Introduction
Head and neck squamous cell carcinoma (HNSCC) is a
common type of cancer and represents ~6% of all diagnosed cases of
cancer (1). Oral squamous cell
carcinoma (OSCC) is associated with a high rate of morbidity and
mortality, with few therapeutic options (2). The survival rate for patients with OSCC
has not improved in recent decades despite the development of novel
therapies (3). It has been reported
that >90% of cancer-associated mortalities are the result of
metastasis (4). Understanding
underlying molecular mechanisms involved in the metastasis of OSCC
is therefore a priority (4).
Zinc-finger protein 750 (ZNF750) is a putative C2H2
zinc finger protein and is typically expressed in keratinocytes
(5). It is an essential regulator of
epidermal differentiation; it binds and activates the epidermal
differentiation genes, including late cornified envelope 3A (LCE3A)
and small proline-rich protein 1A (SPRR1a), and represses epidermal
progenitor genes, including matrix metalloproteinase (MMP) 28
(6). Epidermal differentiation
involves the repression of progenitor genes, which participate in
cell proliferation and adhesion to the underlying basement membrane
(7). A loss in cell-cell adhesion and
an increase in cell motility are prerequisites for cancer
metastasis (7).
ZNF750-repressed genes have previously been enriched
for terms relevant to cell proliferation (6). Previous studies revealed that ZNF750 is
typically mutated or deleted in squamous cell carcinoma (8,9). The loss
of ZNF750 is associated with impaired differentiation and failure
to fully repress the proliferative genetic program, both of which
are key hallmarks of cancer (8).
ZNF750-driven epidermal differentiation occurs partially through
the induction of Kruppel-like factor 4 (KLF4), a transcription
factor that activates late epidermal differentiation-associated
genes and is critical for the epithelial-mesenchymal transition
(EMT) (10,11). Deletion of KLF4 has been reported to
be sufficient to initiate tongue carcinoma development (2).
At present, the biological function of ZNF750 in the
pathogenesis of OSCC is unknown. The present study aimed to
investigate the effect of overexpressed ZNF750 on cell viability,
invasion, migration and expression of EMT-associated genes in OSCC.
Clarification of the role of ZNF750 in OSCC may contribute to the
development of novel treatment strategies.
Materials and methods
Reagents
The lentiviral packaging plasmids psPax2, pRSV-Rev
and VSV-G were provided by Dr Padraig Strappe (Central Queensland
University, North Rockhampton, Australia). The pLVX-PGK-Puro
lentiviral vector backbone and the ZNF750 lentiviral vector
pLVX-hZNF750-PGK-Puro were purchased from Biowit Technologies
(Nanshan, China). Matrigel was obtained from BD Biosciences
(Franklin Lakes, NJ, USA).
Cell culture and treatment
The OSCC cell line CAL-27 and 293T packaging cell
line (purchased from the American Type Culture Collection,
Manassas, VA, USA) were cultured in Dulbecco's modified Eagle's
medium (DMEM) with 10% fetal bovine serum (FBS) (both from HyClone;
GE Healthcare, Chicago, IL, USA) and 1% penicillin/streptomycin
under standard cell culture conditions at 37°C with 5%
CO2. CAL-27 cells growing in the exponential phase were
randomly divided into the following groups: Control, PGK (negative
control, transduced with pLVX-PGK-Puro lentivirus) and ZNF750
groups (transduced with pLVX-hZNF750-PGK-Puro lentivirus).
Lentiviral packaging and CAL-27 cell
transduction
Lentiviral vector packaging and transduction was
performed as described previously with minor modifications
(12). For the generation of LV-PGK
and LV-ZNF750 lentivirus, 7 µg PGK or ZNF750 lentiviral vector
plasmid together with packaging plasmids (7 µg psPax2, 3 µg
pRSV-Rev and 3 µg VSV-G) were co-transfected into 70–80% confluent
293T cells, using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Lipofectamine®
2000/DNA complexes were added to 293T cells in DMEM with 10% FBS
and 1% penicillin/streptomycin to which caffeine (4 mM) and sodium
butyrate (1 mM) were added to increase the lentiviral titer
(13). At 48 and 72 h
post-transfection, the virus particles present in the cell
supernatant were harvested and centrifuged at 5,000 × g for 30 min
to remove cell debris, then filtered through a Steriflip-HV 0.45 µm
polyvinylidene fluoride (PVDF) filter unit (EMD Millipore,
Billerica, MA, USA) and concentrated using PEG-it virus
precipitation solution (System Biosciences, Inc., Palo Alto, CA,
USA) to obtain virus particles. The CAL-27 cells were transduced
with the LV-PGK or LV-ZNF750 lentiviral vectors (at a multiplicity
of infection of 10) after the cells reached 60–70% confluence. The
cells were allowed to recover for 48 h and puromycin (2 µg/ml;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was added into the
culture medium for stable cell line selection.
