Introduction
Mastectomy may negatively affect the quality of life
for women with breast cancer (1). As
the psychological benefits of breast reconstruction are
increasingly acknowledged, reconstruction has become an important
part of the management of breast cancer. (2) There are generally two options for breast
cancer reconstruction: The autologous option involves the transfer
of tissues from other regions (donor sites) of the body to the
chest wall, whereas the prosthetic option involves the placement of
synthetic implants under the chest wall (3). Both procedures have various advantages
and disadvantages (4). Breast
reconstruction with autologous tissue should contain no tumor cell
infiltration. Recent advances in tissue engineering enable the use
of composite tissues for breast reconstruction with stem cells and
scaffolds, which has brought new hope that tissue regeneration can
be achieved (5).
Adult mammary stem cells (AMSCs) are long-lived,
generally quiescent, and capable of differentiating into any cell
type associated with the mammary gland, depending on which signals
are received (6). The first evidence
for the presence of mammary stem cells emerged in 1959 when DeOme
et al (7) demonstrated that
the transplantation of small portions of normal mammary tissue from
donor mice into epithelium-free fat pads of syngeneic recipient
mice led to the development of fully functional mammary outgrowths,
containing ductal, alveolar, and myoepithelial cells. The study
also established the in vivo transplantation model, which
remains the gold standard assay for testing stem or progenitor cell
capacity. Since the DeOme study, significant understanding of AMSCs
has been acquired, partially through the application of various
markers to isolate and enrich these rare cells (8).
The longevity of mammary stem cells increases the
chance that they could undergo tumorigenic changes and transform
into cancer stem cells (CSCs). Although the origin of breast CSCs
has yet to be identified, the similarity between CSCs and normal
stem cells, including their self-renewal and differentiating
capabilities, and evidence that differentiation is irreversible,
strongly support the conclusion that normal stem cells are the
source of CSCs (9).
As breast cancer is an acquired, multigenic disease
that occurs primarily in middle-aged and older individuals, the
application of prospectively isolated embryonic stem cells for
breast reconstruction is not possible. Thus, AMSCs are a promising
cell source; however, the close anatomical similarity between
normal stem cells and CSCs poses an apparent safety issue if
applying AMSCs for tissue regeneration (10). The majority of biomarkers used to
isolate mammary stem cells cannot distinguish between normal and
CSCs (11), inevitably leading to the
question of whether AMSCs from breast cancer patients can safely be
used as an autologous source in breast reconstruction.
In the present study, to address this question a
population of AMSCs were isolated from the periphery of breast
cancer tissues. These cells were characterized for their
differentiation ability, their gene expression profile and their
potential to form tumors in vivo. Gene expression profiles
revealed the downregulation of a group of oncogenes and
upregulation of tumor suppressor genes within the AMSCs relative to
autologous cancer cells. The in vivo tumorigenesis assay
demonstrated that the AMSCs were non-tumorigenic, suggesting that
the AMSCs adjacent to breast cancer are likely to be suitable as
cell sources for breast reconstruction.
Materials and methods
Human tissue samples
The Ethics Committee of Jilin University (Changchun,
China) approved this study, and all patients provided informed
written consent. A total of 9 female patients diagnosed with breast
cancer were recruited for the study (Table I). Fresh mammary tissue samples,
including cancerous and adjacent normal tissues, were obtained
during surgical resection.
 | Table I.Clinicopathological characteristics
of the 9 female patients recruited into the present study. |
Table I.
Clinicopathological characteristics
of the 9 female patients recruited into the present study.
| Patient no. | Age (years) | Histopathological
classification | Tumor stage |
|---|
| 1 | 40 | Simple
carcinoma | T1N1M0 |
| 2 | 36 | Invasive ductal
carcinoma | T3N2M0 |
| 3 | 27 | Pupillary
carcinoma | T2N1M0 |
| 4 | 36 | Mucinous
carcinoma | T2N1M0 |
| 5 | 29 | Invasive lobular
carcinoma | T3N1M0 |
| 6 | 33 | Intraductal
carcinoma | T1N0M0 |
| 7 | 47 | Invasive ductal
carcinoma | T2N1M0 |
| 8 | 36 | Simple
carcinoma | T2N1M0 |
| 9 | 28 | Invasive ductal
carcinoma | T1N1M0 |
Isolation of AMSCs and breast cancer
cells
AMSCs were isolated as previously described
(12), with modifications. Briefly,
normal breast tissues, as confirmed by pathological examination,
were dissected from ≥2 cm away from the periphery of breast tumors
and digested with collagenase IV (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) overnight at 37°C. Collagenase
solution was removed by centrifugation at 1,200 × g and 37°C for 5
min, and the cell pellet was cultured in a 1:1 mixture (v/v) of
Dulbecco's modified Eagle's medium and Ham's F12 nutrient mixture
(DMEM/F12; Invitrogen; Thermo Fisher Scientific, Inc.) containing
10% fetal calf serum (FCS; Hyclone; GE Healthcare Life Sciences,
Logan, UT, USA); 50 µg/ml penicillin and 0.1 mg/ml streptomycin
(Beyotime Institute of Biotechnology, Haimen, China); 2.5 µg/ml
amphotericin-B, 1 µg/ml minocylin, 1 µg/ml insulin, 1 µg/ml
hydrocortisone and 10 µg/ml transferrin (Shanghai HyperHeal Biology
Co. Ltd., Shanghai, China); 11 µg/ml ethanolamine (Fusheng,
Shanghai, China); 50 ng/ml cholera toxin (List Biological
Laboratories, Inc., Campbell, CA, USA); and 10 ng/ml epithelial
growth factor (Invitrogen; Thermo Fisher Scientific, Inc.). After
14 days, cells grown in primary culture were detached with trypsin
and subjected to sequential immunomagnetic isolation with
anti-epithelial specific antigen (anti-ESA; cat. no. MS-675-P;
Invitrogen; Thermo Fisher Scientific, Inc.; dilution: 1:1,000;
incubation at 4°C overnight) and anti-sialomucin (anti-MUC; cat.
no. 35-4900; Invitrogen; Thermo Fisher Scientific, Inc.; dilution:
1:1,000; incubation at 4°C overnight) antibodies followed by goat
anti-mouse IgG microbeads (cat. no. SC-53808; Miltenyi Biotec GmbH,
Bergisch Gladbach, Germany; dilution: 1:1,000; incubation at 37°C
for 2 h). ESA+/MUC− cells were collected and
cultured in the aforementioned growth medium for future
experiments.
