Introduction
Primary liver cancer is one of the most frequently
occurring malignancies worldwide, and therefore a major public
health challenge (1). The development
of liver cancer is a complex, multistep process that is derived
from a series of genetic and epigenetic alternations. The
inactivation of tumor suppressor genes as a result of aberrant DNA
methylation and histone modification is a characteristic step in
tumor development and progression (2). Furthermore, the downregulation of the
epigenetic regulator led to epigenetic alternations and contributed
to abnormal inactivation of tumor suppressor genes in numerous
types of human cancer (3,4). Accordingly, improved understanding the
molecular basis of liver cancer may enhance the development of
novel strategies to improve the treatment of liver cancer.
The four and a half LIM domains (FHL) family has an
important role in regulating cell proliferation, differentiation
and apoptosis, and the member FHL1, located on human chromosome
Xq26, functions in skeletal and cardiac muscle growth (5,6). Numerous
studies of clinical samples have shown that FHL1 expression was
downregulated in multiple types of malignancy, including lung
(7), gastric (8,9), breast,
kidney and prostate cancers (10).
Notably, FHL1 as tumor suppressor gene on chromosome X has a high
risk to be affected as a single hit of genetic and/or epigenetic
abnormality on only one active allele could lead to complete
inactivation of FHL1 (11). FHL1
exerts a tumor suppressor effect via multiple mechanisms, including
the activation of the transforming growth factor-β-like and
mitogen-activated protein kinase signaling pathways and protein
interaction with zonula occludens-1, hypoxia-inducible factor 1-α
and estrogen receptor α (7,9,12,13). However, the role of FHL1 in liver
cancer has not been revealed and the mechanisms associated with
FHL1 downregulation remain unknown.
Enhancer of zeste homolog 2 (EZH2) is a frequently
elevated epigenetic regulator as the catalytic subunit of polycomb
repressive complex 2 (PRC2) in multiple types of human cancer
(14–16). As a histone methyltransferase, EZH2
specifically catalyzes histone H3 lysine 27 tri-methylation
(H3K27me3), a repressive histone modification, to epigenetically
control gene transcription (17).
EZH2 serves an oncogenic role in different types of human cancer,
primarily through the epigenetic silencing of tumor suppressor
genes: For example, EZH2-mediated trimethylation of histone H3 at
lysine 27 (H3K27me3) epigenetically silenced chromodomain helicase
DNA binding protein 5, which serves as a tumor suppressor in HCC
cells (18). Overexpression of EZH2
may lead to hypermethylation of p16 INK4a promoter, followed by a
decreased expression of p16 INK4a in the multi-step
cholangiocarcinogenesis (19). To
address the mechanism of FHL1 downregulation during HCC genesis,
the present study investigated the epigenetic dysregulation and
related effects of FHL1. The data demonstrated that FHL1 was
synergistically silenced by DNA methylation and histone
modification, and revealed the epigenetically regulatory mechanisms
by which FHL1 was inactivated during hepatocarcinogenesis.
Materials and methods
Cell lines and tissue specimens
The human HCC-derived MHCC-97L and Hep3B, human
liver LO-2 and HepG2, which was re-identified as a human
hepatoblastoma cell line (20), cell
lines were obtained from the Cell Bank of Chinese Academy of
Sciences (Shanghai, China). All cells were cultured routinely in
Dulbecco's modified Eagle's medium (DMEM) supplemented with
heat-inactivated fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA), penicillin (100 U/ml) and
streptomycin (100 µg/ml) at 37°C in a 5% CO2-humidified
chamber for 24 h. The tissue samples used in the present study were
obtained from the First Affiliated Hospital of Soochow University
(Suzhou, China). All experimental protocols were approved by
Soochow University Ethics Committee (Suzhou, China) and all
patients provided written informed consent.
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the Hep3B, HepG2 cells
and liver cancer tissues using TRIzol reagent (Thermo Fisher
Scientific, Inc.) and reverse transcribed using a Superscript III
kit (Thermo Fisher Scientific, Inc.). RT-qPCR was performed using
an SYBR-Green qPCR Master mix (Thermo Fisher Scientific, Inc.). An
initial denaturation was performed for 5 min at 94°C, and 35 cycles
were performed with the following PCR program: Denaturation at 94°C
for 30 sec, annealing at 55°C for 30 sec for FHL1 and 55°C for 30
sec for β-actin and elongation at 72°C for 30 sec. The upstream and
downstream FHL1 primers were 5′-ACAATCCTGGCACGACTA-3′ and
5′-AAAATGGGAGAAAAGACG-3′, respectively. Housekeeping gene β-actin
was used as reference gene, its upstream and downstream primers
were 5′-TCACCAACTGGGACGACA-3′ and 5′-TGCAAAGAACACGGCTAA-3′. Gene
expression levels were normalized to β-actin, and the fold change
of target genes was calculated using 2−ΔΔCq (21).
Western blotting
Total proteins were extracted using
radioimmunoprecipitation lysis buffer [150 mM NaCl, 1% NP40, 0.5%
sodium deoxycholate, 0.1% SDS, 50 mM Tris (pH 7.9), 10 mM NaF, PMSF
and protease inhibitors (Complete Cocktail tablets, Roche
Diagnostics, Basel, Switzerland)]. Protein concentrations were
measured using Pierce™ BCA Protein Assay kit (Thermo Fisher
Scientific, Inc.). These extracts (50 µg) were subjected to
electrophoresis by 10% SDS-PAGE and then transferred onto Hybrid-P
polyvinylidene difluoride membrane (Merck KGaA, Darmstadt,
Germany). Following blocking with PBS containing 5% skimmed milk
powder and 0.1% Tween-20 for 2 h at room temperature, the blot was
incubated for immunoblotting analysis with antibodies against FHL1
(cat. no. sc-374246; 1:1,000 dilution; Santa Cruz Biotechnology,
Inc., Dallas, TX, USA), H3K27me3 (cat. no. 9733S; 1:500 dilution;
Cell Signaling Technology, Inc., Danvers, MA, USA) or β-actin (cat.
no. sc-47778; 1:2,000 dilution; Santa Cruz Biotechnology, Inc.,)
overnight at 4°C. Peroxidase-conjugated secondary antibodies (cat.
nos. SAB3701171 and SAB3700831; 1:5,000 dilution; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) were used, and membranes were
developed using the enhanced chemiluminescent immunoassay (Thermo
Fisher Scientific, Inc.) for the detection of antigens.