Cell viability analysis
Each group of cells was seeded in a 96-well plate at
a density of 5×104 cells/well. Cell viability following
overexpression of ZNF750 was evaluated using a Cell Counting Kit-8
(CCK-8; Beyotime Institute of Biotechnology, Haimen, China)
according to the manufacturer's protocol. The counting reagent (50
µl) was added to each well and incubated for 2 h. The absorbance
was measured at 450 nm using a STAKMAX™ microplate reader
(Molecular Devices, LLC, Sunnyvale, CA, USA). Experiments were
performed in triplicate at least three times.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RT-qPCR was performed to examine expression of the
epidermal progenitor genes MMP28 and the epidermal differentiation
genes LCE3A and SPRR1a. According to the manufacturer's protocol,
total RNA from cells was extracted with TRIzol reagent (Thermo
Fisher Scientific, Inc.). RNA (1 µg) was converted to complementary
DNA (cDNA) using a PrimeScript® RT kit (Takara
Biotechnology Co., Ltd., Dalian, China), and SYBR® Green
qPCR amplifications were performed in an ABI 7500 Sequence
Detection system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) using AceQ qPCR SYBR® Green Master Mix (Vazyme,
Piscataway, NJ, USA). The thermocycling conditions for the PCR were
as follows: 95°C for 5 min followed by 40 cycles of 95°C for 10 sec
and 57°C for 30 sec. The primer sequences were as follows: MMP28
forward, 5′-GAGCGTTTCAGTGGGTGTC-3′ and reverse,
5′-CCATTTGTTACCTTGCTTTGC-3′; LCE3A forward,
5′-AGCACAGTGTCTGCCTCCA-3′ and reverse, 5′-GGCATCTGTGGTGACTCAGG-3′;
SPRR1a forward, 5′-AGCAGCAGCAGGTGAAACA-3′ and reverse,
5′-GCTGGAGTGACCGTTGAAG-3′; GAPDH forward,
5′-TGCACCACCAACTGCTTAGC-3′ and reverse,
5′-GGCATGGACTGTGGTCATGAG-3′. The fold change amplification for
MMP28, LCE3A and SPRR1a was normalized to the GAPDH housekeeping
gene using the 2−ΔΔCq method (14). Each experiment was evaluated with
three PCR reactions and each experiment was repeated three
times.
Western blotting
Protein was extracted using radioimmunoprecipitation
lysis buffer, including phenylmethylsulfonate fluoride, and the
concentration was measured using a BCA protein assay kit (all from
Beyotime Institute of Biotechnology). An equal amount of protein
(15 µg) from each group were separated on a 10% SDS-PAGE gel and
transferred onto PVDF membranes (EMD Millipore). The membranes were
blocked using Tris-buffered saline with Tween-20 (20 mM Tris-HCl,
pH 7.5, 150 mM NaCl, 0.05% Tween-20) with 5% non-fat milk at 37°C
for 1 h and probed with primary antibodies: Mouse monoclonal
anti-cyclin B1 (cat no. 4135), rabbit polyclonal anti-neural
(N)-cadherin (cat no. 4061) (both from Cell Signaling Technology,
Inc., Danvers, MA, USA) and anti-KLF4 antibodies (cat no. ab106629;
Abcam, Cambridge, UK) at a dilution of 1:1,000 overnight at 4°C.
Following incubation with a goat anti-mouse or goat anti-rabbit
immunoglobulin G/horseradish peroxidase conjugated secondary
antibody (dilution, 1:1,000; cat no. AA128; Beyotime Biotechnology,
Jiangsu, China) at 37°C for 1 h, membranes were visualized using an
enhanced chemiluminescence reagent from Beyotime Institute of
Biotechnology. Densitometry analysis was performed using AlphaView
analysis software (Alphalmager® 2200; ProteinSimple;
Bio-Techne, Minneapolis, MN, USA).
Cell scratch, invasion and migration
assay
Cells were seeded on 6-well plates and grown to 80%
confluence. The cell monolayer was gently scraped with a sterile
200 µl pipette tip and the wells were washed twice with PBS to
remove the cell debris. The width of the scratch was determined by
images taken under light microscopy at 0 and 24 h after creating
the wound. The cells in three wells of each group were quantified.
Three independent experiments were performed.
