 is a major downstream molecular target of p53
that mediates the apoptotic effect of p53 through the suppression
of DNA replication and G1 stage cell cycle arrest
(20). The results of the present
study demonstrate that the presence of p21 was less common in SCC,
as compared with in IP cells, but miRNA-296-3p may not regulate p21
expression in head and neck cancer cells. In cancer tissue, p21
expression is not always associated with p53 expression as p21
expression levels are low in tissues containing p53 harboring a
functional mutation (20). The
prognosis and aggressiveness of head and neck cancer were revealed
to be associated with the expression of PTEN, but not with p53 and
p21 (21,22). In the present study, PTEN and p21
expression were positively correlated with one another, and these
results indicate that the trigger for the malignant transformation
of IPs to SCCs may underlie the activation of the PI3K/Akt
signaling pathway via the loss of PTEN and the subsequent
degradation of the p53-p21 pathway.
Several studies have demonstrated the overexpression
of miRNA-296 in breast cancer cells (23), glioma cells (24) and esophageal cancer (25). In these three studies, miR-296 was
used to represent miRNA-296-5p+ (miRNA-296 from the 5′ end), and
its target genes were distinctly different from the targets of
miRNA-296-3p. A previous study determined that miRNA-296-3p
elevates and regulates intercellular adhesion molecule 1, in order
to promote prostate cancer metastasis by possibly enhancing the
survival of natural killer cell-resistant circulating tumor cells
(26). In silico analysis has
demonstrated miR-296-3p has 66 target genes, including
carcinogenesis related-gene as zinc finger protein 629 and
claudin-2. These genes may be involved in the attainment of
invasion and metastases in the malignant transformation of IP. The
present study focused on miR-296-3p, but the expression of 200
miRNAs were revealed to vary by >2-fold during malignant
transformation. Each miRNA exhibiting differential expression may
perform a role in carcinogenesis; however, not all miRNAs are
hypothesized to be associated with it. To identify the function of
miRNAs in carcinogenesis, further comprehensive biological
molecular studies are required. Recent developments in omics
research as an analysis tool for the transcriptome and proteome
have made it possible to identify cause and effect associations
(27,28). Furthermore, exome studies using
next-generation sequencers have been developed (15,16), and
the association between genetic alterations and miRNA expression
may now be investigated (16).
In conclusion, the present results indicate that
miRNA expression varies during the malignant transformation of
sinonasal IPs, and that miR-296-3p, which is overexpressed in SCCs,
regulates PTEN expression. The findings of the present study have
significant implications regarding the development of
carcinogenesis biomarkers, as well as preventative and therapeutic
strategies for SCCs arising from IP via the targeting of miRNAs,
and the PI3K/Akt and p53-p21 signaling pathways.
Acknowledgements
The present study was supported by a KAKENHI
Grant-in-Aid for Scientific Research from the Ministry of
Education, Science and Culture (grant no. 26462591), a Health and
Labor Sciences Research Grant for Clinical Cancer Research from the
Ministry of Health, Labor and Welfare of Japan (grant no. H26-141)
and the National Cancer Center Research and Development Fund of
Japan (grant no. 26-A-4).
References
|
1
|
Nakamaru Y, Furuta Y, Takagi D, Oridate N
and Fukuda S: Preservation of the nasolacrimal duct during
endoscopic medial maxillectomy for sinonasal inverted papilloma.
Rhinology. 48:452–456. 2010.PubMed/NCBI
|
|
2
|
Myers EN, Fernau JL, Johnson JT, Tabet JC
and Barnes EL: Management of inverted papilloma. Laryngoscope.
100:481–490. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Furuta Y, Shinohara T, Sano K, Nagashima
K, Inoue K, Tanaka K and Inuyama Y: Molecular pathologic study of
human papillomavirus infection in inverted papilloma and squamous
cell carcinoma of the nasal cavities and paranasal sinuses.
Laryngoscope. 101:79–85. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kinoshita T, Hanazawa T, Nohata N, Okamoto
Y and Seki N: The functional significance of microRNA-375 in human
squamous cell carcinoma: Aberrant expression and effects on cancer
pathways. J Hum Genet. 57:556–563. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Christensen BC, Moyer BJ, Avissar M,
Ouellet LG, Plaza SL, McClean MD, Marsit CJ and Kelsey KT: A let-7
microRNA-binding site polymorphism in the KRAS 3′UTR is associated
with reduced survival in oral cancers. Carcinogenesis.
30:1003–1007. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hatakeyama H, Cheng H, Wirth P, Counsell
A, Marcrom SR, Wood CB, Pohlmann PR, Gilbert J, Murphy B, Yarbrough
WG, et al: Regulation of heparin-binding EGF-like growth factor by
miR-212 and acquired cetuximab-resistance in head and neck squamous
cell carcinoma. PLoS One. 5:e127022011. View Article : Google Scholar
|
|
8
|
Syrjänen S: Human papillomavirus (HPV) in
head and neck cancer. J Clin Virol. 32 Suppl 1:S59–S66. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
zur Hausen H: Papillomaviruses causing
cancer: Evasion from host-cell control in early events in
carcinogenesis. J Natl Cancer Inst. 92:690–698. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Syrjänen K and Syrjanen S: Detection of
human papillomavirus in sinonasal carcinoma: Systematic review and
meta-analysis. Hum Pathol. 44:983–991. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nagata Y, Lan KH, Zhou X, Tan M, Esteva
FJ, Sahin AA, Klos KS, Li P, Monia BP, Nguyen NT, et al: PTEN
activation contributes to tumor inhibition by trastuzumab and loss
of PTEN predicts trastuzumab resistance in patients. Cancer Cell.