Adenovirus preparation
The recombinant adenovirus carrying FHL1 (pAd-FLH1)
or green fluorescent protein (GFP) was generated using Ad-Easy
system according to the manufacturer's protocol (Stratagene;
Agilent Technologies, Inc., Santa Clara, CA, USA). For packaging of
the adenoviruses expressing FHL1 or GFP, pAd-FHL1 and pAd-GFP was
transfected into AD293 cells using Lipofectamine 2000®
reagent (Thermo Fisher Scientific Inc.). The adenoviruses were
amplified in AD293, purified, titrated and stored at −80°C until
use.
Bisulfite sequencing
Genomic DNA was isolated using DNeasy Tissue kit
(Qiagen, Inc., Valencia, CA, USA), and bisulfite modification was
performed with the EpiTect Bisulfite kit (Qiagen, Inc.) according
to the manufacturer's protocol. CpG enrichment region of the FHL1
promoter was analyzed by Cpgplot (http://www.ebi.ac.uk/Tools/seqstats/emboss_cpgplot/).
The PCR products were cloned into pMD 18-T vector (Takara Bio,
Inc., Otsu, Japan), and 8 clones were randomly selected for each
specimen DNA sequencing.
Chromatin immunoprecipitation (ChIP)
assay
A ChIP assay was performed using an EZ ChIP kit (EMD
Millipore, Billerica, MA, USA) according to the manufacturer's
protocol, with slight modifications. The Hep3B and HepG2 cells were
cross-linked in 1% formaldehyde and quenched with the addition of
125 mM glycine. Subsequent to washing twice in ice-cold PBS
containing a protease inhibitor cocktail, the cell lysates were
harvested in ChIP lysis buffer [50 mM HEPEs (pH 7.5), 1 mM EDTA (pH
8.0), 150 mM sodium chloride, 0.1% sodium deoxycholate, 0.1% SDS,
1% Triton X-100 and complete phenylmethylsulfonyl fluoride].
Subsequently, chromatin was sheared to fragments of 300–500 bp by
sonication 9 times for 10–20 sec at 80% setting using VibraCell
Sonicator (Sonics & Materials, Newtown, CT, USA) at a frequency
of 20 kHz. The lysates were pre-cleared with Salmon Sperm
DNA/Protein G Agarose (Roche Diagnostics) for 2 h. Samples were
centrifuged at 12,000 × g for 15 min at 4°C, and then the
supernatant was used for immunoprecipitation at 4°C overnight with
5 µg anti-H3K27me3 (cat. no. 9733S; 1:50 dilution) or anti-mouse
IgG (cat. no. 5415; 1:50 dilution; both from Cell Signaling
Technology, Inc.) as a negative control. Immunoprecipitated
crosslinking DNA fragments were reversed by pronase and
subsequently incubated at 42°C for 2 h and 68°C for 8 h. The FHL1
promoter DNA in the immunoprecipitates was detected by qPCR using
the following primers: Forward, 5′-ACCGAGTGAGAAAAGCCAAT-3′ and
reverse, 5′-TCACCATTGGCAACCACTGAT-3′. The FHL1 signals were
normalized to GAPDH (forward, 5′-TACTAGCGGTTTTACGGGCG-3′ and
reverse, 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′) using the
2−ΔΔCq method (21) to
determine whether the immunoprecipitate was enriched. qPCR protocol
and reactions were performed according to the protocol of the
EZ-ChIP Kit (EMD Millipore). Specifically, 25 µl PCR reaction
system including 2 µl of immunoprecipitate samples, 12.5 µl
SYBR-Green Master mix, 1 µl of primer mix and 9.5 µl
ddH2O. Two-step qPCR parameters were set as follows:
Initial denaturation at 94°C for 10 min, then denaturation at 94°C
for 20 sec, then two-step annealing/extension for 1 min at
60°C.
In vitro epigenetic drug
treatment
The 3-deanzaneplanocin A (DZNep; Cayman Chemical
Company, Ann Arbor, MI, USA) was dissolved in dimethyl sulfoxide
(DMSO) and 5-Aza-2′ deoxycytidine (5-Aza-dC; Sigma-Aldrich; Merck
KGaA) was dissolved in 50% acetic acid. Trichostatin A (TSA;
Sigma-Aldrich; Merck KGaA) was dissolved in ethanol. The solvents
(DMSO, acetic acid, and ethanol) were used as controls in the
corresponding treatment. For the DZNep treatment, DZNep (10 µM) was
added to the culture medium for 48 or 72 h. For the 5-Aza-dC
treatment, 5-Aza-dC (10 µM) was replenished daily for 72 h. For the
TSA treatment, TSA (0.25 µg/ml) was only added to the cells in the
last 24 h of the experiment.
Colony formation assay
A total of 5×104 Hep3B cells were
cultured in DMEM media supplemented with 10% heat-inactivated fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.),
penicillin (100 U/ml) and streptomycin (100 µg/ml) at 37°C in a 5%
CO2-humidified chamber in 10 cm dishes in triplicate. A
total of 3 weeks later, the anchorage-dependent colonies were
washed twice with PBS and stained with crystal violet (0.5%
w/v).
Transwell migration assay
In total, 1×105 Hep3B cells were seeded
in triplicate Boyden chambers with an 8 µm-pore sized membrane in
the top chamber (BD Biosciences, Franklin Lakes, NJ, USA) in
serum-free DMEM media (Thermo Fisher Scientific, Inc.). Media
containing 10% FBS (Thermo Fisher Scientific, Inc.) was used in the
bottom chambers. Subsequent to 48 h incubation, the cells in the
upper chamber were wiped, and the migrated cells on the lower
surface of the membrane were fixed, stained with 0.1% crystal
violet and photographed using a light microscope (magnification,
×20).