For the cell invasion assay, Corning
Transwell® chambers with polycarbonate membrane (8 µm
pore size) were used to examine the effect of ZNF750 on CAL-27 cell
invasion. Matrigel (BD Biosciences) was used as the substrate for
invasion as previously described (15). Briefly, cells (1×105) were
dispersed onto Matrigel-coated polycarbonate membranes at a
dilution of 1:6 in serum-free culture medium in the upper chamber.
DMEM containing 10% FBS was added to the lower ch-amber. Invasion
was allowed to proceed for 24 h at 37°C in 5% CO2. Cells
remaining attached to the upper surfaces of the Matrigel-coated
polycarbonate membrane (non-invading cells) were carefully removed
with cotton swabs. Cells that had invaded to the lower surfaces of
the membrane were fixed with 4% formaldehyde for 15 min, stained
with 0.1% crystal violet for 10 min and visualized under a light
microscope (CKX71; Olympus Corporation, Tokyo, Japan). Assays were
performed in triplicate and the average number of invaded cells per
field was assessed using Image-Pro® Plus 6 software
(Media Cybernetics, Inc., Rockville, MD, USA). The cell migration
assay was performed with a similar protocol to the invasion assay,
but without a Matrigel coating.
Statistical analysis
Values are expressed as the mean ± standard
deviation. The data were analyzed by using a one-way analysis of
variance followed by a Student-Newman-Keuls-q test. P<0.05 was
considered to indicate a statistically significant difference.
Results
ZNF750 decreases CAL-27 cell
viability
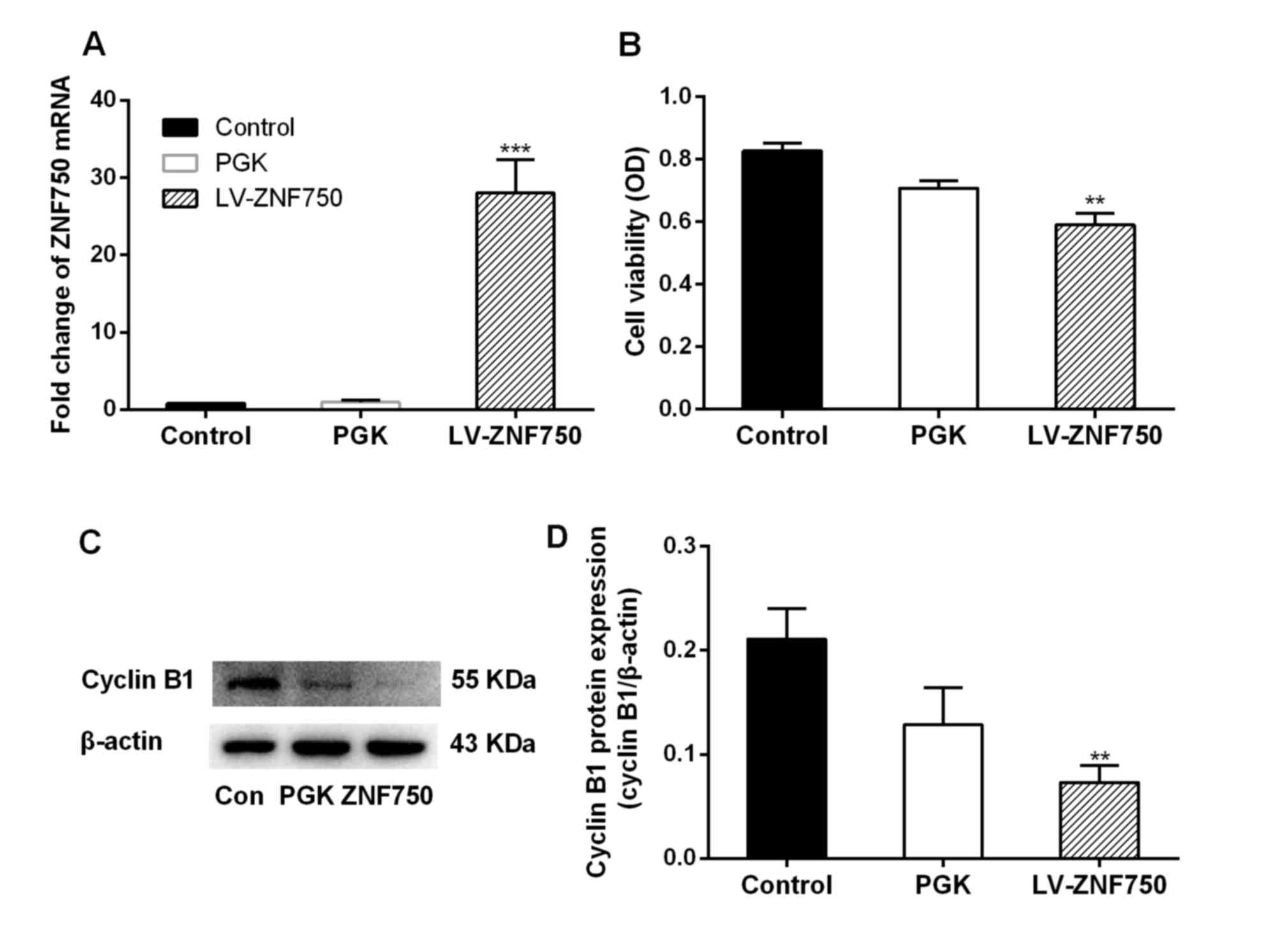
ZNF750 mRNA expression was significantly increased
(~28-fold) in the LV-ZNF750 group compared with the control and PGK
groups (P<0.001; Fig. 1A). In
addition, a significant decrease in cell viability and cyclin B1
expression was observed following ZNF750 overexpression compared
with the control groups (both P<0.01; Fig. 1B and C).