To isolate breast cancer cells, the breast tumor
tissue samples were minced into small pieces with a scalpel and
digested with collagenase IV overnight at 37°C. The collagenase
solution was then removed by centrifugation and the cell pellet was
seeded in the same growth medium as for AMSC isolation. AMSCs grew
as adherent cells and thus were isolated by removing the
non-adherent cells. Upon culture in the aforementioned growth
medium, adherent AMSCs between passages two and four were used for
further experiments.
For treatment with basic fibroblast growth factor
(bFGF; cat. no. CYT-557; ProSpec-Tany, Rehovot, Isreal), cells were
cultured in M199 medium (Invitrogen; Thermo Fisher Scientific,
Inc.) containing 15% fetal bovine serum, 100 U/ml penicillin, 100
µg/ml streptomycin, and 10 ng/ml bFGF at 37°C for 24 h.
Immunocytochemistry
Cells in the logarithmic growth phase were fixed
with 10% neutral buffered formalin and stained with anti-ESA (cat.
no. MS-675-P, Invitrogen, Thermo Fisher Scientific, Inc.; dilution:
1:1,000; incubation at 4°C overnight), anti-common acute
lymphoblastic leukemia antigen (anti-CALLA; cat. no. SC46656, Santa
Cruz Biotechnology, Santa Cruz, CA, USA; dilution: 1:100;
incubation at 4°C overnight), and anti-keratin-19 antibodies
(anti-K-19; cat. no. SC6278, Santa Cruz Biotechnology, Santa Cruz,
CA, USA; dilution: 1:100; incubation at 4°C overnight) using an
indirect immunoperoxidase avidin-biotinylated enzyme complex method
that involved the avidin/biotin blocking kit (cat. no. SP-2001;
Vector Laboratories, Burlingame, CA, USA;), followed by
species-specific biotinylated secondary antibodies targeting the
corresponding primary antibodies (dilution: 1:100; incubation at
37°C for 1 h), and detected using the Vectastain Elite ABC HRP
reagent (cat. no. PK-7100) in accordance with the manufacturer's
protocol.
Microarray analysis
The gene expression profiles of the isolated AMSCs
and breast cancer cells of the 9 patients were determined using a
CapitalBio Human Genome Oligo Array (22 K) (cat. no. 400010;
CapitalBio Corporation, Beijing, China) of 21,552 human genes,
according to the manufacturer's instructions. Briefly, total RNA
was extracted from isolated cells using TRIzol (Invitrogen; Thermo
Fisher Scientific, Inc.) in accordance with the manufacturer's
protocol. Total RNA was reversed transcribed into the first-strand
cDNA using the First Strand Enzyme Mix and Buffer Mix included in
the kit at 42°C for 2 h. Subsequently, the synthesis of
second-strand cDNA was achieved using the Second Strand Master Mix
at 16°C for 1 h followed by 65°C for 10 min. cDNA was then
transcribed into cRNA in a reaction mixture containing 4 µl
nuclease water, 20 µl T7 Buffer Mix (cat. no. HZ101-975; Shanghai
Wei Zhen Industrial Co., Ltd., Shanghai, China) and 6 µl T7 enzyme
mix (cat. no. AM2719G1; TideRadar Technology Co., Ltd., Beijing,
China) and allowed to interact at 40°C for 8–14 h. cRNA (5 µg in
7.5 µl nuclease free water) was hybridized to random primers (4 µl)
at 65°C for 5 min, followed by incubation on ice for 5 min, and
reversed transcribed into cDNA in a Master Mix containing 5 µl of
4xCbcScriptII Buffer (CapitalBio Corporation, Beijing, China), 2 µl
0.1 M DTT, 1.5 µl CbcScriptII at 25°C for 10 min followed by at
37°C for 1.5 h. Following the addition of Termination Buffer (cat.
no. BTN130614; Beijing Baiaolaibo Science and Technology Ltd.,
Beijing, China) at 65°C for 10 min and, Neutralization Solution
(cat. no. A7132; Promega Corporation, Madison, WI, USA) was added
to the reaction mixture at room temperature for 5 min. cDNA was
then purified and labeled with Cy5-dCTP (for breast cancer cells)
or Cy3-dCTP (for AMSCs; GE Healthcare Life Sciences) in a reaction
mixture containing 4 µl random primers, 5 µl 5X Klenow Buffer, 1 µl
Cy5-dCTP (PA55031; GE Healthcare Life Sciences) or Cy3-dCTP (cat.
no. PA55031; GE Healthcare Life Sciences) and 1.2 µl Klenow
Fragment (cat. no. D7035; Beyotime Institute of Biotechnology) at
37°C for 1.5 h, followed by 70°C for 5 min.
The labeled cDNA was then hybridized to the
microarray chip in 81.6-µl hybridization mixture containing 40 µl
of fluorescently labeled cDNA and 41.6 µl of hybridization buffer
[25% formamide, 3X saline sodium citrate buffer, 5X Denhardt's
solution (Beijing Dingguo Changsheng Biotechnology Co., Ltd.,
Beijing, China), 0.2X SDS]. After a 3-min incubation at 95°C, the
chip containing the hybridization mixture was placed on ice, sealed
with Lifterslip Coverslip (Thermo Fisher Scientific) and incubated
at 42°C overnight, scanned using a LuxScan 10KA scanner, and
processed using LuxScan 3.0 imaging analysis software (CapitalBio).
Following background subtraction and normalization, genes
exhibiting an experiment/control signal ratio of <0.5 were
considered to be downregulated, and those with a ratio of >2.0
were considered to be upregulated.
Semi-quantitative
reverse-transcription PCR (RT-PCR)
Total RNA was extracted using a TriPure™ reagent
(Boehringer; Roche Applied Science, Mannheim, Germany) in
accordance with the manufacturer's protocol. The extracted RNA was
treated with DNase I and further purified with phenol chloroform.
Total RNA (1 µg) was reversed transcribed using Superscript II
reverse transcriptase (cat. no. 18064071, Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's
instructions.
The following primers were used for PCR: MYC
forward, 5′-CCC AGC GAG GAC ATC TGG AAG AA-3′; MYC reverse,
5′-GAG AAG CCG CTC CAC ATG CAG TC-3′ (amplicon size, 268 bp);
H-RAS forward, 5′-AAG CTT GTG GTG GTG GGC GCT AAA GGC-3′;
H-RAS reverse, 5′-CTT TCA CCC GCT TGA TCT GCT CCC TGT ACT-3′
(amplicon size, 300 bp); ErbB receptor tyrosine kinase 2
(ERBB2, forward, 5′-GCC CTG GAC ACC TAC AAC AC-3′;
ERBB2 reverse, 5′-TCC GGC AGA AAT GCC AGG CT-3′ (amplicon
size, 329 bp); β-actin (ACTB) forward, 5′-CCT GTA CGC CTC
TGG CCG TAC CAC T-3′; and ACTB reverse, 5′- CTG TAG CCG CGC
TCG GTG AGG ATC T-3′ (amplicon size, 174 bp).
