Introduction
Lung cancer is one of the most common types of
cancer and is the leading cause of cancer-associated mortality
worldwide, with an estimated of >200,000 new cases every year
(1). Non-small cell lung cancer
(NSCLC), the most common type of lung cancer, which accounts for
85% of all lung cancer cases, includes four subtypes:
Adenocarcinoma, squamous cell carcinoma, adenosquamous cell
carcinoma and large cell carcinoma (2). In China, the incidence of NSCLC is still
increasing in urban and rural areas (3). For NSCLC patients with early-stage or
locally advanced lung cancer, surgery is the most effective
treatment, followed by chemotherapy and radiotherapy. For stage
III/IV NSCLC, platinum-based combined chemotherapy is the current
standard of treatment (4,5). In recent years, great advancement has
been made in the management of NSCLC. However, the prognosis of
NSCLC remains unfavorable, with a 5-year overall survival rate of
~16% (6,7). Therefore, fully understanding of the
molecular mechanisms underlying NSCLC carcinogenesis and
progression is necessary for improving the diagnosis, prevention
and treatment of patients with NSCLC.
MicroRNAs (miRNAs) are a large family of small,
non-protein-coding and single-stranded RNA molecules that range
from 19 to 25 nucleotides in length (8). miRNAs have important roles in gene
expression through interacting with complementary nucleotide
sequences in the 3′ untranslated regions (3′UTRs) of target mRNAs,
leading to degradation or translational suppression of their target
mRNAs (9). A single miRNA can
regulate the expression of a wide variety of target genes, which
contain target binding sites that interact with miRNAs.
Additionally, a single mRNA can be regulated by a number of
different miRNAs (10). More than 60%
of human genes have been predicted to be regulated by miRNAs, and
therefore miRNAs are involved in various biological processes,
including development, cell proliferation, cell cycle, apoptosis,
differentiation, metastasis and resistance to chemotherapeutics
(11–13). An increasing number of studies
reported that miRNAs are frequently dysregulated in a large number
of types of human cancer and have important roles in the regulation
of carcinogenesis and disease progression (9,14). Certain
miRNA may act as an oncogene or a tumor suppressor gene, depending
on the roles of their target genes (15). Based on these findings, miRNAs may be
investigated as a therapeutic targets in cancer diagnosis,
prevention and therapy.
In the present study, the expression levels of
miR-301a in NSCLC tissues and cell lines were investigated, and its
roles on NSCLC cell proliferation, migration and invasion were
evaluated. The results of the present study will provide further
insights into the molecular mechanisms responsible for NSCLC
carcinogenesis and progression, and identify miR-301a as a
potential therapeutic target in NSCLC treatment.
Materials and methods
Clinical samples
A total of forty-two paired NSCLC tissues and
adjacent non-tumor lung tissues were obtained from patients (29
males, 13 females; age range, 47–79 years; mean age, 58 years)
receiving surgical treatment at The First Affiliated Hospital of
Zhengzhou University between February 2012 and August 2014. All
patients did not receive chemotherapy or radiotherapy prior to
surgery. All tissues were excised, immediately snap-frozen in
liquid nitrogen and then transferred to −80°C refrigerator until
use. The present study was approved by the Ethical Committee of The
First Affiliated Hospital of Zhengzhou University (Zhengzhou,
China), and informed consent was obtained from all patients.
Cell culture and transfection
A total of five NSCLC cell lines (NCI-H1975, H1299,
SPC-A-1, H520 and A549), a normal human bronchial epithelial cell
line (16HBE) and 293 were purchased from Institute of Biochemistry
and Cell Biology of the Chinese Academy of Sciences (Shanghai,
China). These cell lines were cultured in Dulbecco's modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) or RPMI 1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.), 2 µM glutamine (Gibco;
Thermo Fisher Scientific, Inc.), as well as 100 U/ml penicillin and
100 µg/ml streptomycin (Gibco; Thermo Fisher Scientific, Inc.). All
cells were kept in a humidified incubator at 37°C with 5%
CO2.