ZNF750 induces the expression of
differentiation-associated genes and reduces the expression of
progenitor genes in CAL-27 cells
Following ZNF750 overexpression, MMP28 was
significantly downregulated (~2-fold; P<0.001; Fig. 2A); however, the expression of the
differentiation genes LCE3A and SPRR1a was significantly increased
(~5- and 3-fold, respectively; P<0.001) compared with the
control groups as measured by RT-qPCR (Fig. 2A). The expression of the terminal
epidermal differentiation driver KLF4 was significantly increased
at the mRNA and protein level following ZNF750 overexpression
compared with the control groups (P<0.001 and P<0.01,
respectively; Fig. 2). There was a
2-fold increase in KLF4 protein expression in the ZNF750
overexpression group compared with the control groups (Fig. 2B).
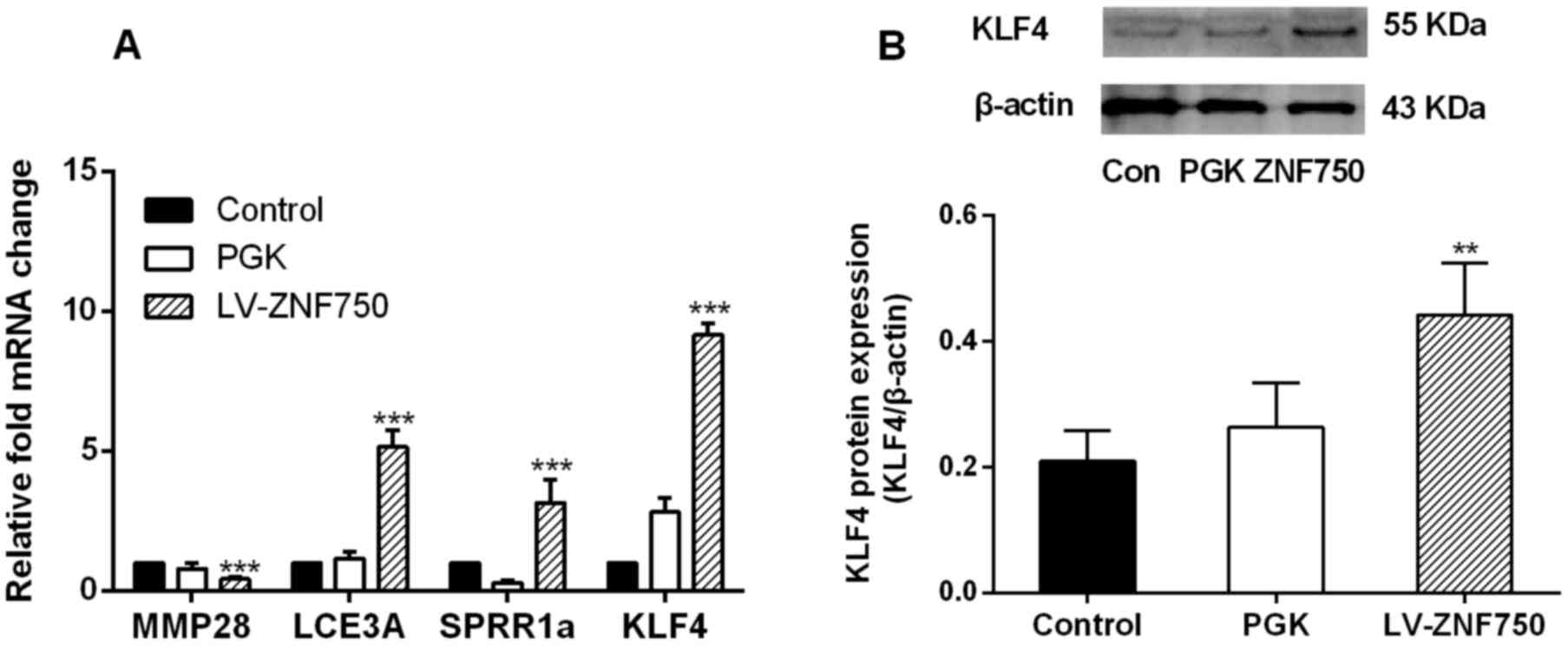 | Figure 2.ZNF750 reduces the expression of the
progenitor gene MMP28 but reduces the expression of
differentiation-associated genes. (A) mRNA expression of MMP28,