The PCR conditions for the amplification of
MYC were 28 cycles of 94°C for 1 min, 60°C for 1 min and
72°C for 1 min, followed by a 5-min final extension at 72°C. PCR
amplification of H-RAS comprised denaturation at 94°C for 4
min; 30 cycles of 94°C for 1 min, 62°C for 1 min and 72°C for 1
min; followed by an elongation 72°C for 10 min. PCR amplification
of ERBB2 consisted of denaturation at 94°C for 2 min; then
35 cycles at 94°C for 45 sec, 55°C for 45 sec and 72°C for 1 min;
followed by a 5-min final extension at 72°C. PCR amplification of
ACTB included denaturation at 95°C for 3 min; followed by 40
cycles at 95°C for 30 sec, 57°C for 40 sec and 72°C for 45 sec;
followed by a 6-min final extension at 72°C. The PCR products were
loaded onto ethidium bromide-stained 1% agarose gel and imaged
using a Gel Doc XR+ System (Bio-Rad Laboratories, Inc., Hercules,
CA, USA). The band density was quantified using Quantity One
software (version 4.52, Bio-Rad Laboratories, Inc.) and presented
as a ratio of target gene to β-actin (internal control).
Western blot analysis
Whole cell extracts from AMSCs and breast CSCs were
prepared by lysing cells in lysis buffers [300 mM NaCl, 50 mM
Tris-HCl (pH 8.0), 25 mM EDTA, 20% (v/v) SDS, and 0.2 mg/ml
proteinase K]. Insoluble debris was removed by centrifugation at
16,000 × g for 2 min. Protein concentration in whole cell extracts
was determined using a Bio-Rad Protein Assay (Bio-Rad Laboratories,
Inc.). From each sample, 20–30 µg of whole cell extract was
resolved by SDS-PAGE, transferred to an Immobilon-P polyvinylidene
fluoride membrane (EMD Millipore, Billerica, MA, USA) and probed
with the following primary antibodies: anti-ErbB2 (cat. no. 2244S;
dilution: 1:1,000; incubation at 4°C overnight), anti-Ras (cat. no.
3339S; dilution: 1:1,000; incubation at 4°C overnight), anti-PTEN
(cat. no. 9549L; dilution: 1:1,000; incubation at 4°C overnight),
and anti-β-actin (internal control; cat. no. 8844S; dilution:
1:500; incubation at 4°C overnight) antibodies (all from Cell
Signaling Technology, Inc., Danvers, MA, USA) followed by
horseradish peroxidase (HRP)-conjugated species-specific secondary
antibodies targeting the corresponding primary antibodies at room
temperature for 2 h. Protein bands were visualized using an
Amersham ECL Western Blotting Detection kit (GE Healthcare
Bio-Sciences, Pittsburgh, PA, USA).
Culture of MCF-7 cells
MCF-7 cells were purchased from the Cell Bank of
Type Culture of Chinese Academy of Sciences (Shanghai, China) and
cultured in DMEM containing 10% FCS, 50 µg/ml penicillin, 0.1 mg/ml
streptomycin, 1.0 µg/ml sodium pyruvate, 10 µg/ml insulin, 0.1 mM
non-essential amino acids, and 1.5 g/l sodium bicarbonate at 37°C
in a sterile atmosphere containing 5% CO2.
In vivo tumorigenesis assay
The Institutional Animal Care and Use Committee of
Jilin University approved all animal experiments. A total of 28
female non-obese diabetic/severe combined immunodeficiency
(NOD/SCID) mice (age, 4–6 weeks; weight, 19–22 g) were purchased
from the Shanghai Laboratory Animal Center (Shanghai, China) and
housed in a pathogen-free facility, with water and food provided
ad libitum.
The mice were randomly divided into an experimental
group (n=14) and a control group (n=14), and were individually
injected subcutaneously with AMSCs (2×106) or MCF-7
cells (2×106), respectively, in the right dorsolateral
area. The growth of subcutaneous tumors was monitored by evaluating
the length (L) and width (W) of the tumors using a caliper every
two days. The tumor volume (V) was calculated as V=1/2 xL
xW2. At 30 days after the injection for the experimental
group and 14 days for the control group, all mice were sacrificed
by cervical dislocation and tissues from the injected region were
isolated. The isolated tissues were fixed in formalin, embedded in
paraffin, stained with hematoxylin and eosin, and observed under a
microscope.