The miR-301a inhibitor, negative control (NC)
inhibitor, DLC1 small-interfering RNA targeting DLC1 (DLC1 siRNA)
and NC siRNA were purchased from Shanghai GenePharma Co., Ltd.,
(Shanghai, China). The DLC1 siRNA sequence was
5′-TACGTCCAAGGTCGGGCAGGAAGA-3′ and the NC siRNA sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. H1299 and A549 cells were seeded in
six-well plates to 70% confluence and transfected with miRNA
inhibitor (100 pmol) or siRNAs (100 pmol) using Lipofectamine 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. This assay did not use untransfected
cells. After transfection 48 h, RT-qPCR was performed to detect
miR-301a expression. CCK-8 and cell migration and invasion assays
were carried out at 24 and 48 h following transfection. Western
blotting analysis was carried out at 72 h following
transfection.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from tissues and cells (H1299 and A549)
was isolated using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocols. For
miR-301a expression, RT-qPCR was performed using the
SYBR® Green PCR kit (Toyobo Life Science, Osaka, Japan)
on an Applied Biosystems Realtime 7500 Sequence Detection system
(Applied Biosystems; Thermo Fisher Scientific Inc.). The primer
sequences for miR-301a and U6 were as follows: miR-301a forward,
5′-ACACTCCAGCTGGGCAGTGCAATAGTATTGTC-3′ and reverse,
5′-CTCAACTGGTGTCGTGGA-3′; and U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′. For DLC1 mRNA detection, reverse
transcription was performed using the PrimeScript RT reagent kit
(Takara Biotechnology Co., Ltd., Dalian, China) followed by qPCR
using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.). The
primer sequences were as follows: DLC1 forward
5′-CCGCCTGAGCATCTACGA-3′ and reverse, 5′-TTCTCCGACCACTGATTGACTA-3′;
β-actin forward, 5′-CCTGGCACCCAGCACAAT-3′ and reverse,
5′-GCTGATCCACATCTGCTGGAA-3′; miR-301a forward,
5′-ACACTCCAGCTGGGCAGTGCAATAGTATTGTC-3′ and reverse,
5′-CTCAACTGGTGTCGTGGA-3′; and U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′. miR-301a expression was normalized
to U6 expression, and β-actin was used as an internal control for
DLC1 mRNA expression. The relative expression was analyzed with the
2−∆∆Cq method (16).
Cell counting Kit-8 (CCK-8) assay
To evaluate the effects of miR-301a on the
proliferation of NSCLC cells, the CCK8 kit (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) was used. For this assay,
transfected cells were collected and seeded into 96-well plates at
an initial density of 3,000 cells/well. Cell proliferation was
detected every 24 h until 96 h according to the manufacturer's
protocol. At each time points, 10 µl CCK8 solution was added to
each well and cultured for 2 h at 37°C. Optical density (OD) was
measured at a wavelength of 450 nm using a microplate reader. Each
experiment was performed in three replicates and repeated three
times.
Cell migration and invasion assay
To assess the effects of miR-301a on migration and
invasion of NSCLC cells, cell migration and invasion assays were
performed using 24-well Transwell chambers (8 mm pore size; BD
Biosciences, Franklin Lakes, NJ, USA). For cell migration assays,
transfected cells were collected at 48 h post-transfection,
suspended into FBS-free culture medium and seeded in the upper
chambers at an initial density of 5×104 cells/well. The
lower chambers were filled with 500 µl DMEM containing 20% FBS as a
chemoattractant. Following incubation at 37°C for 48 h, the
non-migrated cells were carefully removed with a cotton swab. The
migrated cells were fixed with 95% methanol at room temperature for
15 min, stained with 0.1% crystal violet at room temperature for 15
min, and captured with the IX71 inverted microscope (Olympus
Corporation, Tokyo, Japan). Cell invasion assays were similar to
the cell migration assays, except that the Transwell chambers were
coated with Matrigel (BD Biosciences). Cell numbers were calculated
in five random fields for each Transwell chamber.
Western blot assay
Total proteins were isolated from transfected cells
using RIPA buffer containing a protease inhibitor cocktail (Roche,
Mannheim, Germany). Concentration of total proteins was quantified
with BCA Protein assay (Pierce; Thermo Fisher Scientific). Equal
amounts of protein were separated on a 10% SDS-PAGE and then
transferred to polyvinylidene fluoride membranes (PVDF; Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Non-specific binding sites
of PVDF membranes were saturated with 5% skimmed milk in TBS
solution containing 0.1% Tween 20 (TBST). Subsequently, the
membranes were probed with primary antibodies, mouse anti-human
monoclonal DLC1 antibody (1:1,000; catalog no. sc-271915; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA) and mouse anti-human
monoclonal GADPH antibody (1:1,000; catalog no. sc-166574; Santa
Cruz Biotechnology), at 4°C overnight. After three washes with
TBST, the membranes were incubated with corresponding horseradish
peroxidase (HRP)-conjugated secondary antibody (1:10,000; Santa
Cruz Biotechnology, Inc.) for 2 h at room temperature, washed with
TBST for three times and detected using an enhanced
chemiluminescence detection system (ECL; GE Healthcare, Chicago,
IL, USA). Relative protein levels of DLC1 were normalized to
GAPDH.