LCE3A, SPRR1a and KLF4 and (B) protein expression of KLF4 following
overexpression of ZNF750. **P<0.01, ***P<0.001 compared with
the control group. All experiments were performed in triplicate at
least three times. ZNF750, zinc-finger protein 750; MMP28, matrix
metalloproteinase; LCE3A, late cornified envelope 3A; SPRR1a, small
proline rich protein 1A; KLF4, Kruppel-like factor 4. PGK, negative
control transduced with pLVX-PGK-Puro lentivirus; LV-ZNF750,
transduced with pLVX-hZNF750-PGK-Puro lentivirus. |
ZNF750 overexpression inhibits cell
migration and invasion
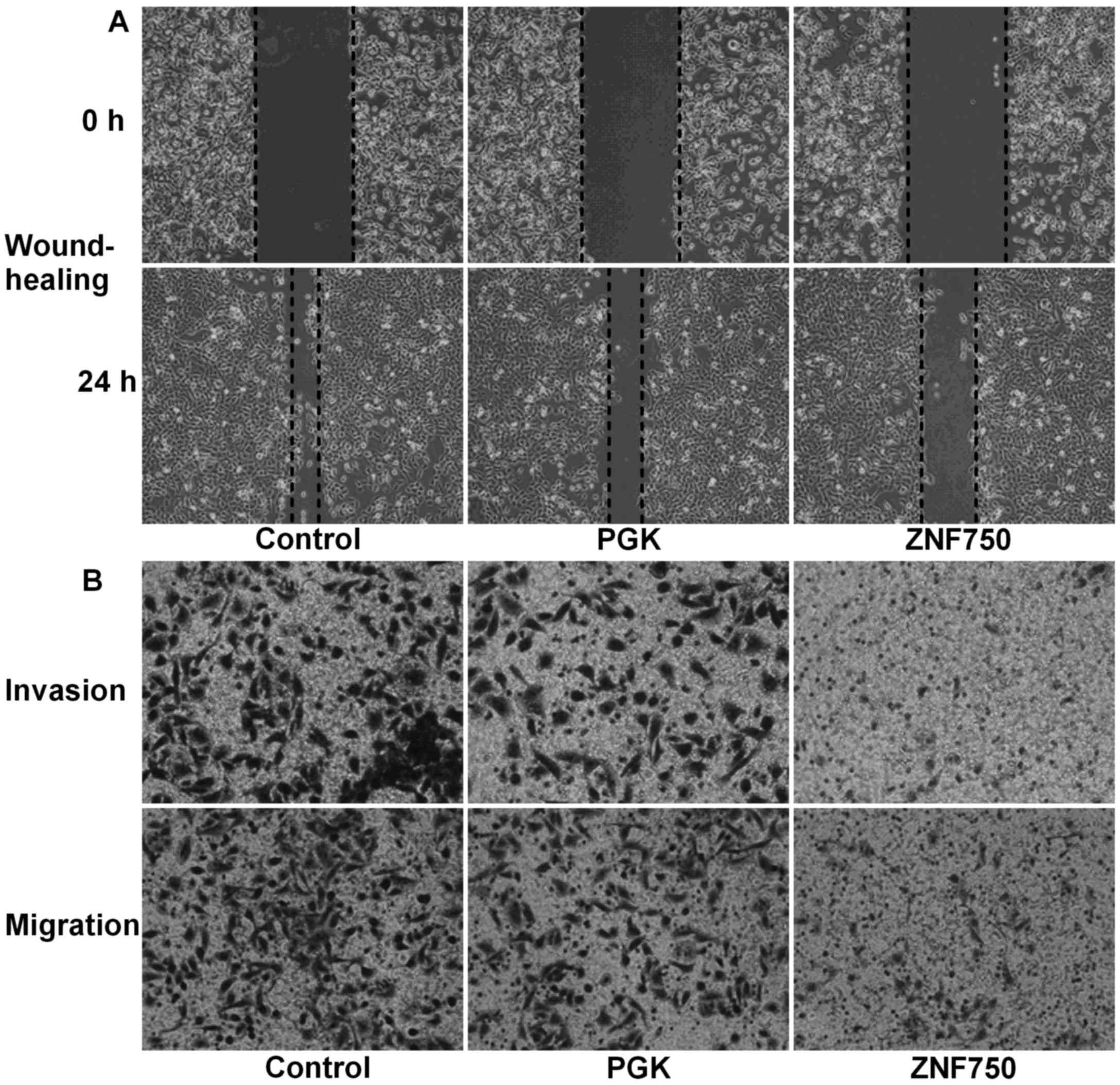
In order to estimate the metastatic ability of
CAL-27 cells, cell invasion and migration were analyzed in addition
to the expression of N-cadherin. A wound healing assay demonstrated
that the migration distance covered by cells overexpressing ZNF750
was markedly decreased at 24 h (cells migrated only 25–30% into the
scratch) compared with that covered by cells in the control and PGK
groups, where the cells had migrated 75–85% into the scratch