Results
AMSCs isolated from normal
tumor-adjacent mammary tissue exhibit stem cell features
To characterize AMSCs from breast cancer patients,
tumor-free mammary tissue was extracted from ≥2 cm away from the
tumor periphery in patients with breast cancer, and confirmed by
pathological examination. ESA+/MUC− cells
were purified through immunomagnetic isolation, which has been
demonstrated to separate a cell population into luminal and
myoepithelial cells (13). Under
non-differentiating conditions, these cells attached to the plate
and became round or polygonal, with an indistinct cell boundary
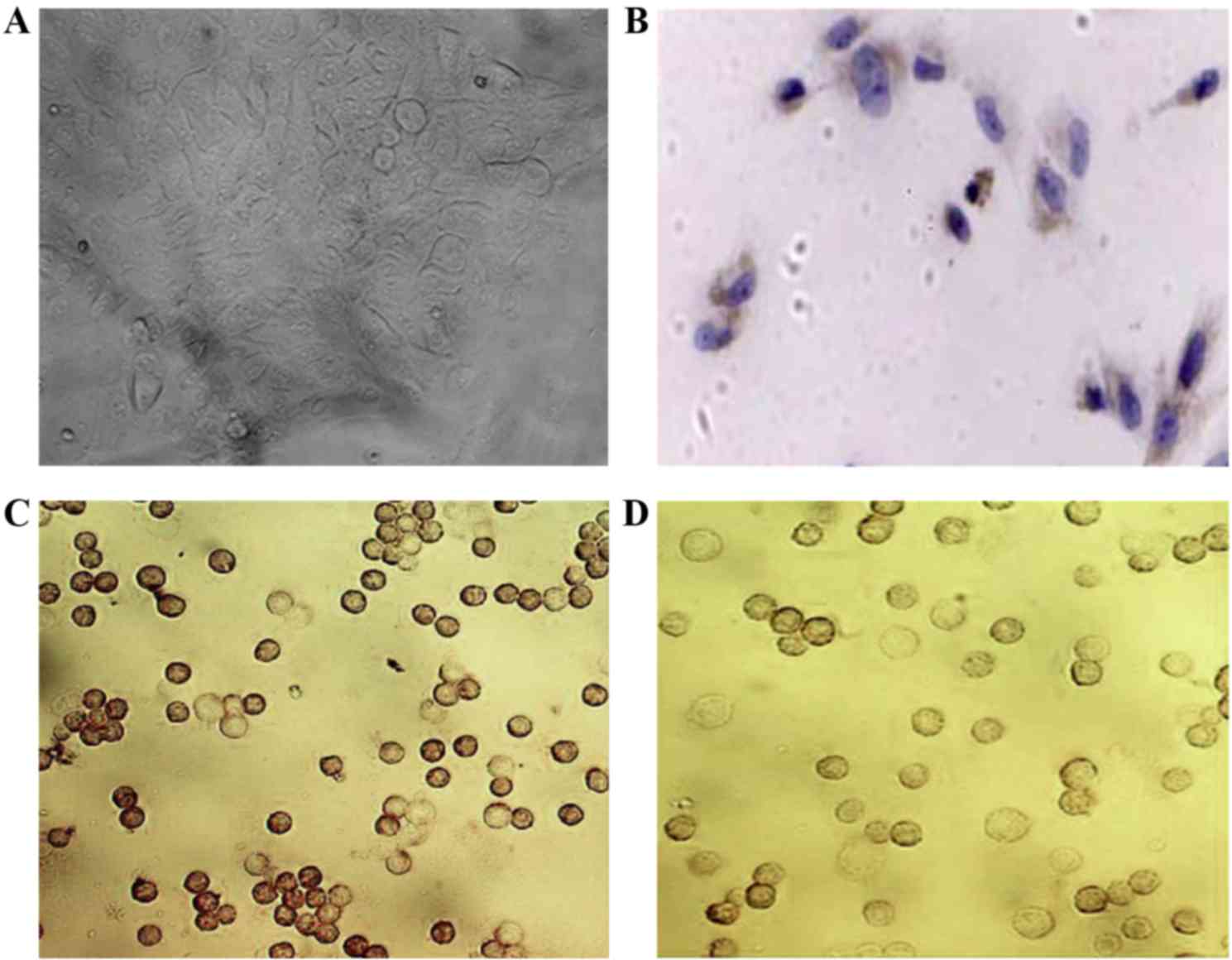
(Fig. 1A). Using immunocytochemical
staining, these cells were identified as positive for ESA on the
cellular membrane and partially in the cytoplasm (Fig. 1B) and for K-19 (Fig. 1C), and were positive for CALLA
following bFGF treatment (Fig.
1D).
AMSCs exhibit low expression levels of
oncogenes and high expression levels of tumor suppressor genes
To characterize the molecular features associated
with the isolated AMSCs, we performed a gene microarray analysis
and compared the gene expression profiles of paired AMSCs and
cancer cells extracted from patients (Fig. 2A). From a total of 21,522 genes
examined, 798 differed >2.5-fold between the groups, with 174
downregulated and 624 upregulated in AMSCs compared with cancer
cells. From those differentially expressed genes, 16 have a clearly
demonstrated clinical association with human breast cancer (as
defined by the CapitalBio Gene Ontology Database (www.capitalbio.com); Table
II), including 10 upregulated tumor suppressor genes and 6
downregulated oncogenes.
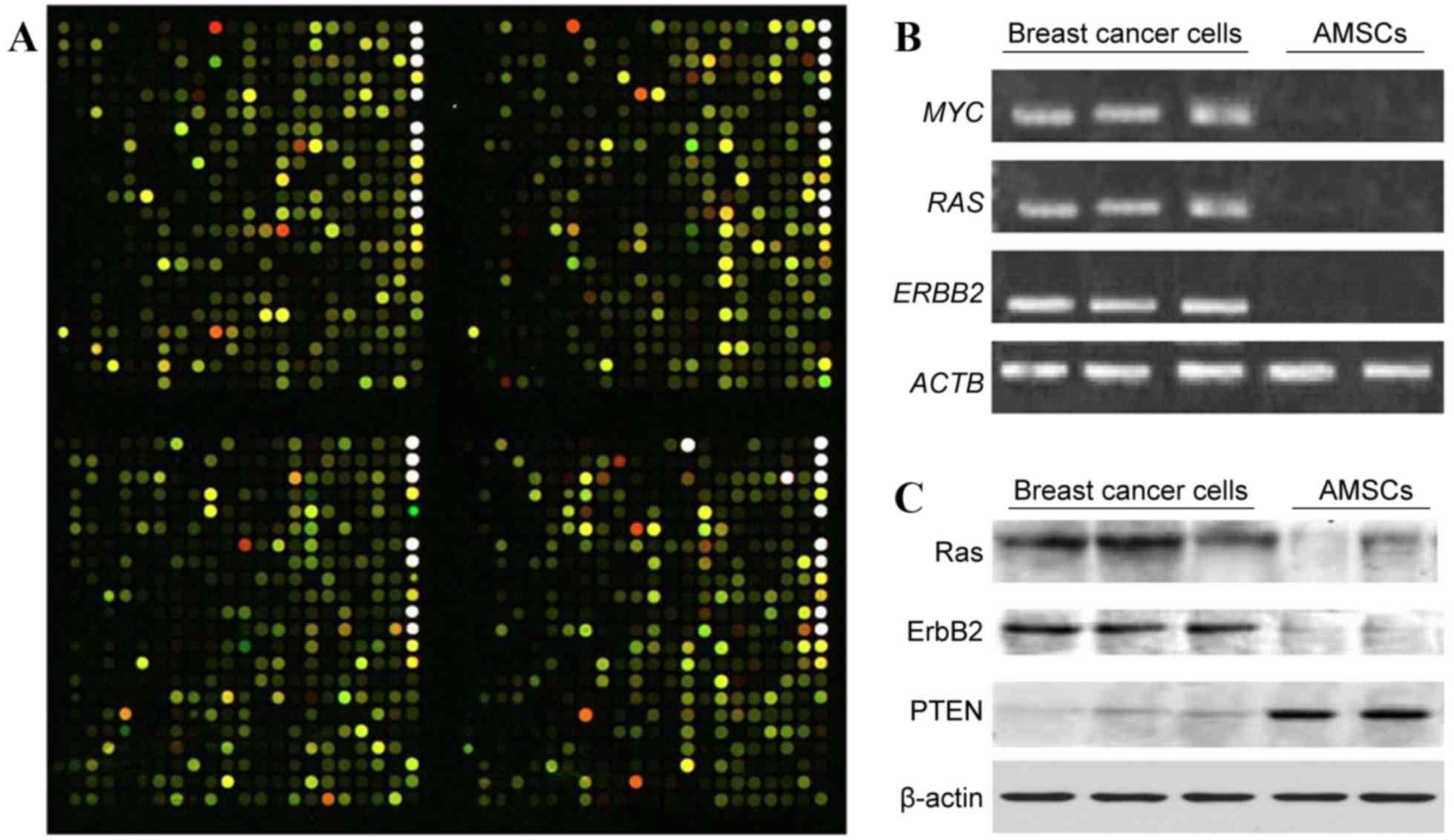 | Figure 2.AMSCs exhibit differential gene
expression compared with autologous breast cancer cells. (A)
Representative image of four blocks following the hybridization of
Cy3-labeled AMSC sample (green signal) and Cy5-labeled breast
cancer sample (red signal). The CapitalBio Human Genome Oligo Array
(22K) includes 21,522 70-mer oligo probes spotted on a 75×25-mm,
chemically modified glass slide. The green color indicates a
stronger signal from the AMSC sample, whereas red indicates a
stronger signal from the breast cancer sample, and yellow indicates
a similar signal from the two tissue types. (B) Gel electrophoretic
analyses of RT-PCR products. Total RNA was extracted from isolated
breast cancer cell samples and AMSC samples selected at random from
the 9 patients with breast cancer, and was reverse transcribed. The
relative gene expression levels of MYC, RAS, and ERBB2 were
examined by semi-quantitative PCR, with ACTB used as an internal
control. (C) Western blot analysis of Ras, ErbB2 and PTEN in breast
cancer cell samples and AMSC samples with β-actin used as internal
control. AMSC, adult mammary stem cell; RT-PCR, reverse
transcription-polymerase chain reaction; ErbB2, ErbB receptor
tyrosine kinase 2; ACTB, β-actin; PTEN, phosphatase and tensin
homolog. |
 | Table II.Summary of 16 differentially
expressed genes between AMSCs and cancer cells that have
demonstrated an association with human breast cancer. |
Table II.
Summary of 16 differentially
expressed genes between AMSCs and cancer cells that have
demonstrated an association with human breast cancer.
| A, Genes