Statistical analysis
The differences between two groups in gene
expression, colony number and migrated cell number were evaluated
using Student's t-test. The statistical significance of differences
in multiple groups was determined by analysis of variance with
Bonferroni test. Data in histograms was shown as the mean ±
standard deviation from 3 independent replicates. The comparison of
the methylated CpG percentage between tumor and non-cancerous
specimens was performed by χ2 test. All statistical
analyses were conducted using GraphPad Prism software 5.0 (GraphPad
Software, Inc., La Jolla, CA, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
FHL1 expression was frequently
downregulated in patients with liver cancer
The present study examined the expression of FHL1
mRNA in a cohort of 49 paired specimens from liver cancer patients
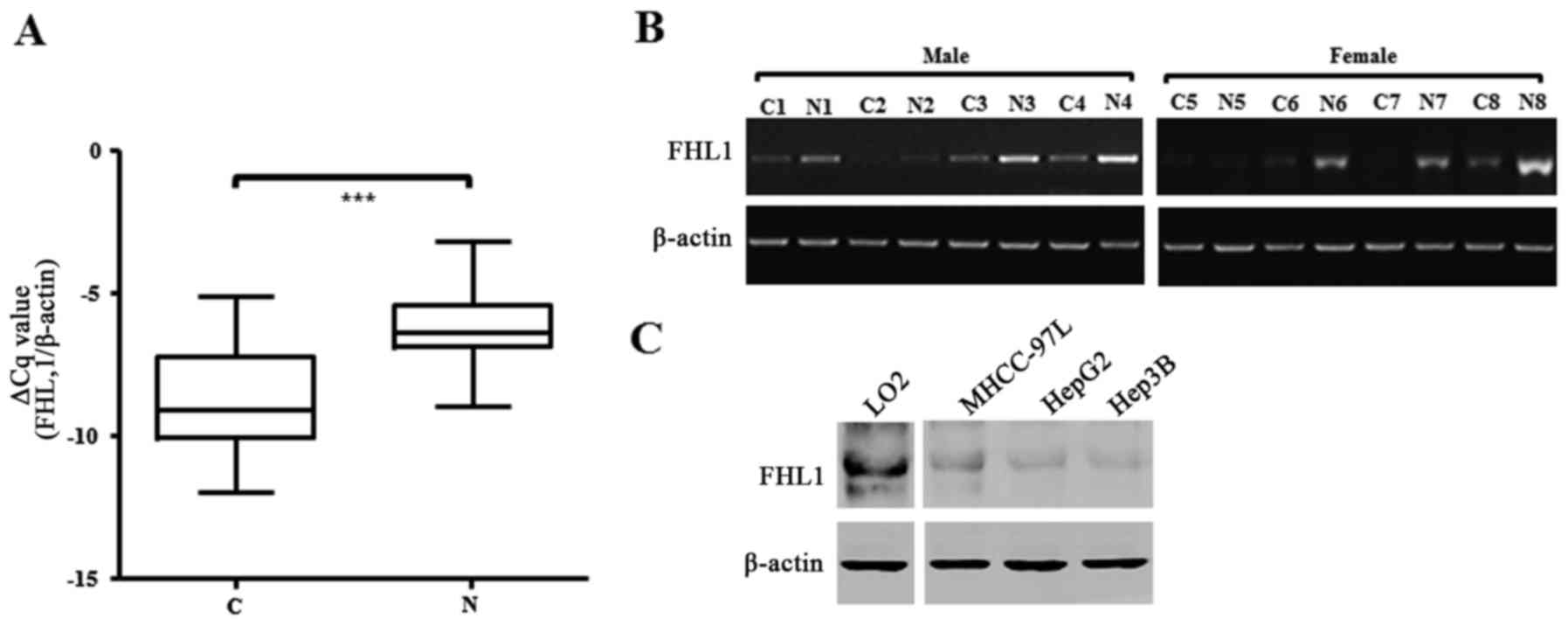
RT-qPCR. The data showed that the FHL1 mRNA level significantly
decreased in tumor tissue compared with the matched non-cancerous
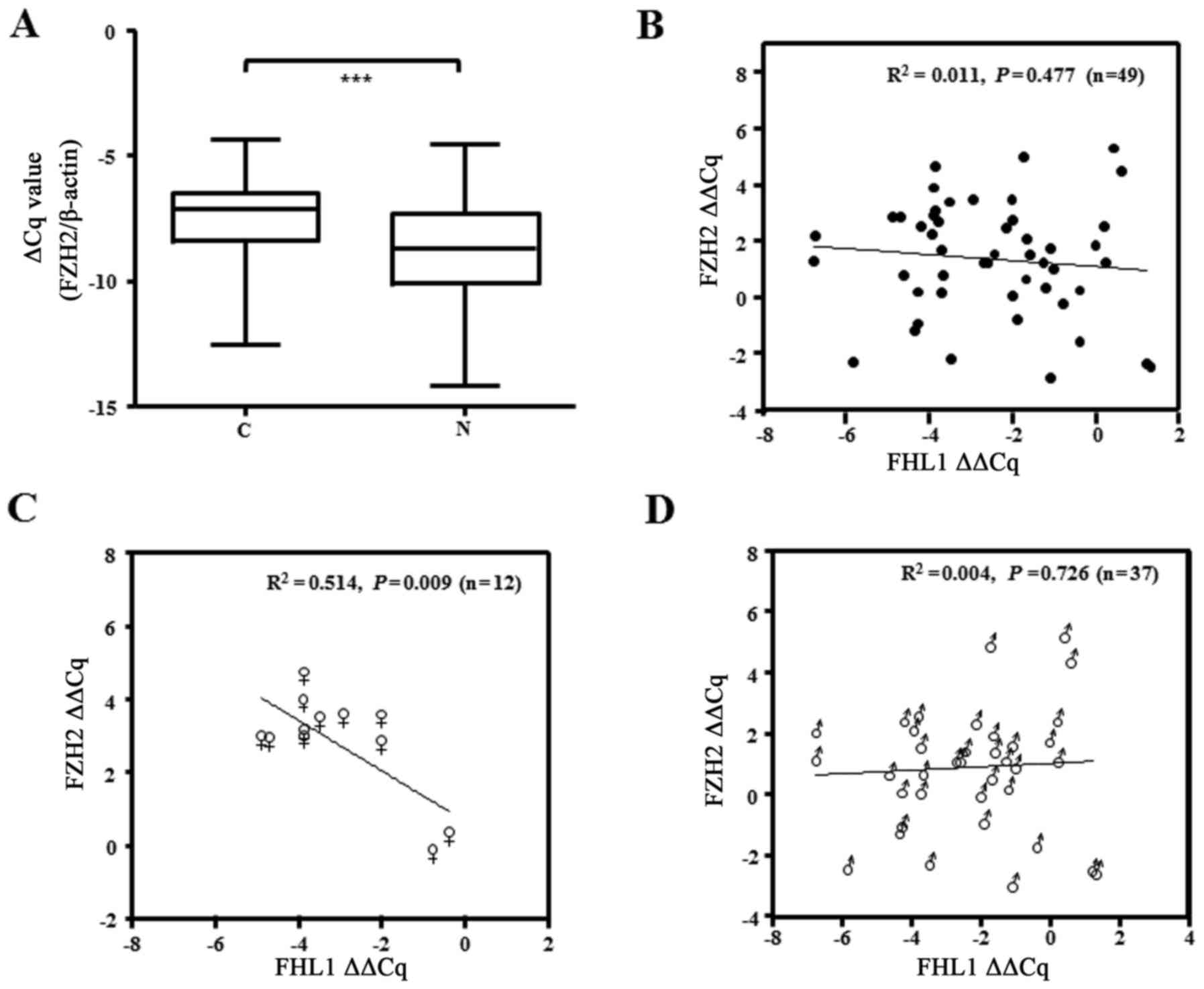
tissues of patients with liver cancer (P<0.001; Fig. 1A). Of the 49 paired liver cancer
specimens examined, 42 (85.7%) exhibited at least a 2-fold
downregulation of FHL1 expression compared with that of the matched
non-cancerous liver tissue (Table I),
and 8 exhibited a marked difference between tumor and matched
non-cancerous tissue, as confirmed by qPCR (Fig. 1B). In addition, the present study
analyzed the association between FHL1 expression and clinical
factors. However, FHL1 downregulation was not significantly
associated with the sex, age, hepatitis B virus, tumor size,