(Fig. 3A). In addition, invasion and
migration assays demonstrated that the number of cells that had
migrated to the lower chamber in the ZNF750 group was markedly
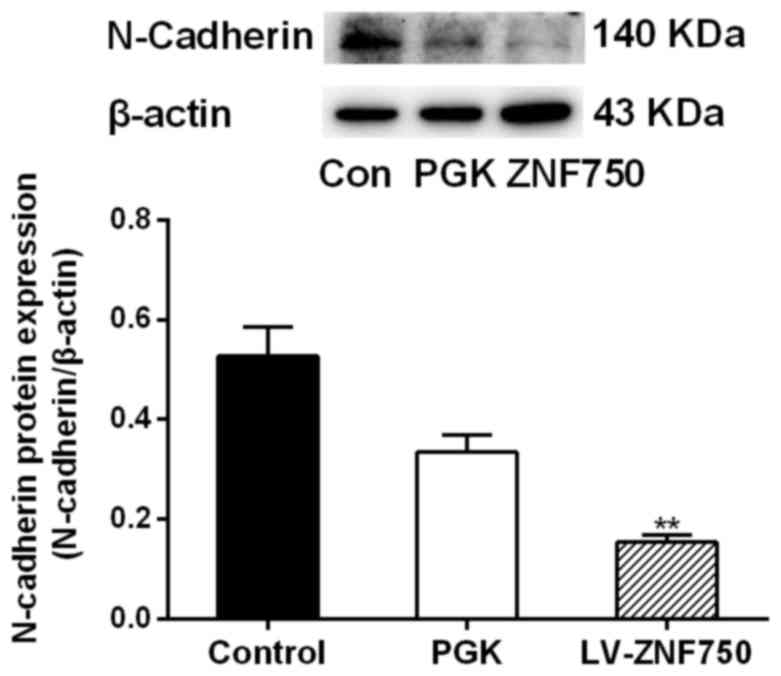
decreased compared with that of the control groups (Fig. 3B). Additionally, N-cadherin protein
expression was significantly decreased in the ZNF750 group compared
with the control group (P<0.01; Fig.
4).
Discussion
ZNF750 is a gene that typically resides in a focal
deletion in the HNSCC genome (16).
Despite previous studies reporting the role of ZNF750 in epidermal
differentiation (7,10,17), the
role of ZNF750 in OSCC has not yet been documented. To understand
the potential anticancer effects of ZNF750 on OSCC, the role of
ZNF750 in cell proliferation, differentiation, invasion and
migration was investigated. The data from the present study
indicated that ZNF750 is a tumor suppressor gene in OSCC. ZNF750
overexpression inhibited CAL-27 invasion and migration, suggesting
that ZNF750 may inhibit cell metastasis during cancer
progression.