Bioinformatic analysis
To predict the potential targets of miR-301a,
bioinformatic analysis was performed using TargetScan version 6.2
http://www.targetscan.org/vert_71/).
Dual-luciferase reporter assays
The predicted and mutated sequences targeting the
3′UTR of DLC1 were both amplified and cloned into pmirGLO.
pmirGLO-DLC1-3′UTR Wt (wild-type) and pmirGLO-DLC1-3′UTR Mut
(mutated) were synthesized by Shanghai GenePharma Co., Ltd. 293
cells were seeded in 24-well plates at a density of
1×105 cells per well, and co-transfected with miR-301a
inhibitor (20 pmol) or NC inhibitor (20 pmol) and
pmirGLO-DLC1-3′UTR Wt (0.2 µg) or pmirGLO-DLC1-3′UTR Mut (0.2 µg)
by using Lipofectamine 2000. The transfected cells were harvested,
and the relative luciferase activities were determined using
Dual-Luciferase Reporter Assay system (Promega Corporation,
Madison, WI, USA) 48 h post-transfection. All experiments were
performed in triplicate and repeated three times. Renilla
luciferase activity was normalized to firefly luciferase
activity.
Statistical analysis
Results were presented as the mean ± standard
deviation. SPSS (version 13.0; SPSS Inc., Chicago, IL, USA) was
used for statistical analysis with Student's t-tests or one-way
analysis of variance (ANOVA) for comparisons of multiple groups.
SNK test was used as a post hoc test following ANOVA. P<0.05 was
considered statistically significant.
Results
miR-301a was upregulated in NSCLC
tissues and cell lines
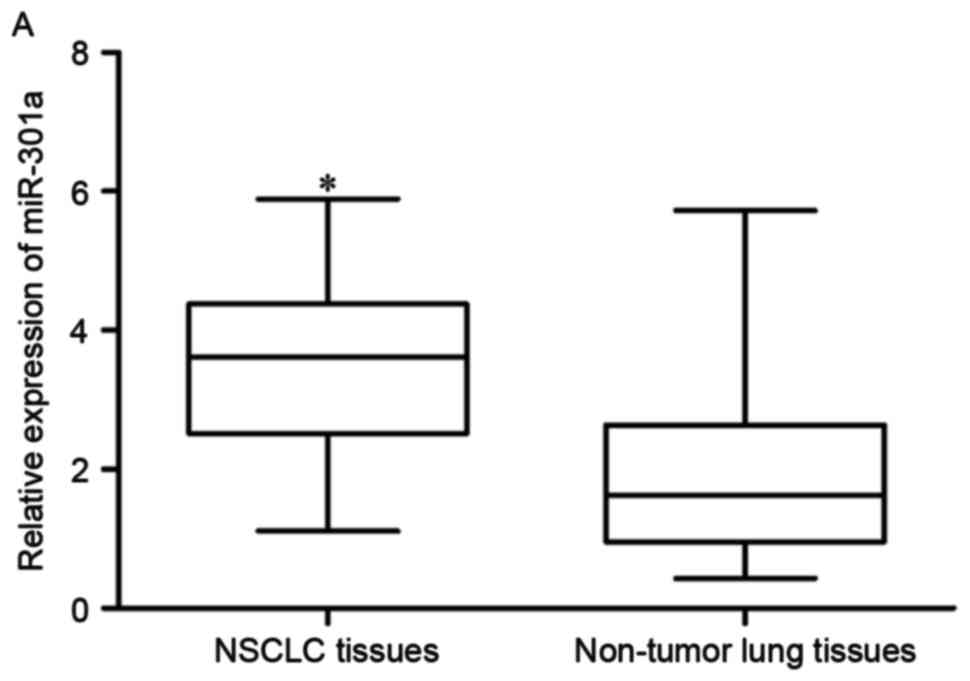
In order to investigate whether miR-301a was
differentially expressed in NSCLC, RT-qPCR was performed to analyze
miR-301a expression levels in NSCLC tissues and adjacent non-tumor
lung tissues. The results indicated that miR-301a was significantly
upregulated in NSCLC tissues compared with adjacent non-tumor lung
tissues (P<0.05; Fig. 1A).
miR-301a expression in NSCLC cell lines (NCI-H1975,
H1299, SPC-A-1, H520 and A549) and a normal human bronchial
epithelial cell line (16HBE) was also evaluated. Compared with
16HBE, all five NSCLC cell lines expressed higher levels of
miR-301a (P<0.05; Fig. 1B), which
was in accordance with the data from tissue samples (P<0.05;
Fig. 1A). The high expression levels
of miR-301a in NSCLC tissues and cell lines indicated that miR-301a
may act as an oncogene in NSCLC.