metastasis and Edmondson-Steiner grading system (22). The results of western blotting showed
that FHL1 exhibited less expression in three liver cancer cell
lines, consisting of MHCC-97L, Hep3B and HepG2, one of the most
popular hepatoblastoma cell lines (20), than the immortal liver LO2 and WRL68
cell lines. These data showed that FHL1 was downregulated in human
liver cancer, which is consistent with the observations of previous
studies (12).
 | Table I.Association between
clinicopathological characteristics and FHL1 expression in 49 HCC
specimens. |
Table I.
Association between
clinicopathological characteristics and FHL1 expression in 49 HCC
specimens.
|
| FHL1 expression |
|
|---|
|
|
|
|
|---|
| Characteristics | Decrease | No change | P-value |
|---|
| Sex |
|
| 1.00 |
| Male | 32 | 5 |
|
|
Female | 10 | 2 |
|
| Age |
|
| 0.45 |
|
<40 | 6 | 0 |
|
|
40–50 | 11 | 3 |
|
|
>50 | 25 | 4 |
|
| HBV |
|
| 0.34 |
|
Positive | 37 | 7 |
|
|
Negative | 5 | 0 |
|
| Tumor size (T) |
|
| 0.15 |
|
T1+T2 | 38 | 6 |
|
|
T3+T4 | 4 | 1 |
|
| Distant metastases
(M) |
|
| 0.47 |
| M0 | 39 | 7 |
|
| M1 | 3 | 0 |
|
| Edmondson |
|
| 1.00 |
|
I+II | 6 | 1 |
|
|
III+IV | 36 | 6 |
|
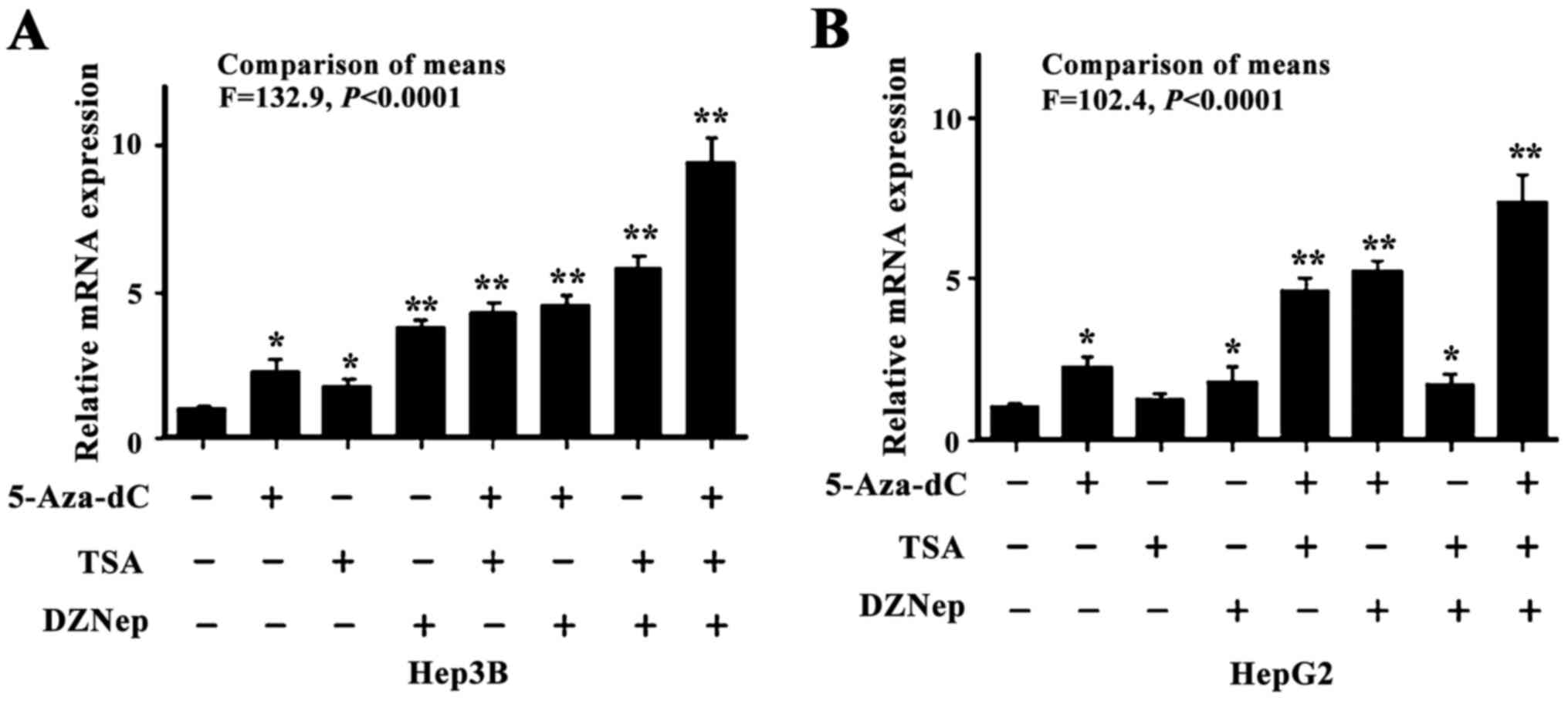
FHL1 expression was synergistically by
DNA methylation and histone modification
Since DNA methylation and histone modifications are
closely associated with respect to establishing a less permissive
chromatin status to suppress gene transcription, the present study
sought to reveal the combined effect of different epigenetic
machineries associated with FHL1 downregulation in liver cancer.