A previous study indicated that normal expression of
ZNF750 leads to reduced expression of the progenitor gene MMP28,
and increased expression of the differentiation genes LCE3A and
SPRR1a mRNA in primary human undifferentiated keratinocytes
(7). Loss of ZNF750 has been reported
to result in impaired differentiation and enhanced proliferation of
cancer cells (8). ZNF750 interacts
with the terminal epidermal differentiation factor KLF4 to induce
the expression of differentiation-associated genes (7), and KLF4 deletion is sufficient to impair
squamous cell differentiation and initiate tongue carcinoma
development (2). In line with these
previous reports, the present study revealed that ZNF750
upregulates the differentiation genes LCE3A and SPRR1a, and
increases the expression of KLF4, while reducing the expression of
MMP28. Although KLF4 protein expression was not increased compared
with its mRNA expression, KLF4 protein expression may promote the
effects of ZNF750. The results from the present study demonstrated
that the overexpression of ZNF750 in CAL-27 cells results in
decreased cell viability and decreased expression of the cell cycle
regulator cyclin B1. The cyclin B1 gene is maximally active during
the G2/M and M phases of the cell cycle. Downregulation
of cyclin B1 induces G2/M cell cycle arrest (18). Increased G2/M arrest has
been associated with enhanced apoptosis (19). The present study suggested that ZNF750
may be important for the control of cell-cycle phase progression
and inhibition of cancer proliferation.
Data from the present study suggested that ZNF750
inhibits cancer cell metastasis. A wound healing assay demonstrated
that the migratory capacity of CAL-27 cell was inhibited by ZNF750,
confirming the anti-metastatic characteristics of ZNF750. In the
control group, cells had migrated into 75–85% of the scratch at 24
h, while the ZNF750 overexpressing cells, migrated only 25–30% into
the scratch following 24 h incubation. This assay may reflect the
inhibited migratory activity of ZNF750-overexpressing cells. In
addition, ZNF750 overexpression resulted in markedly decreased
numbers of invaded/migrated cells compared with the control groups.
ZNF750 overexpression resulted in decreased expression of the
mesenchymal marker N-cadherin. N-cadherin expression is associated
with the EMT phenotype and with enhanced invasion in cancer
(20). N-cadherin serves an important
role in the malignant behaviors of HNSCC (21). EMT is a crucial process involved in
the initiation and progression of metastasis, regulating the
detachment of cancer cells from the epithelium and facilitating
their invasion into stromal tissues (22). A major hallmark of the EMT process is
the loss or reduction of epithelial markers, including E-cadherin,
accompanied by the gain of mesenchymal markers, including
N-cadherin, a phenomenon known as ‘cadherin switching’ leading to
the loss of cell-to-cell adhesion, promotion of invasion, and
subsequent migration to distant regions (20). In the present study, N-cadherin
protein expression was associated with the malignant behavior of
OSCC cells. N-cadherin expression was decreased in ZNF750 groups
compared with the control groups, suggesting that ZNF750 restrains
the invasive potency of CAL-27 cells. The data from the present
study suggest that ZNF750 inhibits the metastatic properties of
CAL-27 cells.
In conclusion, the data from the present study
indicate that ZNF750 induces anticancer/anti-metastatic effects by
promoting cell differentiation-associated gene expression, and by
inhibiting cell progenitor gene expression and the metastatic
characteristics of CAL-27 cells. These findings highlight the role
of ZNF750 as a tumor suppressor gene for OSCC. Further studies are
required to investigate the potential of ZNF750 as a therapeutic
target for cancer treatment.
Acknowledgements
The present study was supported by a grant from the
Shandong Province Natural Science Foundation, China (grant nos.
ZR2015HL093, ZR2013HL015), the National Natural Science Foundation
of China (grant nos. 81272958, 81472530 and 81471048), and Shandong
Province Higher Educational Science and Technology Program (grant
no. J12LK01).
References
|
1
|
Sivanantham B, Sethuraman S and Krishnan
UM: Combinatorial effects of curcumin with an anti-neoplastic agent
on head and neck squamous cell carcinoma through the regulation of
EGFR-ERK1/2 and apoptotic signaling pathways. ACS Comb Sci.
18:22–35. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Abrigo M, Alvarez R, Paparella ML, Calb
DE, Bal de Kier Joffe E, Gutkind JS and Raimondi AR: Impairing
squamous differentiation by Klf4 deletion is sufficient to initiate
tongue carcinoma development upon K-Ras activation in mice.
Carcinogenesis. 35:662–669. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fribley AM, Miller JR, Brownell AL,
Garshott DM, Zeng Q, Reist TE, Narula N, Cai P, Xi Y, Callaghan MU,
et al: Celastrol induces unfolded protein response-dependent cell
death in head and neck cancer. Exper Cell Res. 330:412–422. 2015.