Downregulation of miR-301a inhibited
proliferation, migration and invasion of NSCLC cells
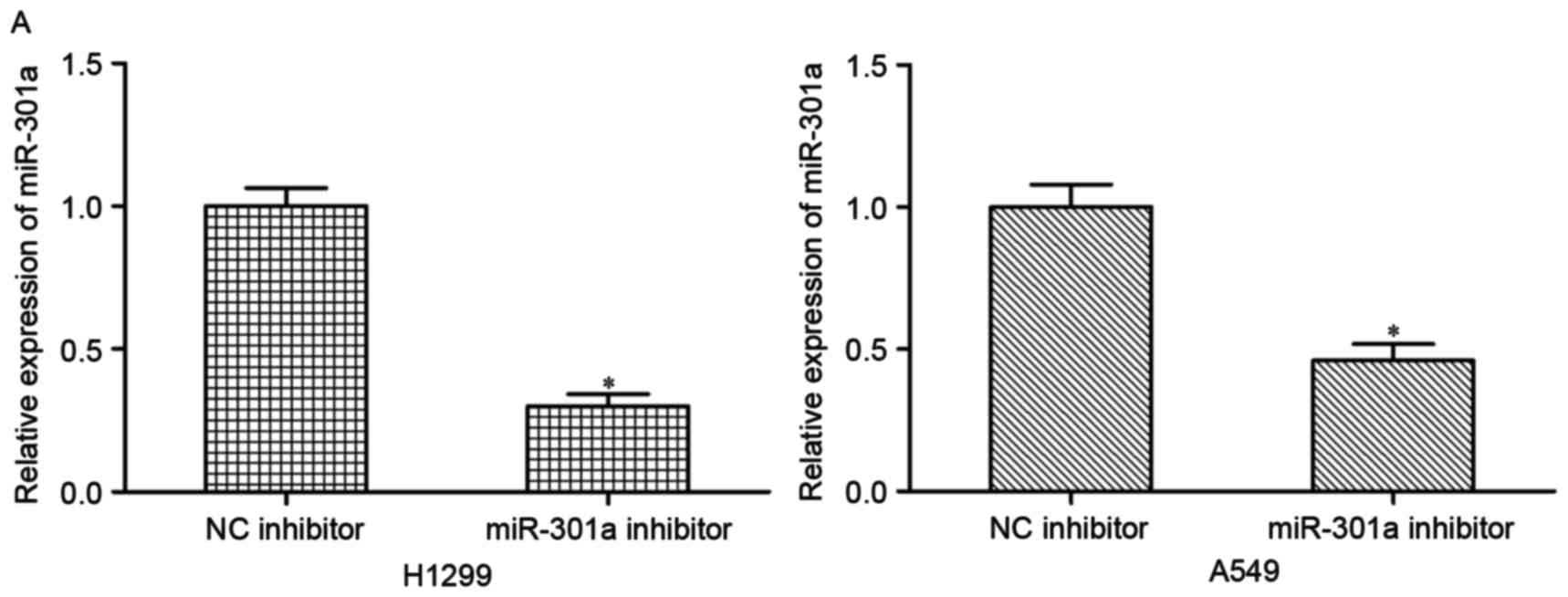
To investigate the effects of miR-301a on NSCLC
progression, H1299 and A549 cells were transfected with miR-301a
inhibitor or NC inhibitor. The decrease in the expression of
miR-301a in H1299 and A549 cells following transfection with
miR-301a inhibitor was confirmed by RT-qPCR (P<0.05; Fig. 2A).
Following transfection with miR-301a inhibitor or NC
inhibitor, a series of biological experiments were performed in
vitro, including the CCK-8 assay, cell migration and invasion
assays. CCK-8 assay results suggested that miR-301a downregulation
significantly inhibited proliferation of H1299 and A549 cells
compared with cells transfected with the NC inhibitor (P<0.05;
Fig. 2B,). In addition, the cell
migration assays showed that transfection of the miR-301a inhibitor
markedly decreased migratory ability of H1299 and A549 cells
(P<0.05; Fig. 2C). Furthermore,
cell invasion assays revealed that the invasive capacity of H1299
and A549 cells transfected with the miR-301a inhibitor was
significantly lower compared with the NC inhibitor groups
(P<0.05; Fig. 2D,). These data
demonstrated that downregulation of miR-301a significantly
inhibited the proliferation, migration and invasion of NSCLC
cells.
DLC1 is a target gene of miR-301a in
NSCLC
To investigate the molecular mechanisms by which
miR-301a downregulation inhibited proliferation, migration and
invasion of NSCLC cells, bioinformatic analysis was performed to
search for potential target genes of miR-301a using TargetScan 6.2.