upregulated in AMSCs relative to cancer cells |
|---|
|
|---|
| Gene symbol | Gene locus | Gene name and
general functions in breast cancer | Ratio (AMSCs/cancer
cells) | (Refs.) |
|---|
| RB1 | 13q14.2 | RB transcriptional
corepressor 1; regulates cell cycle progression | 19.1491 | (25) |
| PTEN | 10q23.3 | Phosphatase and
tensin homolog; phosphatase involved in multiple signaling
pathways, including phosphoinositide 3-kinase/protein kinase B,
cyclin D1 and p53 signaling | 2.0883 | (26) |
| ADRB2 | 5q31-q32 | Adrenoceptor β 2;
regulates growth and migration of breast cancer cells | 2.1505 | (27) |
| CDKN2A | 9p21 | Cyclin-dependent
kinase inhibitor 2A; controls cell cycle progression and
senescence | 2.6221 | (28) |
| CLCA2 | 1p31-p22 | Chloride channel
accessory 2; induced by p53 to inhibit breast cancer cell
proliferation | 25.5938 | (29) |
| SOD2 | 6q25.3 | Superoxide
dismutase 2, mitochondrial; neutralizes reactive oxygen species and
may be epigenetically silenced in breast cancer cells | 4.6025 | (30) |
| NFKBIA | 14q13 | NFκB inhibitor α;
regulates NFκB signaling in breast cancer | 2.6952 | (31) |
| MMP12 | 11q22.3 | Matrix
metalloproteinase 12; inhibits tumor vascularization | 13.0442 | (32) |
| ITGB3 | 17q21.32 | Integrin subunit β
3; regulates tumor progression, metastasis and stem cell
properties | 5.6503 | (33) |
| IL1B | 2q14 | Interleukin 1 β;
pro-inflammatory cytokine regulating breast cancer progression | 10.4994 | (34) |
|
| B, Genes
downregulated in AMSCs relative to cancer cells |
|
| Gene symbol | Gene locus | Gene name and
general functions in breast cancer | Ratio (AMSCs/cancer
cells) | (Refs.) |
|
| ERBB2 | 17q21 | Erb-B2 receptor
tyrosine kinase 2; regulates multiple functions, including
apoptosis, proliferation, adhesion, motility and vascularization,
to promote breast cancer progression. | 0.3866 | (35) |
| HRAS | 11p15.1-p15.4 | HRas
proto-oncogene, GTPase; proto-oncogene regulating intracellular
signaling pathways to promote tumorigenesis. | 0.4989 | (36) |
| MYC | 8q24.12-q24.13 | V-Myc avian
myelocytomatosis viral oncogene homolog; proto-oncogene regulating
cell proliferation, apoptosis and stem/progenitor cell
function. | 0.3921 | (37,38) |
| GHR | 5p13-p12 | Growth hormone
receptor; mediates growth hormone signaling. | 0.3815 | (39) |
| H19 | 11p15.5 | H19, imprinted
maternally expressed transcript (non-protein coding); encodes an
untranslated RNA that is overexpressed in breast cancer and
promotes cellular transformation/in vivo tumorigenesis. | 0.3125 | (40) |
|
ADAMTSL1 | 9p22.1-p22.2 | ADAMTS-like 1; a
secreted molecule resembling members of the ADAMTS family of
proteases with important functions regulating extracellular
matrix. | 0.4506 | (41) |
To validate the microarray data, semi-quantitative
RT-PCR was performed on certain targets identified by microarray,
including MYC, RAS and ERBB2. Consistent with
the microarray analysis, there were higher mRNA levels of all three
of these genes in the isolated cancer cells than in the AMSCs
(Fig. 2B). The levels of RAS,
ERBB2, and phosphatase and tensin homolog (PTEN)
proteins in these cells were also evaluated. As presented in
Fig. 2C, the levels of RAS and
ERBB2 were higher in breast cancer cells than in the AMSCs,
whereas AMSCs had a higher level of PTEN than the cancer
cells.