View Article : Google Scholar
|
|
4
|
Marx V: Tracking metastasis and tricking
cancer. Nature. 494:133–136. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Birnbaum RY, Zvulunov A, Hallel-Halevy D,
Cagnano E, Finer G, Ofir R, Geiger D, Silberstein E, Feferman Y and
Birk OS: Seborrhea-like dermatitis with psoriasiform elements
caused by a mutation in ZNF750, encoding a putative C2H2 zinc
finger protein. Nat Genet. 38:749–751. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Boxer LD, Barajas B, Tao BS, Zhang J and
Khavari PA: ZNF750 interacts with KLF4 and RCOR1, KDM1A, and
CTBP1/2 chromatin regulators to repress epidermal progenitor genes
and induce differentiation genes. Genes Dev. 28:2013–2026. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen IC, Chiang WF, Huang HH, Chen PF,
Shen YY and Chiang HC: Role of SIRT1 in regulation of
epithelial-to-mesenchymal transition in oral squamous cell
carcinoma metastasis. Mol Cancer. 13:1–19. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lin DC, Hao JJ, Nagata Y, Xu L, Shang L,
Meng X, Sato Y, Okuno Y, Varela AM, Ding LW, et al: Genomic and
molecular characterization of esophageal squamous cell carcinoma.
Nat Genet. 46:467–473. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang L, Zhou Y, Cheng C, Cui H, Cheng L,
Kong P, Wang J, Li Y, Chen W, Song B, et al: Genomic analyses
reveal mutational signatures and frequently altered genes in
esophageal squamous cell carcinoma. Am J Hum Genet. 96:597–611.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sen GL, Boxer LD, Webster DE, Bussat RT,
Qu K, Zarnegar BJ, Johnston D, Siprashvili Z and Khavari PA: ZNF750
is a p63 target gene that induces KLF4 to drive terminal epidermal
differentiation. Dev Cell. 22:669–677. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tiwari N, Meyer-Schaller N, Arnold P,
Antoniadis H, Pachkov M, van Nimwegen E and Christofori G: Klf4 is
a transcriptional regulator of genes critical for EMT, including
Jnk1 (Mapk8). PLoS One. 8:e573292013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen H, Yang H, Xu C, Yue H, Xia P,
Strappe PM and Wang L, Pan L, Tang W, Chen S and Wang L: Gene
expression profiling of common signal transduction pathways
affected by rBMSCs/F92A-Cav1 in the lungs of rat with pulmonary
arterial hypertension. Biomed Pharmacother. 83:100–106. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ellis BL, Potts PR and Porteus MH:
Creating higher titer lentivirus with caffeine. Human Gene Ther.
22:93–100. 2011. View Article : Google Scholar
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen HY, Wang JM, Wang HY, Zhang YX, Liu
W, Pan L, Wang WH, Chen SF, Jin WG and Wang L: Effect of short
hairpin RNA-induced CXCR4 silence on ovarian cancer cell. Biomed
Pharmacother. 66:549–553. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lawrence MS, Stojanov P, Mermel CH,
Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander
ES and Getz G: Discovery and saturation analysis of cancer genes
across 21 tumour types. Nature. 505:495–501. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cohen I, Birnbaum RY, Leibson K, Taube R,
Sivan S and Birk OS: ZNF750 is expressed in differentiated
keratinocytes and regulates epidermal late differentiation genes.
PLoS One. 7:e426282012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mollah MI, Dong KP and Park HJ: Cordyceps
militaris grown on germinated soybean induces G2/M Cell cycle
arrest through downregulation of cyclin B1 and Cdc25c in human
colon cancer HT-29 cells. Evid Based Complement Alternat Med.
2012:2492172012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dipaola RS: To arrest or not to G2-M
cell-cycle arrest. Clin Cancer Res. 8:3512–3519. 2002.PubMed/NCBI
|
|
20
|
Zhang X, Liu G, Kang Y, Dong Z, Qian Q and
Ma X: N-cadherin expression is associated with acquisition of EMT
phenotype and with enhanced invasion in erlotinib-resistant lung
cancer cell lines. Plos One. 8:e576922012. View Article : Google Scholar
|
|
21
|
Nguyen PT, Kudo Y, Yoshida M, Kamata N,
Ogawa I and Takata T: N-cadherin expression is involved in
malignant behavior of head and neck cancer in relation to
epithelial-mesenchymal transition. Histol Histopathol. 26:147–156.
2011.PubMed/NCBI
|
|
22
|
Osborne LD, Li GZ, How T, O'Brien ET,
Blobe GC, Superfine R and Mythreye K: TGF-β regulates LARG and
GEF-H1 during EMT to affect stiffening response to force and cell
invasion. Mol Biol Cell. 25:3528–3540. 2014. View Article : Google Scholar : PubMed/NCBI
|