TargetScan 6.2 analysis indicated that DLC1 is a major target of
miR-301a (Fig. 3A). DLC1 was chosen
for further confirmation because that DLC1 is aberrantly lowly
expressed in NSCLC and contributes to NSCLC formation and
progression (17–19).
To identify whether DLC1 was a target of miR-301a,
dual-luciferase reporter assays were performed. pmirGLO-DLC1-3′UTR
Wt or pmirGLO-DLC1-3′UTR Mut, and miR-301a inhibitor or NC
inhibitor were co-transfected into 293T cells. As indicated in
Fig. 3B, the knockdown of miR-301a
increased the luciferase activity compared with cells
co-transfected with pmirGLO-DLC1-3′UTR Wt and NC inhibitor. In the
cells transfected with pmirGLO-DLC1-3′UTR Mut, there was no
significance difference in luciferase activity between cells
transfected with NC inhibitor and miR-301a inhibitor
(P>0.05).
Subsequently, RT-qPCR and western blotting was
performed to investigate the ability of miR-301a to regulate the
expression of DLC1 at mRNA and protein levels, respectively. The
results indicated that miR-301a downregulation markedly increased
DLC1 expression in H1299 and A549 cells at mRNA (P<0.05;
Fig. 3C) and protein (P<0.05;
Fig. 3D) levels. Collectively, the
findings indicated that DLC1 is a direct target gene of miR-301a in
NSCLC.
Knockdown of DLC1 partly reverses the
effects of miR-301a downregulation on NSCLC cells
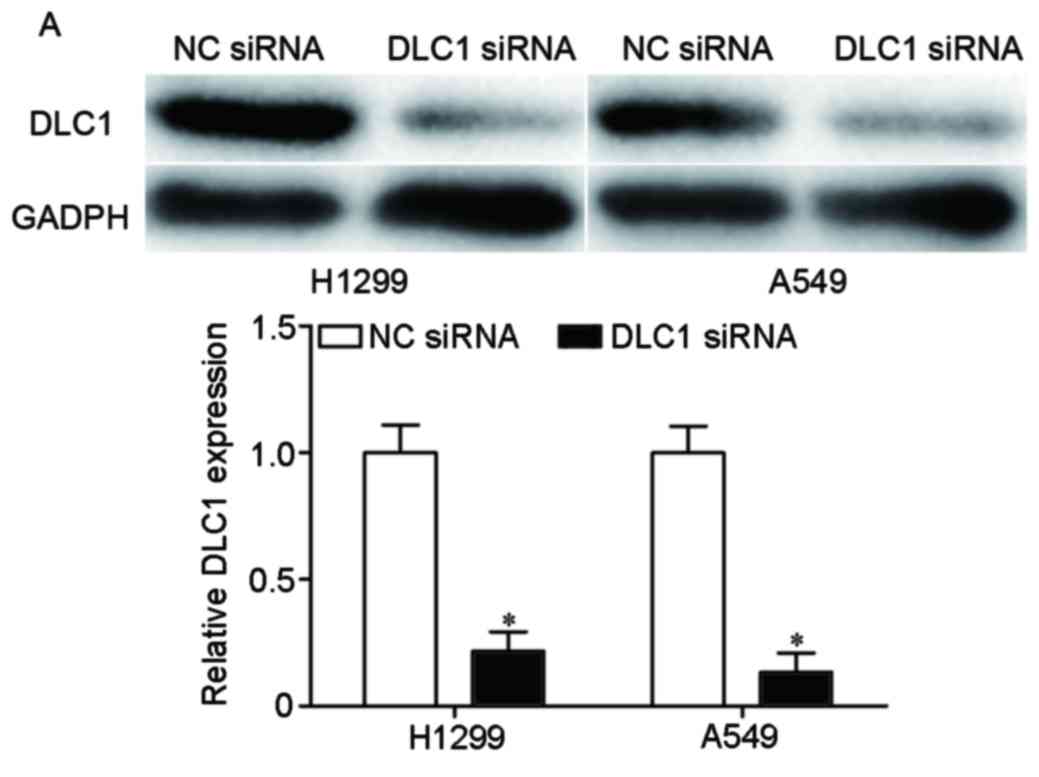
To confirm the roles of DLC1 in NSCLC, H1299 and
A549 cells were transfected with DLC1 siRNA or NC siRNA. The
downregulation of DLC1 in H1299 and A549 cells was determined by
western blotting (P<0.05; Fig.
4A).
Next, it was examined whether DLC1 knockdown was
able to rescue the carcinogenic effects of miR-301a in NSCLC. The
results indicated that downregulation of DLC1 was able to rescue
cell proliferation (P<0.05; Fig.
4B), migratory (P<0.05; Fig.