Two types of liver cancer cell lines were used, consisting of the
hepatocellular carcinoma-derived Hep3B cell line and the
hepatoblastoma-derived HepG2 cell line, to investigate the
epigenetic effects on FHL1. The two cell lines were treated with
DZNep, a small molecular EZH2 inhibitor, and 5-Aza-dC and
Trichostatin A TSA, well characterized DNA methylation and histone
acetylation inhibitors. As expected, the individual or combined
treatments significantly led to increased FHL1 expression
(P<0.05). While treatment of DZNep, 5-Aza-dC and TSA
individually elevated expression FHL1, combined treatment of the 3
drugs synergistically restored FHL1 expression in Hep3B cells
(Fig. 2A). Additionally, only TSA
treatment alone did not restore FHL1 expression and the
co-treatment did not induce a further increase in FHL1 expression
compared with 5-Aza-dC alone in HepG2 cells (Fig. 2B), suggesting that DNA methylation and
histone methylation have a crucial epigenetic role in mediating
FHL1 downregulation.
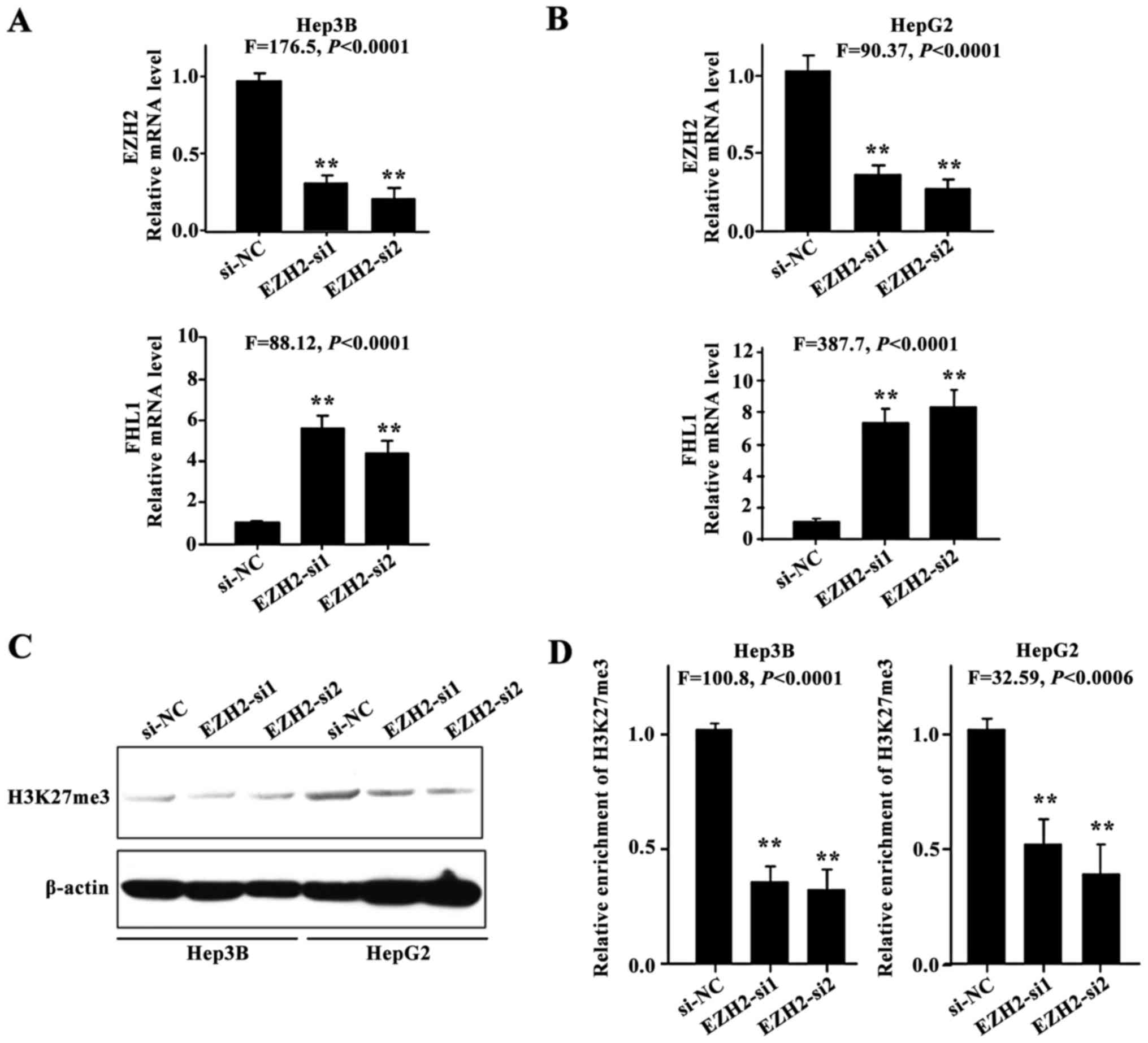
EZH2 knockdown restored FHL1
expression
The present study hypothesized that aberrant histone
methylation may contribute to FHL1 silencing in HCC cell lines. To
investigate whether FHL1 expression could be restored subsequent to
the knockdown of EZH2, 2 siRNAs against EZH2 were employed to
silence endogenous EZH2 expression. FHL1 was transcriptionally
induced in Hep3B (P<0.01; Fig. 3A)
and HepG2 cells (P<0.01; Fig. 3B),
indicating that EZH2-mediated H3K27me3 contributes to the
suppression of FHL1 in Hep3B and HepG2 cells. As expected, EZH2
knockdown resulted in a reduction in the level of H3K27me3
(Fig. 3C), and a ChIP assay was
performed to assess the enrichment of transcriptional repressive
histone modifications H3K27me3 on the FHL1 promoter in Hep3B and
HepG2 cells (P<0.01; Fig. 3D). The
findings indicated that the epigenetic silencing of FHL1 by
EZH2-mediated H3K27me3 is an important mechanism in human liver
cancer.