AMSCs do not generate tumors in nude
NOD/SCID mice, and form regularly arranged duct-like
structures
To assess the properties and tumorigenic nature of
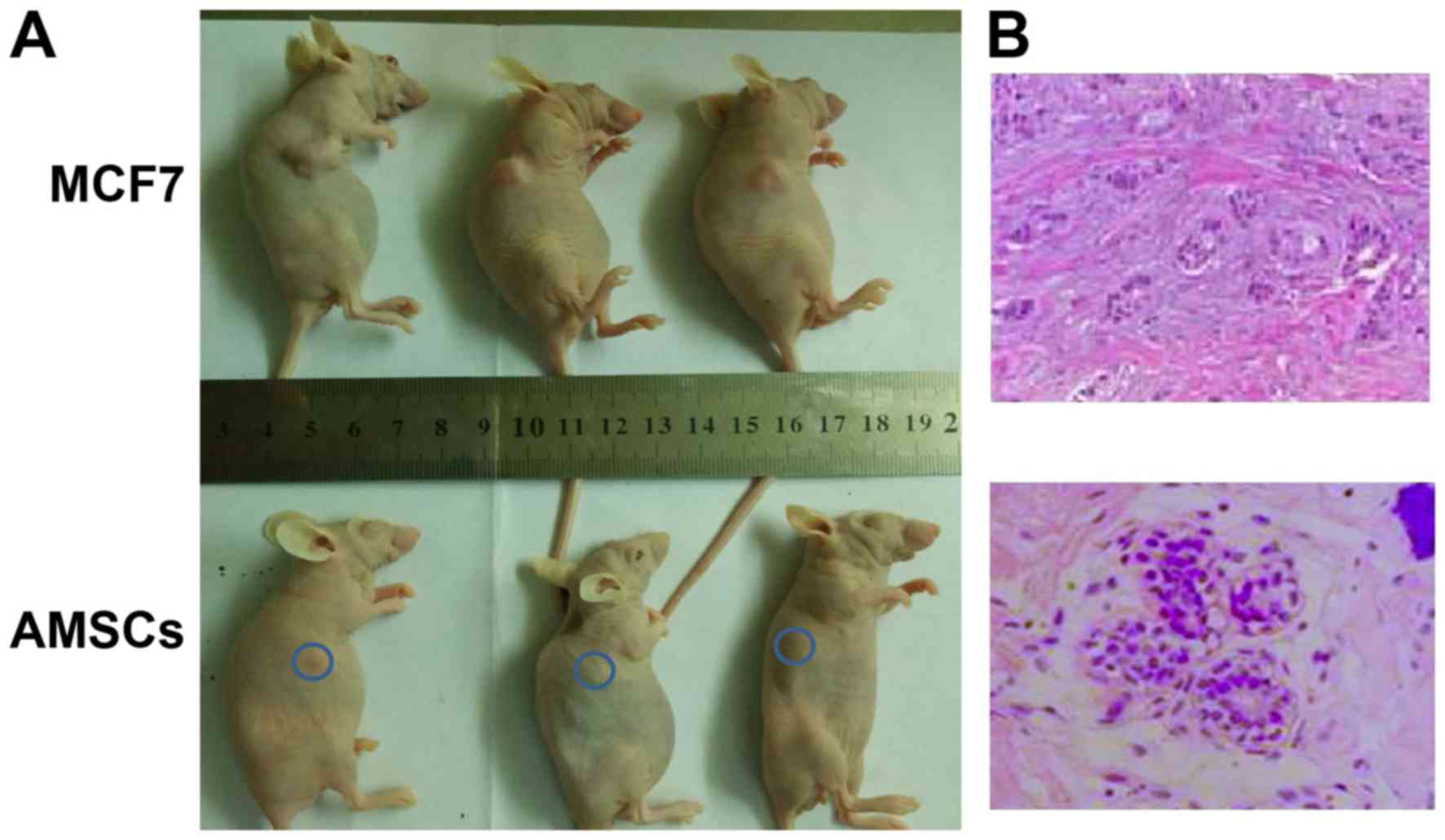
the isolated AMSCs, an in vivo analysis was conducted by
injecting the AMSCs into immunodeficient mice, then monitoring the
growth of tumors in vivo and histologically. As a control,
MCF-7 cells were used, which are breast cancer cells of low
malignancy that exhibit an early-stage cancer phenotype. In the
AMSC-injected mice, over the 30-day observation period, the lump
under the skin resulting from subcutaneous injection did not
increase in size, while injection with MCF-7 cells led to prominent
outgrowth following 14 days (Fig.
3A). Histological examination revealed that the subcutaneous
lump from AMSC injection contained duct-like structures composed of
an inner layer of cuboidal luminal epithelial cells with an outer
layer of squamous cells (Fig. 3B,
left). By contrast, the outgrowth from MCF-7 cells exhibited a
glandular-like arrangement of hyperplastic cells of various sizes
(Fig. 3B, right).
Discussion
In the present study, the phenotype of AMSCs was
characterized, AMSC gene expression profiles were compared with
breast cancer cells, and tumorigenic potential in vivo was
assessed. It was demonstrated that AMSCs exhibited the
characteristics of normal, non-malignant stem cells, in that they
presented the morphology of luminal epithelial cells in
non-differentiating culture, exhibited downregulated expression of
oncogenes and upregulated expression of tumor suppressor genes, and
formed normal mammary structures upon xenograft injection into nude
mice.
In 2006, two groups independently reported the
generation of functional mammary glands from a single AMSC isolated
from mice (14,15). In the same year, Proia and Kuperwasser
(16) published a detailed protocol
for reconstructing human mammary tissues in a mouse model. These
studies may contribute advancements towards the use of AMSCs for
breast reconstruction subsequent to mastectomy, as is typically
performed on female patients with breast cancer.
Multiple markers, individually or combined, have
been applied to isolate AMSCs (17).
In the current study, the results confirmed those of Gudjonsson
et al (18), who demonstrated
that there were generally two luminal epithelial cell populations:
A major population co-expressing MUC and ESA
(MUC+/ESA+), and a minor population positive
only for ESA (MUC−/ESA+).
MUC+/ESA+ cells are differentiated, acinar
and restricted to the luminal epithelium, without stem cell
properties. MUC−/ESA+ cells are suprabasally
localized in situ, retain the potential to differentiate
into MUC+/ESA+ luminal epithelial cells or
Thy-1+/α-smooth muscle actin+ myoepithelial
cells, and give rise to elaborate terminal ductal lobular units in
three-dimensional cultures (18).
Consistent with that study, it was identified that the
MUC−/ESA+ cells isolated by immunomagnetic
sorting exhibited the morphology of luminal epithelial cells in
non-differentiating culture in the present study.