4C) and invasive (P<0.05; Fig.
4D) abilities of H1299 and A549 cells, which were previously
inhibited by miR-301a inhibitor. These findings further verified
that DLC1 is a direct target gene of miR-301a and that transfection
with miR-301a inhibitor inhibited proliferation, migration and
invasion of NSCLC cells, at least in part by inducing expression of
DLC1.
Discussion
NSCLC is one of the most common malignant diseases
with a high mortality rate (20).
NSCLC carcinogenesis is a multi-stage process, which involves a
broad spectrum of changes in gene expression and physiological
structure (21). The abnormal
expression of genes mainly includes activation of oncogenes and
inactivation of tumor suppressor genes (22). Accumulated evidence have suggested
that miRNA is involved in carcinogenesis as either oncogene or
tumor suppressor and the functions of many cancer-associated miRNAs
have been identified (23,24). Moreover, dysregulation of miRNAs
expression has been indicated to be potential sensitive and
accurate biomarkers for cancer diagnosis and prognosis of human
cancer (25,26). Therefore, understanding the roles of
miRNAs in NSCLC might provide valuable information for therapeutic
strategies in the therapy for patients with NSCLC. The present
study focused on the roles of miR-301a in NSCLC. The present study
investigated the expression and biological functions of miR-301a in
NSCLC as well as the potential mechanism by which miR-301a acts as
an oncogene in NSCLC initiation and progression. The findings
indicated that: i) miR-301a was significantly upregulated in NSCLC
tissues and cell lines compared with adjacent non-tumor lung
tissues and normal human bronchial epithelial cell line,
respectively; ii) miR-301a downregulation inhibited the
proliferation, migration and invasion of NSCLC cells; iii)
bioinformatic analysis showed that DLC1 is a putative target of
miR-301a; iv) miR-301a downregulation increased the expression
levels of DLC1 by directly targeting its 3′UTR; v) DLC1 knockdown
partially reverses the inhibition of proliferation, migration and
invasion induced by downregulation of miR-301a in NSCLC cells.
Previous studies have shown that miR-301a was
closely associated with multiple malignant phenotypes of human
cancer. miR-301a was reported to be upregulated in numerous types
of human cancer, including Ewing's sarcoma (27), laryngeal squamous cell carcinoma
(28), pancreatic cancer (29), colorectal cancer (30), gastric cancer (31) and hepatocellular carcinoma (32). Furthermore, the levels of miR-301a
expression were demonstrated to be correlated with
clinicopathological features in cancer patients. Xu et al
(31) reported that in gastric
cancer, miR-301a was associated with tumor size, invasion depth,
lymph node metastasis and tumor-node-metastasis (TNM) stage. Yu
et al (33) reported that the
expression level of miR-301a in triple-negative breast cancer was
positively correlated with tumor size, depth of invasion, TNM stage
and lymph node metastasis. In addition, multivariate analysis
suggested that miR-301a expression was an independent prognostic
factor for the survival of patients with triple-negative breast
cancer (33). Xia and colleagues
(29) showed that higher expression
of miR-301a was observed in pancreatic cancer patients with lymph
node metastasis and advanced pathological stages. miR-301a was also
identified as an independent prognostic factor for worse survival
(29). These findings suggested that
miR-301a was upregulated in various types of cancer and may act as
diagnostic and prognostic biomarkers.
Furthermore, studies have also revealed that
miR-301a is able to act as an oncogene in carcinogenesis and
progression of human cancer. For example, in Ewing's sarcoma,
knockdown of miR-301a suppressed proliferation and cell cycle
progression (27). Furthermore,
downregulation of miR-301a in Ewing's sarcoma cells significantly
suppressed tumor growth in vivo (27). Xia et al indicated that
upregulation of miR-301a promoted pancreatic cancer cell colony
formation, invasive and migratory abilities in vitro as well
as tumorigenicity in vivo (29). Fang et al found that miR-301a
acts as an oncogene in colorectal cancer by increasing
proliferation, migration and invasion as well as tumor growth
(30). In breast cancer, exogenous
expression of miR-301a expression increased cell migration,
invasion and metastasis both in vitro and in vivo
(34). Zhou et al (32) showed that miR-301a increased
proliferation, migration and invasion of hepatocellular carcinoma
cells and inhibited apoptosis. In accordance with these results,
the findings of the present study indicated that inhibition of
miR-301a expression decreased proliferation, migration and invasion
of NSCLC cells. These findings suggested that miR-301a may be a
therapeutic target in NSCLC.