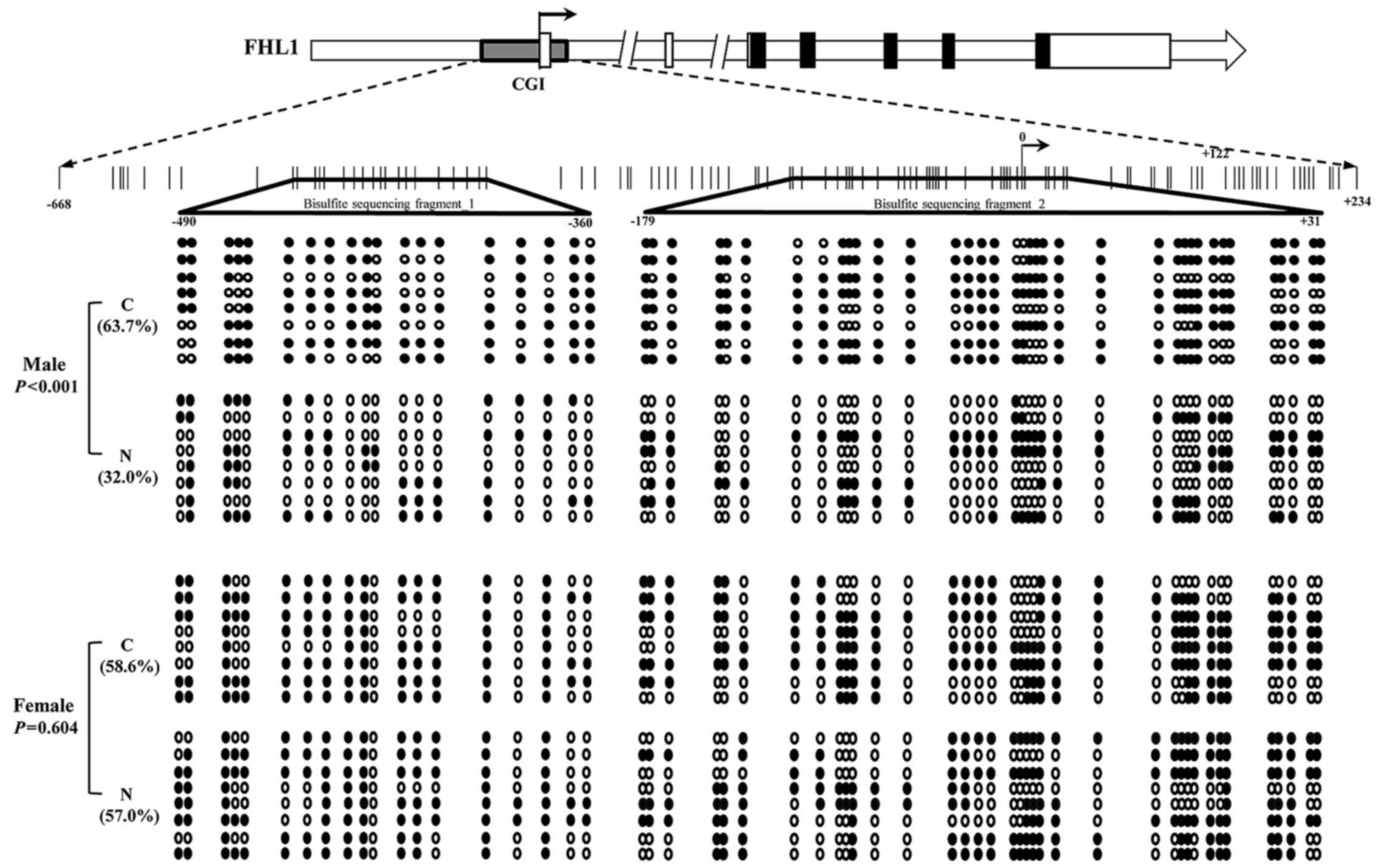
Methylation of FHL1 promoter in liver
cancer specimens
To further assess the association between the FHL1
downregulation and the methylation status of the potential
methylation positions of the FHL1 gene using CpG plot arithmetic.
As a result, a typical CpG island (−668 to +234) was found within
the promoter and exon of the FHL1 gene. A total of 2 DNA fragments
located on the CpG island were amplified, and bisulfite sequencing
was performed to analyze the methylation status in the 8 paired HCC
specimens with FHL1 downregulation. The results indicated that the
methylation level of the CpG island was significantly enriched in
the 4 male HCC specimens compared with the matched non-tumorous
liver tissue (P<0.001; Fig. 4).
However, no significant difference was identified between tumors
and matched non-tumorous specimens in the 4 female patients
(P=0.604; Fig. 4). The data propose a
hypothesis that other epigenetic mechanisms contribute to FHL1
deregulation in female patients with HCC.
Association between FHL1 and EZH2
expression in human liver cancer tissues
To determine the association between FHL1 and EZH2,
the present study examined EZH2 mRNA expression in 49 paired tissue
samples. The results showed that EZH2 was significantly upregulated
in the aforementioned cohort of samples with FHL1 downregulation
(Fig. 5A). The present study then
additionally analyzed the association between FHL1 downregulation
and EZH2 upregulation, and failed to identify a significant
association between the expression of the 2 genes (P=0.477;
Fig. 5B). Notably, an inverse
correlation between FHL1 and EZH2 was observed in the samples of
the female patients with HCC (P=0.009; Fig. 5C), while this was not observed the
samples from male patients (P=0.726; Fig.
5D), suggesting the role of EZH2 upregulation in suppressing
FHL1 expression in female patients with liver cancer.
