Introduction
Osteosarcoma (OS), which is one of the common
primary bone tumors in children and adolescents is highly
aggressive and metastatic (1); it
comprises ~5% of all pediatric tumors and patients with pulmonary
metastasis present a poor prognosis (2). There are definite differences in the
mechanism between primary and metastatic OS, and numerous previous
studies have been performed to expound them (3–5).
Multiple signaling pathways, including the Notch
signaling pathway, are involved in cancer migration initiation
through the extracellular matrix (ECM) towards the bloodstream
(6). Survival of cancer cells in the
bloodstream is mediated by the integrin signaling pathway to
activate the phosphatidylinositol 3-kinase/protein kinase B
survival pathway (7). Signaling
pathways, including those of Src and Wnt, are involved in apoptosis
resistance throughout metastasis (8,9). In
addition, the immune system is also invaded and modulated for
survival advantage (10).
Subsequently, cell adhesion is arrested, and the cells extravasate
to their new environment and re-establish cell-cell adhesions
(11). There are numerous various
viewpoints regarding the mechanism underlying metastatic tumor cell
arrest to distant organ sites (12,13).
Angiogenesis is necessary for cancer cell proliferation and
metastasis (12); however, there
remain a number of unknown mechanisms, including the precise
mechanism underlying how cells alternate between exhibiting primary
and metastatic properties.
Therefore, in the present study, gene expression
profiles were used to further investigate the metastatic mechanism
of OS. The GSE49003 dataset considered in the present study was
previously analyzed by Diao et al (14) to identify differentially expressed
genes (DEGs) between metastatic and non-metastatic patients with
OS, and crucial microRNAs associated with OS metastasis, by merging
data from different metastatic or non-metastatic OS cell lines.
However, different types of metastatic OS may be regulated by
different molecular mechanisms. In addition, transcription factors
(TFs) may also serve a vital role in this pathomechanism.
Therefore, the dataset was reanalyzed in the present study to
emphasize the different mechanisms of different metastatic OS cell
lines.
Materials and methods
Gene expression profile data
The raw expression data (dataset GSE49003;
http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49003),
as provided by Endo-Munoz et al on July 18, 2013, were used
in the present study. The microarray expression profile was
obtained from two metastatic OS cell lines and two non-metastatic
OS cell lines. The metastatic KHOS and KRIB cell lines and the
non-metastatic HOS cell line were used with three-duplicated
samples, and gene expression data from each of these cell lines
were used. The platform of this dataset was GPL6847 Illumina
HumanHT-12 V3.0 expression beadchip (Illumina Inc., San Diego, CA,
USA).
Data preprocessing and DEG
screening
The Limma package (15) in Bioconductor was used to add probe
annotation files for each Illumina chip in order to preprocess the
expression profile. Background correction, quantile normalization
and probe summarization were performed to generate the gene
expression data matrix.
DEGs between KHOS and HOS and between KRIB and HOS
were determined using the Limma package. The differential
expression of genes were analyzed by Student's t-test, and those
with a false discovery rate adjusted P-value of <0.01 and |fold
change| ≥2 were screened.
Functional enrichment analysis of
DEGs
The Gene Ontology (GO) (16) project was established for gene
classifications by molecular function, biological process (BP) and
cellular component. DEGs of KHOS vs. HOS and KRIB vs. HOS were
functionally enriched by GO. In addition, the Kyoto Encyclopedia of
Genes and Genomes (KEGG) (17), which
has a become a major database resource for understanding high-level
functions of genes, was utilized. The default threshold of
P<0.01 was selected for the hypergeometric enrichment test.
Protein-protein interaction (PPI)
network construction and subnetwork mining
The Search Tool for the Retrieval of Interacting
Genes/Proteins is a biological database that provides known and
predicted PPIs (13). The tool was
applied in the present study to identify interacting protein pairs
between DEGs with a PPI score of 0.9. Subsequently, Cytoscape
software version 2.8.0 (18) was used
to visualize the constructed PPI network. Subnetworks (modules)
with a hypergeometric P-value <0.05 were identified by the
ClusterONE plugin (19) from
Cytoscape. Furthermore, the Database for Annotation, Visualization
and Integrated Discovery (20,21) was
utilized to perform the KEGG pathway cluster analyses of DEGs in
modules with P<0.05.