Due to their inherent ability to differentiate into
a variety of cell types, AMSCs have become a key component in
regenerative medicine. Their longevity, however, also makes them
candidates for CSC generation, by increasing the likelihood that
they will acquire all the changes necessary for tumorigenesis. To
characterize the potential cancerous nature of AMSCs, two assays
were performed. Initially, the gene expression profiles of AMSCs
were compared with breast cancer cells isolated from the same
patients. From the 798 differentially expressed genes, ≥16 genes
were previously associated with breast cancer. Of these, genes with
tumor-inhibitory functions, including RB transcriptional
corepressor 1, PTEN, adrenoceptor β2, cyclin-dependent
kinase inhibitor 2A, NFκB inhibitor α, calcium-activated chloride
channel-2, and mitochondrial superoxide dismutase, were present at
enhanced levels in AMSCs compared with cancer cells. By contrast,
several oncogenes, including ERBB2, MYC, RAS,
and growth hormone receptor were upregulated in cancer cells
relative to the AMSCs. This suggests that isolated AMSCs have not
undergone oncogenic transformation to the same extent as cancer
cells from the same source.
This conclusion is consistent with the concept of
field cancerization proposed by Slaughter et al (19) in 1953 when, through extensive
histological examinations, they identified histologically abnormal,
though not cancerous, tissue surrounding oral squamous cell
carcinoma. This phenomenon has also been observed in other human
cancers, including breast cancer (20). Therefore, although the tissues used
for AMSC isolation may be confirmed by pathological examination to
be tumor-free and exhibit a gene expression profile corresponding
to a normal phenotype, AMSCs may still contain oncogenic changes
that are not detected by microarray analysis which, nonetheless,
indicate the early events of carcinogenesis. To address this
possibility, these cells were further characterized with functional
assays.
An in vivo tumorigenesis assay was employed
to assess the AMSCs, since this is considered the gold standard for
the analysis of normal cells and CSCs. When compared with the
low-malignancy MCF-7 cells, even a high number of injected AMSCs
(2×106) did not form any tumors; however, over a 12-week
observation period, the AMSCs formed ductal structures containing
luminal epithelial and myoepithelial cells resembling normal
mammary parenchyma. By contrast, the MCF-7 cells readily formed
well-differentiated tumors with glandular structures. This data
further corroborated the multipotency of AMSCs; although they
exhibited a luminal epithelial phenotype in vitro, they
differentiated into luminal and myoepithelial cells in
vivo.
Although microarray analysis revealed marked
differences between AMSCs and autologous breast cancer cells, it is
desirable to compare the gene expression profile between AMSCs and
autologous CSCs. A recent study has demonstrated that CSCs share
major somatic mutations with bulk cancer cells (21); it would be interesting and potentially
more accurate for future studies to compare the genetic mutations
between AMSCs and autologous CSCs or cancer cells for any
underlying carcinogenic potential of the AMSCs.
Besides the potential stem cells residing in the
mammary epithelial compartment, the mesenchymal compartment within
or outside the mammary gland may also contain stem cells that can
be transdifferentiated into mammary epithelial components. Using
the Cre-LoxP recombination system, Morroni et al (22) observed a reversible
transdifferentiation between mammary adipocytes and epithelium in
adult mice during pregnancy and lactation. Furthermore, De Matteis
et al (23) observed that
white adipose tissue from the dorso-lumbar fat depot in the fourth
right mammary glands of Rosa26 mice (males or virgin females) gave
rise to epithelial lobuloalveolar glands that tested positive for
immunocytochemical markers of milk-secreting glandular cells,
following implantation under the capsule of the fourth right
mammary gland of pregnant and lactating wild-type females.
Compared with mesenchymal stem or precursor cells,
mammary epithelial stem cells are more readily differentiated to
other epithelial components. However, their ability to
differentiate into stromal components, which are important for
mammary structure and function, may be limited (24). Consistently, the present study
demonstrated that isolated AMSCs primarily formed epithelial ductal
structures, which were distinct from the mesenchymal compartment,
suggesting that the further characterization of mesenchymal stem
cells may have great significance with regard to constructing a
more functionally intact mammary gland.
In summary, in the present study preliminary
evidence that AMSCs isolated from normal tissue adjacent to breast
cancer have the phenotypes of normal stem cells, rather than CSCs,
is provided. Therefore, AMSCs are a potential cell source for
autologous breast reconstruction.
Acknowledgements
The authors would like to thank Medjaden Bioscience
Limited (Hong Kong, China) for assisting in the preparation of this
manuscript. This study was supported by National Natural Science
Foundation of China (grant no. 30300336), the Jilin Provincial
Science and Technology Department of China (grant no. 200705134)
and Bethune Project B of Jilin University (grant no. 2012217).
References
|
1
|
Thomas-MacLean R: Beyond dichotomies of
health and illness: Life after breast cancer. Nurs Inq. 12:200–209.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Roje Z, Roje Z, Janković S and Ninković M:
Breast reconstruction after mastectomy. Coll Antropol. 34 Suppl
1:S113–S123. 2010.
|
|
3
|
Rozen WM, Rajkomar AK, Anavekar NS and
Ashton MW: Post-mastectomy breast reconstruction: A history in
evolution. Clin Breast Cancer. 9:145–154. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Djohan R, Gage E and Bernard S: Breast
reconstruction options following mastectomy. Cleve Clin J Med. 75
Suppl 1:S17–S23. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Techanukul T and Lohsiriwat V: Stem cell
and tissue engineering in breast reconstruction. Gland Surg.
3:55–61. 2014.PubMed/NCBI
|
|
6
|
Holland MS and Holland RE: The cellular
perspective on mammary gland development: Stem/progenitor cells and
beyond. J Dairy Sci. 88 Suppl 1:E1–E8. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Deome KB, Faulkin LJ Jr, Bern HA and Blair
PB: Development of mammary tumors from hyperplastic alveolar
nodules transplanted into gland-free mammary fat pads of female C3H
mice. Cancer Res. 19:515–520. 1959.PubMed/NCBI
|
|
8
|
Ercan C, van Diest PJ and Vooijs M:
Mammary development and breast cancer: The role of stem cells. Curr
Mol Med. 11:270–285. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gupta PB, Chaffer CL and Weinberg RA:
Cancer stem cells: Mirage or reality? Nat Med. 15:1010–1012. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li L and Neaves WB: Normal stem cells and
cancer stem cells: The niche matters. Cancer Res. 66:4553–4557.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen X, Liu Q and Song E: Mammary stem
cells: Angels or demons in mammary gland? Signal Transduction
Targeted Therapy. 2:160382017. View Article : Google Scholar
|
|
12
|
Kao CY, Nomata K, Oakley CS, Welsch CW and
Chang CC: Two types of normal human breast epithelial cells derived
from reduction mammoplasty: Phenotypic characterization and
response to SV40 transfection. Carcinogenesis. 16:531–538. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ponti D, Zaffaroni N, Capelli C and
Daidone MG: Breast cancer stem cells: An overview. Eur J Cancer.