6:117–127. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Courthod G, Franco P, Palermo L, Pisconti
S and Numico G: The role of microRNA in head and neck cancer:
Current knowledge and perspectives. Molecules. 19:5704–5716. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stambolic V, MacPherson D, Sas D, Lin Y,
Snow B, Jang Y, Benchimol S and Mak TW: Regulation of PTEN
transcription by p53. Mol Cell. 8:317–325. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pedrero JM, Carracedo DG, Pinto CM,
Zapatero AH, Rodrigo JP, Nieto CS and Gonzalez MV: Frequent genetic
and biochemical alterations of the PI 3-K/AKT/PTEN pathway in head
and neck squamous cell carcinoma. Int J Cancer. 114:242–248. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
da Costa AA, D'Almeida Costa F, Ribeiro
AR, Guimarães AP, Chinen LT, Lopes CA and de Lima VC: Low PTEN
expression is associated with worse overall survival in head and
neck squamous cell carcinoma patients treated with chemotherapy and
cetuximab. Int J Clin Oncol. 20:282–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stransky N, Egloff AM, Tward AD, Kostic
AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C,
McKenna A, et al: The mutational landscape of head and neck
squamous cell carcinoma. Science. 333:1157–1160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Agrawal N, Frederick MJ, Pickering CR,
Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et
al: Exome sequencing of head and neck squamous cell carcinoma
reveals inactivating mutations in NOTCH1. Science. 333:1154–1157.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bian Y, Hall B, Sun ZJ, Molinolo A, Chen
W, Gutkind JS, Waes CV and Kulkarni AB: Loss of TGF-β signaling and
PTEN promotes head and neck squamous cell carcinoma through
cellular senescence evasion and cancer-related inflammation.
Oncogene. 31:3322–3332. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Squarize CH, Castilho RM, Abrahao AC,
Molinolo A, Lingen MW and Gutkind JS: PTEN deficiency contributes
to the development and progression of head and neck cancer.
Neoplasia. 15:461–471. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Schwerer MJ, Sailer A, Kraft K, Baczako K
and Maier H: Patterns of p21 (waf1/cip1) expression in
non-papillomatous nasal mucosa, endophytic sinonasal papillomas and
associated carcinomas. J Clin Pathol. 54:871–876. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sham CL, To KF, Chan PK, Lee DL, Tong MC
and van Hasselt CA: Prevalence of human papillomavirus,
Epstein-Barr virus, p21, and p53 expression in sinonasal inverted
papilloma, nasal polyp, and hypertrophied turbinate in Hong Kong
patients. Head Neck. 34:520–533. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim SG, Lee OY, Choi JW, Park YH, Kim YM,
Yeo MK, Kim JM and Rha KS: Pattern of expression of cell
cycle-related proteins in malignant transformation of sinonasal
inverted papilloma. Am J Rhinol Allergy. 25:75–81. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yoon AR, Gao R, Kaul Z, Choi IK, Ryu J,
Noble JR, Kato Y, Saito S, Hirano T, Ishii T, et al: MicroRNA-296
is enriched in cancer cells and downregulates p21WAF1 mRNA
expression via interaction with its 3′ untranslated region. Nucleic
Acids Res. 39:8078–8091. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Würdinger T, Tannous BA, Saydam O, Skog J,
Grau S, Soutschek J, Weissleder R, Breakefield XO and Krichevsky
AM: miR-296 regulates growth factor receptor overexpression in
angiogenic endothelial cells. Cancer Cell. 14:382–393. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hong L, Han Y, Zhang H, Li M, Gong T, Sun
L, Wu K, Zhao Q and Fan D: The prognostic and chemotherapeutic
value of miR-296 in esophageal squamous cell carcinoma. Ann Surg.
251:1056–1063. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu X, Chen Q, Yan J, Wang Y, Zhu C, Chen
C, Zhao X, Xu M, Sun Q, Deng R, et al: MiRNA-296-3p-ICAM-1 axis
promotes metastasis of prostate cancer by possible enhancing
survival of natural killer cell-resistant circulating tumour cells.
Cell Death Dis. 4:e9282013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chung CH, Parker JS, Karaca G, Wu J,
Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X,
et al: Molecular classification of head and neck squamous cell
carcinomas using patterns of gene expression. Cancer Cell.
5:489–500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hatakeyama H, Kondo T, Fujii K, Nakanishi
Y, Kato H, Fukuda S and Hirohashi S: Protein clusters associated
with carcinogenesis, histological differentiation and nodal
metastasis in esophageal cancer. Proteomics. 6:6300–6316. 2006.
View Article : Google Scholar : PubMed/NCBI
|