FHL1 inhibits cell proliferation and
migration in vitro
The present study then investigated the effect of
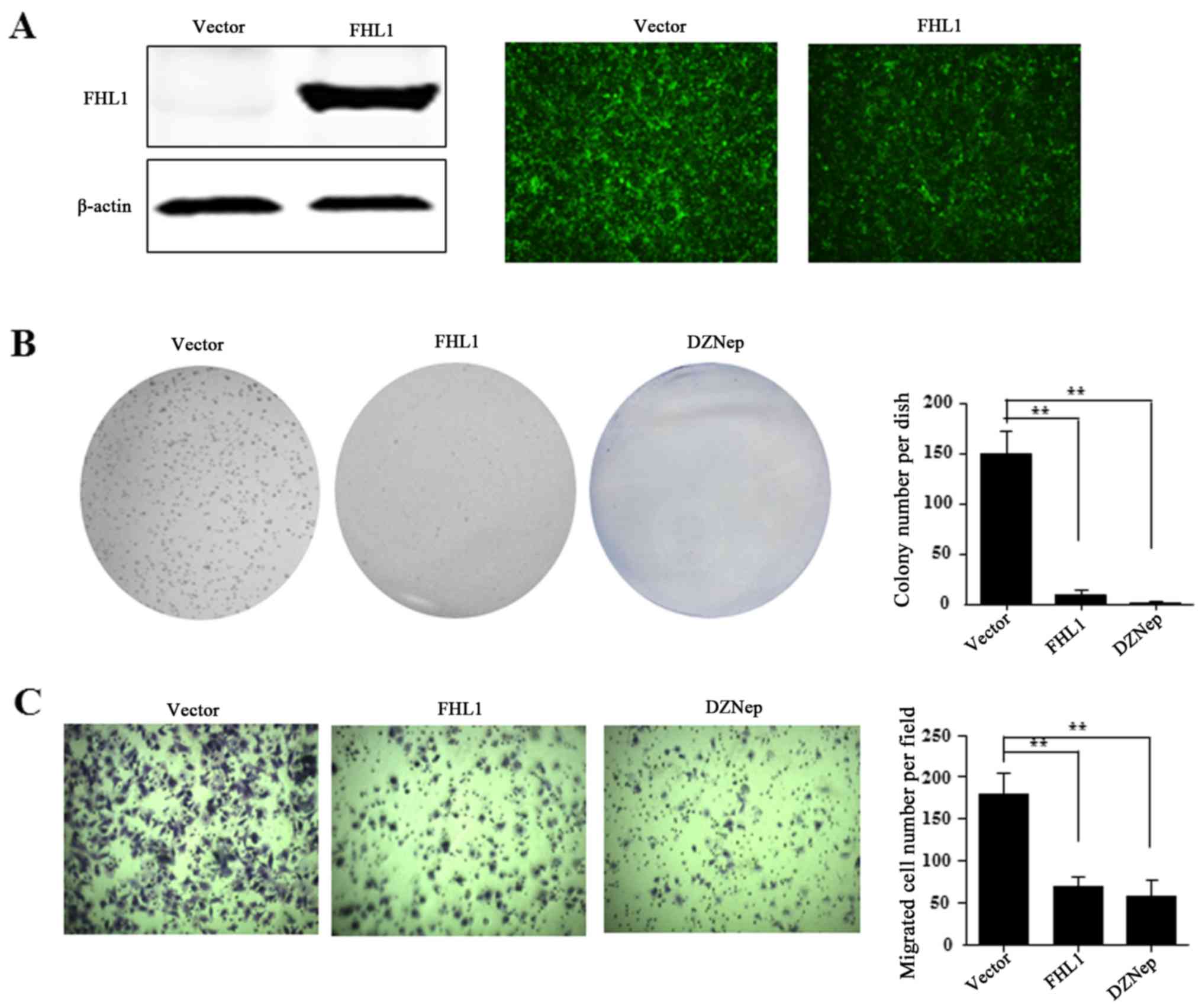
FHL1 overexpression on HCC cell growth. For FHL1 overexpression,
the recombinant adenovirus Ad-FHL1 tagged with GFP was transduced
into Hep3B cells. After 3 days, FHL1 was overexpressed and almost
100% transduction efficiency was observed, as indicated by western
blotting assay and GFP protein observation in Hep3B cells (Fig. 6A). To reveal the role of FHL1, the
present study also performed anchorage-dependent colony formation
and migration assays. The results demonstrated that FHL1
overexpression and DZNep treatment significantly inhibited the
level of cell colony formation of Hep3B compared to the Ad-GFP
empty vector control (Fig. 6B).
Furthermore, the cell migration assay indicated that FHL1
overexpression or DZNep treatment significantly suppressed cell
migration ability compared with the control cells (Fig. 6C). These findings suggest that EZH2 is
involved in suppressing FHL1.
Discussion
FHL1 has a tumor suppressive role in a number of
types of human cancer (7–9,12). The
findings of the present study reveal that EZH2 may act as a
regulator of FHL1 expression in human liver cancer. FHL1 was
identified as the first member of the fragile tumor suppressor gene
on chromosome X, and is inactivated by DNA methylation (23). Downregulation of FHL1 in tumor samples
has been reported in breast (24),
gastric (8) and lung cancers
(7). A previous study revealed that
EZH2 is a catalytic subunit of the epigenetic regulator PRC2, which
trimethylates Lys27 of histone H3, leading to the silencing of
target genes that are involved in numerous biological processes,
including tumor progression (16).
Overexpression of EZH2 has been detected in a range of types of
cancer, and is associated with tumor malignancy (14,15) via
the epigenetic silencing of tumor and metastasis suppressor genes
(25,26). However, FHL1 downregulation was not
significantly associated with clinicopathological characteristics
of patients applied in the present study. With respect to the role
of FHL1 in tumor initiation and progression, numerous studies
concluded that FHL1 inhibits the growth of cancer cells, transforms
fibroblasts and suppress the migration and invasion of bladder
cancer cells (7,12,13,27). The
data obtained in the present study is in line with previous
reports, and demonstrates that FHL1 overexpression inhibits cell
proliferation and migration in HCC cells.
Primary liver cancer comprises HCC, intrahepatic
cholangiocarcinoma and other rare tumors, notably fibrolamellar
carcinoma and hepatoblastoma (28).
At present, few studies have reported the expression level of FHL1
in ICC, fibrolamellar carcinoma and hepatoblastoma. The present
study was also limited to HCC, due to rare incidence of other
pathological types. However, the epigenetic regulations of FHL1
were investigated in the hepatoblastoma HepG2 cell line, which
indicated that FHL1 was dysregulated by similar epigenetic
mechanisms. In conclusion, the present study showed that DNA
methylation and EZH2-induced H3K27me3 is associated with the
epigenetic repression of the FHL1 tumor suppressor gene in HCC.