42:1219–1224. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shackleton M, Vaillant F, Simpson KJ,
Stingl J, Smyth GK, Asselin-Labat ML, Wu L, Lindeman GJ and
Visvader JE: Generation of a functional mammary gland from a single
stem cell. Nature. 439:84–88. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stingl J, Eirew P, Ricketson I, Shackleton
M, Vaillant F, Choi D, Li HI and Eaves CJ: Purification and unique
properties of mammary epithelial stem cells. Nature. 439:993–997.
2006.PubMed/NCBI
|
|
16
|
Proia DA and Kuperwasser C: Reconstruction
of human mammary tissues in a mouse model. Nat Protoc. 1:206–214.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kalirai H and Clarke RB: Human breast
epithelial stem cells and their regulation. J Pathol. 208:7–16.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gudjonsson T, Villadsen R, Nielsen HL,
Ronnov-Jessen L, Bissell MJ and Petersen OW: Isolation,
immortalization, and characterization of a human breast epithelial
cell line with stem cell properties. Genes Dev. 16:693–706. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Slaughter DP, Southwick HW and Smejkal W:
Field cancerization in oral stratified squamous epithelium;
clinical implications of multicentric origin. Cancer. 6:963–968.
1953. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Försti A, Louhelainen J, Söderberg M,
Wijkström H and Hemminki K: Loss of heterozygosity in
tumour-adjacent normal tissue of breast and bladder cancer. Eur J
Cancer. 37:1372–1380. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Klevebring D, Rosin G, Ma R, Lindberg J,
Czene K, Kere J, Fredriksson I, Bergh J and Hartman J: Sequencing
of breast cancer stem cell populations indicates a dynamic
conversion between differentiation states in vivo. Breast Cancer
Res. 16:R722014. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Morroni M, Giordano A, Zingaretti MC,
Boiani R, De Matteis R, Kahn BB, Nisoli E, Tonello C, Pisoschi C,
Luchetti MM, et al: Reversible transdifferentiation of secretory
epithelial cells into adipocytes in the mammary gland. Proc Natl
Acad Sci USA. 101:pp. 16801–16806. 2004; View Article : Google Scholar : PubMed/NCBI
|
|
23
|
De Matteis R, Zingaretti MC, Murano I,
Vitali A, Frontini A, Giannulis I, Barbatelli G, Marcucci F,
Bordicchia M, Sarzani R, et al: In vivo physiological
transdifferentiation of adult adipose cells. Stem Cells.
27:2761–2768. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mammary stem cells and the differentiation
hierarchy: Current status and perspectives. Genes Dev.
28:1143–1158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Burkhart DL and Sage J: Cellular
mechanisms of tumour suppression by the retinoblastoma gene. Nat
Rev Cancer. 8:671–682. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Blanco-Aparicio C, Renner O, Leal JF and
Carnero A: PTEN, more than the AKT pathway. Carcinogenesis.
28:1379–1386. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Luthy IA, Bruzzone A, Piñero CP, Castillo
LF, Chiesa IJ, Vázquez SM and Sarappa MG: Adrenoceptors: Non
conventional target for breast cancer? Curr Med Chem. 16:1850–1862.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Caldon CE, Daly RJ, Sutherland RL and
Musgrove EA: Cell cycle control in breast cancer cells. J Cell
Biochem. 97:261–274. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Walia V, Ding M, Kumar S, Nie D, Premkumar
LS and Elble RC: hCLCA2 Is a p53-Inducible Inhibitor of Breast
Cancer Cell Proliferation. Cancer Res. 69:6624–6632. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hitchler MJ, Oberley LW and Domann FE:
Epigenetic silencing of SOD2 by histone modifications in human
breast cancer cells. Free Radic Biol Med. 45:1573–1580. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pratt MA, Tibbo E, Robertson SJ, Jansson
D, Hurst K, Perez-Iratxeta C, Lau R and Niu MY: The canonical
NF-kappaB pathway is required for formation of luminal mammary
neoplasias and is activated in the mammary progenitor population.
Oncogene. 28:2710–2722. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Margheri F, Serrati S, Lapucci A,
Anastasia C, Giusti B, Pucci M, Torre E, Bianchini F, Calorini L,
Albini A, et al: Systemic sclerosis-endothelial cell antiangiogenic
pentraxin 3 and matrix metalloprotease 12 control human breast
cancer tumor vascularization and development in mice. Neoplasia.
11:1106–1115. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pontier SM and Muller WJ: Integrins in
mammary-stem-cell biology and breast-cancer progression-a role in
cancer stem cells? J Cell Sci. 122:207–214. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jin L, Yuan RQ, Fuchs A, Yao Y, Joseph A,
Schwall R, Schnitt SJ, Guida A, Hastings HM, Andres J, et al:
Expression of interleukin-1beta in human breast carcinoma. Cancer.
80:421–434. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Freudenberg JA, Wang Q, Katsumata M,
Drebin J, Nagatomo I and Greene MI: The role of HER2 in early
breast cancer metastasis and the origins of resistance to
HER2-targeted therapies. Exp Mol Pathol. 87:1–11. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Moon A: Differential functions of Ras for
malignant phenotypic conversion. Arch Pharm Res. 29:113–122. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jamerson MH, Johnson MD and Dickson RB: Of
mice and Myc: c-Myc and mammary tumorigenesis. J Mammary Gland Biol
Neoplasia. 9:27–37. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stoelzle T, Schwarb P, Trumpp A and Hynes
NE: c-Myc affects mRNA translation, cell proliferation and
progenitor cell function in the mammary gland. BMC Biol. 7:632009.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
van Garderen E and Schalken JA:
Morphogenic and tumorigenic potentials of the mammary growth
hormone/growth hormone receptor system. Mol Cell Endocrinol.
197:153–165. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lottin S, Adriaenssens E, Dupressoir T,
Berteaux N, Montpellier C, Coll J, Dugimont T and Curgy JJ:
Overexpression of an ectopic H19 gene enhances the tumorigenic
properties of breast cancer cells. Carcinogenesis. 23:1885–1895.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hirohata S, Wang LW, Miyagi M, Yan L,
Seldin MF, Keene DR, Crabb JW and Apte SS: Punctin, a novel
ADAMTS-like molecule, ADAMTSL-1, in extracellular matrix. J Biol
Chem. 277:12182–12189. 2002. View Article : Google Scholar : PubMed/NCBI
|