Previously, a number of predicted targets of
miR-301a have been reported in various types of cancer, including
GAX in hepatocellular carcinoma (32), BIM (35)
and SMAD family member 4 (29) in
pancreatic cancer, phosphatase and tensin homolog (34) and runt related transcription factor 3
(36) in gastric cancer, TGFBR2
(37) and suppressor of cytokine
signaling 6 (30) in colorectal
cancer and AMPKα1 in osteosarcoma (38).
To investigate the molecular mechanism by which
miR-301a contributes to proliferation, migration and invasion of
NSCLC, the potential target genes of miR-301a were investigated. In
the present study, DLC1 was identified as a novel target of
miR-301a. Initially, bioinformatic analysis indicated that DLC1 was
a putative target of miR-301a. Next, dual-luciferase reporter
assays showed that miR-301a directly targeted the 3′UTR of DLC1.
Downregulation of miR-301a increased DLC1 expression at mRNA and
protein levels. DLC1 knockdown also partially reversed the
inhibition of proliferation, migration and invasion induced by
miR-301a knockdown in NSCLC cells. All these results indicated that
DLC1 is a direct target gene of miR-301a in NSCLC. Identification
of target genes of miR-301a is important for elucidating the
functions of miR-301a in carcinogenesis and progression of NSCLC,
which may provide promising therapeutic targets for NSCLC.
In conclusion, the present study demonstrated that
miR-301a was upregulated in NSCLC tissues and cell lines compared
with normal adjacent tissues and a normal human bronchial
epithelial cell line. Functional analyses indicated that miR-301a
may promote proliferation, migration and invasion of NSCLC cells by
directly targeting DLC1. Therefore, miR-301a may contribute to the
initiation and progression of NSCLC. These findings may help to
further elucidate the molecular mechanisms underlying the
carcinogenesis and progression of NSCLC, and provide evidence for
the miR-301a/DLC1 pathway as a potential therapeutic target for
patients with NSCLC.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Spira A and Ettinger DS: Multidisciplinary
management of lung cancer. N Engl J Med. 350:379–392. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang L, Parkin DM, Li L and Chen Y: Time
trends in cancer mortality in China: 1987–1999. Int J Cancer.
106:771–783. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pfister DG, Johnson DH, Azzoli CG, Sause
W, Smith TJ, Baker S Jr, Olak J, Stover D, Strawn JR, Turrisi AT,
et al: American Society of Clinical Oncology treatment of
unresectable non-small-cell lung cancer guideline: Update 2003. J
Clin Oncol. 22:330–353. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yilmaz A, Damadoglu E, Salturk C, Okur E,
Tuncer LY and Halezeroglu S: Delays in the diagnosis and treatment
of primary lung cancer: Are longer delays associated with advanced
pathological stage? Ups J Med Sci. 113:287–296. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Verdecchia A, Francisci S, Brenner H,
Gatta G, Micheli A, Mangone L and Kunkler I: EUROCARE-4 Working
Group: Recent cancer survival in Europe: A 2000–02 period analysis
of EUROCARE-4 data. Lancet Oncol. 8:784–796. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Somaiah N and Simon GR: Molecular targeted
agents and biologic therapies for non-small cell lung cancer. J
Thorac Oncol. 5 12 Suppl 6:S434–S454. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Donadeu FX, Schauer SN and Sontakke SD:
Involvement of miRNAs in ovarian follicular and luteal development.
J Endocrinol. 215:323–334. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liwak U, Faye MD and Holcik M: Translation
control in apoptosis. Exp Oncol. 34:218–230. 2012.PubMed/NCBI
|
|
13
|
Rutnam ZJ and Yang BB: The involvement of
microRNAs in malignant transformation. Histol Histopathol.
27:1263–1270. 2012.PubMed/NCBI
|
|
14
|
Rothschild SI: Epigenetic therapy in lung
cancer-role of microRNAs. Front Oncol. 3:1582013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yuan BZ, Jefferson AM, Baldwin KT,
Thorgeirsson SS, Popescu NC and Reynolds SH: DLC-1 operates as a
tumor suppressor gene in human non-small cell lung carcinomas.
Oncogene. 23:1405–1411. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Feng H, Zhang Z, Wang X and Liu D:
Identification of DLC-1 expression and methylation status in
patients with non-small-cell lung cancer. Mol Clin Oncol.
4:249–254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Healy KD, Hodgson L, Kim TY, Shutes A,
Maddileti S, Juliano RL, Hahn KM, Harden TK, Bang YJ and Der CJ:
DLC-1 suppresses non-small cell lung cancer growth and invasion by
RhoGAP-dependent and independent mechanisms. Mol Carcinog.
47:326–337. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu XH, Liu ZL, Sun M, Liu J, Wang ZX and
De W: The long non-coding RNA HOTAIR indicates a poor prognosis and
promotes metastasis in non-small cell lung cancer. BMC Cancer.