Acknowledgements
The present study was supported by Applied Basic
Research Programs of Science and Technology Department of Suzhou
(grant no. SYS201447) and Youth Science and Technology Project of
Suzhou (grant no. kjxw2014001).
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Scaggiante B, Kazemi M, Pozzato G, Dapas
B, Farra R, Grassi M, Zanconati F and Grassi G: Novel
hepatocellular carcinoma molecules with prognostic and therapeutic
potentials. World J Gastroenterol. 20:1268–1288. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hassler MR and Egger G: Epigenomics of
cancer-emerging new concepts. Biochimie. 94:2219–2230. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sandoval J and Esteller M: Cancer
epigenomics: Beyond genomics. Curr Opin Genet Dev. 22:50–55. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ng EK, Lee SM, Li HY, Ngai SM, Tsui SK,
Waye MM, Lee CY and Fung KP: Characterization of tissue-specific
LIM domain protein (FHL1C) which is an alternatively spliced
isoform of a human LIM-only protein (FHL1). J Cell Biochem.
82:1–10. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Morgan MJ and Madgwick AJ: The LIM
proteins FHL1 and FHL3 are expressed differently in skeletal
muscle. Biochem Biophys Res Commun. 255:245–250. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Niu C, Liang C, Guo J, Cheng L, Zhang H,
Qin X, Zhang Q, Ding L, Yuan B, Xu X, et al: Downregulation and
growth inhibitory role of FHL1 in lung cancer. Int J Cancer.
130:2549–2556. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu Y, Liu Z and Guo K: Expression of FHL1
in gastric cancer tissue and its correlation with the invasion and
metastasis of gastric cancer. Mol Cell Biochem. 363:93–99. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sakashita K, Mimori K, Tanaka F, Kamohara
Y, Inoue H, Sawada T, Hirakawa K and Mori M: Clinical significance
of loss of Fhl1 expression in human gastric cancer. Ann Surg Oncol.
15:2293–2300. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li X, Jia Z, Shen Y, Ichikawa H, Jarvik J,
Nagele RG and Goldberg GS: Coordinate suppression of Sdpr and Fhl1
expression in tumors of the breast, kidney, and prostate. Cancer
Sci. 99:1326–1333. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Spatz A, Borg C and Feunteun J:
X-chromosome genetics and human cancer. Nat Rev Cancer. 4:617–629.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ding L, Wang Z, Yan J, Yang X, Liu A, Qiu
W, Zhu J, Han J, Zhang H, Lin J, et al: Human four-and-a-half LIM
family members suppress tumor cell growth through a TGF-beta-like
signaling pathway. J Clin Invest. 119:349–361. 2009.PubMed/NCBI
|
|
13
|
Shen Y, Jia Z, Nagele RG, Ichikawa H and
Goldberg GS: SRC uses Cas to suppress Fhl1 in order to promote
nonanchored growth and migration of tumor cells. Cancer Res.
66:1543–1552. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Geng J, Li X, Zhou Z, Wu CL, Dai M and Bai
X: EZH2 promotes tumor progression via regulating VEGF-A/AKT
signaling in non-small cell lung cancer. Cancer Lett. 359:275–287.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zingg D, Debbache J, Schaefer SM, Tuncer
E, Frommel SC, Cheng P, Arenas-Ramirez N, Haeusel J, Zhang Y,
Bonalli M, et al: The epigenetic modifier EZH2 controls melanoma
growth and metastasis through silencing of distinct tumour
suppressors. Nat Commun. 6:60512015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li LY: EZH2: Novel therapeutic target for
human cancer. Biomedicine (Taipei). 4:12014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cao R, Wang L, Wang H, Xia L,
Erdjument-Bromage H, Tempst P, Jones RS and Zhang Y: Role of
histone H3 lysine 27 methylation in Polycomb-group silencing.
Science. 298:1039–1043. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie CR, Li Z, Sun HG, Wang FQ, Sun Y, Zhao
WX, Zhang S, Zhao WX, Wang XM and Yin ZY: Mutual regulation between
CHD5 and EZH2 in hepatocellular carcinoma. Oncotarget.
6:40940–40952. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sasaki M, Yamaguchi J, Itatsu K, Ikeda H
and Nakanuma Y: Over-expression of polycomb group protein EZH2
relates to decreased expression of p16 INK4a in
cholangiocarcinogenesis in hepatolithiasis. J Pathol. 215:175–183.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Paradis V: Histopathology of
hepatocellular carcinoma. Recent Results Cancer Res. 190:21–32.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Asada K, Ando T, Niwa T, Nanjo S, Watanabe
N, Okochi-Takada E, Yoshida T, Miyamoto K, Enomoto S, Ichinose M,
et al: FHL1 on chromosome X is a single-hit gastrointestinal
tumor-suppressor gene and contributes to the formation of an
epigenetic field defect. Oncogene. 32:2140–2149. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ding L, Niu C, Zheng Y, Xiong Z, Liu Y,
Lin J, Sun H, Huang K, Yang W, Li X and Ye Q: FHL1 interacts with
oestrogen receptors and regulates breast cancer cell growth. J Cell
Mol Med. 15:72–85. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang C, Liu X, Chen Z, Huang H, Jin Y,
Kolokythas A, Wang A, Dai Y, Wong DT and Zhou X: Polycomb group
protein EZH2-mediated E-cadherin repression promotes metastasis of
oral tongue squamous cell carcinoma. Mol Carcinog. 52:229–236.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cao Q, Yu J, Dhanasekaran SM, Kim JH, Mani
RS, Tomlins SA, Mehra R, Laxman B, Cao X, Yu J, et al: Repression
of E-cadherin by the polycomb group protein EZH2 in cancer.
Oncogene. 27:7274–7284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matsumoto M, Kawakami K, Enokida H, Toki
K, Matsuda R, Chiyomaru T, Nishiyama K, Kawahara K, Seki N and
Nakagawa M: CpG hypermethylation of human four-and-a-half LIM
domains 1 contributes to migration and invasion activity of human
bladder cancer. Int J Mol Med. 26:241–247. 2010.PubMed/NCBI
|
|
28
|
Sia D, Villanueva A, Friedman SL and
Llovet JM: Liver cancer cell of origin, molecular class, and
effects on patient prognosis. Gastroenterology. 152:745–761. 2017.
View Article : Google Scholar : PubMed/NCBI
|