13:4642013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen X, Chen S, Hang W, Huang H and Ma H:
MiR-95 induces proliferation and chemo- or radioresistance through
directly targeting sorting nexin1 (SNX1) in non-small cell lung
cancer. Biomed Pharmacother. 68:589–595. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang Y, Zhao Y, Sun S, Liu Z, Zhang Y and
Jiao S: Overexpression of MicroRNA-221 is associated with poor
prognosis in non-small cell lung cancer patients. Tumour Biol.
37:10155–10160. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen X, Gong J, Zeng H, Chen N, Huang R,
Huang Y, Nie L, Xu M, Xia J, Zhao F, et al: MicroRNA145 targets
BNIP3 and suppresses prostate cancer progression. Cancer Res.
70:2728–2738. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu D, Zhou Y, Pan H, Zhou J, Fan Y and Qu
P: microRNA-99a inhibiting cell proliferation, migration and
invasion by targeting fibroblast growth factor receptor 3 in
bladder cancer. Oncol Lett. 7:1219–1224. 2014.PubMed/NCBI
|
|
25
|
Xu T, Liu X, Han L, Shen H, Liu L and Shu
Y: Up-regulation of miR-9 expression as a poor prognostic biomarker
in patients with non-small cell lung cancer. Clin Transl Oncol.
16:469–475. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bai Y, Wang YL, Yao WJ, Guo L, Xi HF, Li
SY and Zhao BS: Expression of miR-32 in human non-small cell lung
cancer and its correlation with tumor progression and patient
survival. Int J Clin Exp Pathol. 8:824–829. 2015.PubMed/NCBI
|
|
27
|
Kawano M, Tanaka K, Itonaga I, Iwasaki T
and Tsumura H: MicroRNA-301a promotes cell proliferation via PTEN
targeting in Ewing's sarcoma cells. Int J Oncol. 48:1531–1540.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu Y, Gao W, Zhang C, Wen S, Huangfu H,
Kang J and Wang B: Hsa-miR-301a-3p acts as an oncogene in laryngeal
squamous cell carcinoma via target regulation of Smad4. J Cancer.
6:1260–1275. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xia X, Zhang K, Cen G, Jiang T, Cao J,
Huang K, Huang C, Zhao Q and Qiu Z: MicroRNA-301a-3p promotes
pancreatic cancer progression via negative regulation of SMAD4.
Oncotarget. 6:21046–21063. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fang Y, Sun B, Xiang J and Chen Z:
MiR-301a promotes colorectal cancer cell growth and invasion by
directly targeting SOCS6. Cell Physiol Biochem. 35:227–236. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Xu XD, He XJ, Tao HQ, Zhang W, Wang YY, Ye
ZY and Zhao ZS: Abnormal expression of miR-301a in gastric cancer
associated with progression and poor prognosis. J Surg Oncol.
108:197–202. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou P, Jiang W, Wu L, Chang R, Wu K and
Wang Z: miR-301a is a candidate oncogene that targets the homeobox
gene Gax in human hepatocellular carcinoma. Dig Dis Sci.
57:1171–1180. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu H, Li H, Qian H, Jiao X, Zhu X, Jiang
X, Dai G and Huang J: Upregulation of miR-301a correlates with poor
prognosis in triple-negative breast cancer. Med Oncol. 31:2832014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma F, Zhang J, Zhong L, Wang L, Liu Y,
Wang Y, Peng L and Guo B: Upregulated microRNA-301a in breast
cancer promotes tumor metastasis by targeting PTEN and activating
Wnt/β-catenin signaling. Gene. 535:191–197. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen Z, Chen LY, Dai HY, Wang P, Gao S and
Wang K: miR-301a promotes pancreatic cancer cell proliferation by
directly inhibiting Bim expression. J Cell Biochem. 113:3229–3235.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang M, Li C, Yu B, Su L, Li J, Ju J, Yu
Y, Gu Q, Zhu Z and Liu B: Overexpressed miR-301a promotes cell
proliferation and invasion by targeting RUNX3 in gastric cancer. J
Gastroenterol. 48:1023–1033. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang W, Zhang T, Jin R, Zhao H, Hu J,
Feng B, Zang L, Zheng M and Wang M: MicroRNA-301a promotes
migration and invasion by targeting TGFBR2 in human colorectal
cancer. J Exp Clin Cancer Res. 33:1132014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhang Y, Duan G and Feng S: MicroRNA-301a
modulates doxorubicin resistance in osteosarcoma cells by targeting
AMP-activated protein kinase alpha 1. Biochem Biophys Res Commun.
459:367–373. 2015. View Article : Google Scholar : PubMed/NCBI
|