Overlapping gene analysis
Overlapping DEGs that were upregulated in KHOS and
KRIB cells compared with HOS cells were identified, and
downregulated DEGs that were common in KHOS and KRIB cells were
identified. Thereafter, KEGG signaling pathways of these two types
of overlapping genes were enriched.
Furthermore, based on the regulatory association
between TFs and target genes recorded in the University of
California Santa Cruz (UCSC) (22)
database, regulatory associations between TFs and their target DEGs
were identified.
Results
DEGs of various groups
A total of 1,552 (711 downregulated and 841
upregulated) and 1,330 DEGs (570 downregulated and 760 upregulated)
were obtained from the KHOS vs. HOS and KRIB vs. HOS comparisons,
respectively.
Functional enrichment analyses of
DEGs
Significant enriched terms of GO and KEGG pathway
enrichment analyses in KHOS and KRIB groups are presented in
Tables I and II, respectively. Upregulated genes of KHOS
were associated with GO-BP terms of anatomical structure
morphogenesis and cellular response to extracellular stimulus, and
the downregulated genes were enriched in BP terms of multicellular
organismal development and anatomical structure morphogenesis.
Using KEGG, focal adhesion, axon guidance and peroxisome
proliferator-activated receptor signaling pathways were
significantly enriched by upregulated genes of KHOS, and pathways
in cancer, tight junctions and cell adhesion molecules were
enriched by downregulated DEGs in KHOS cells (Table I). GO and pathway enrichment analyses
of KRIB were similar to those of KHOS, and the significantly
enriched GO term of KRIB overexpressed genes was anatomical
structure morphogenesis, and the significantly enriched pathway was
focal adhesion (Table II).
 | Table I.Gene ontology and pathway enrichment
analysis of genes in KHOS cells. |
Table I.
Gene ontology and pathway enrichment
analysis of genes in KHOS cells.
| Category
identity | Name | Count | P-value |
|---|
| Upregulated
genes |
|
|
|
|
GO:0009653 | Anatomical structure
morphogenesis | 132 |
8.44×10−7 |
|
GO:0031668 | Cellular response to
extracellular stimulus | 20 |
1.75×10−6 |
|
GO:0031669 | Cellular response to
nutrient levels | 18 |
1.98×10−6 |
|
GO:0071496 | Cellular response to
external stimulus | 24 |
2.72×10−6 |
|
GO:0072358 | Cardiovascular system
development | 61 |
4.90×10−6 |
|
Pathway:4510 | Focal adhesion | 19 |
5.07×10−4 |
|
Pathway:4360 | Axon guidance | 14 |
7.61×10−4 |
|
Pathway:250 | Alanine, aspartate
and glutamate metabolism |
6 |
1.66×10−3 |
|
Pathway:3320 | PPAR signaling
pathway |
9 |
2.08×10−3 |
|
Pathway:4512 | ECM-receptor
interaction | 10 |
2.35×10−3 |
| Downregulated
genes |
|
|
|
|
GO:0007275 | Multicellular
organismal development | 310 | 0.00 |
|
GO:0009653 | Anatomical
structure morphogenesis | 192 | 0.00 |
|
GO:0032502 | Developmental
process | 338 | 0.00 |
|
GO:0044767 | Single-organism
developmental process | 285 | 0.00 |
|
GO:0048468 | Cell
development | 158 | 0.00 |
|
Pathway:5200 | Pathways in
cancer | 35 |
2.61×10−5 |
|
Pathway:5146 | Amoebiasis | 14 |
1.07×10−3 |
|
Pathway:5144 | Malaria |
9 |
1.08×10−3 |
|
Pathway:4530 | Tight junction | 16 |
1.24×10−3 |
|
Pathway:4514 | Cell adhesion
molecules | 16 |
1.34×10−3 |
 | Table II.Gene ontology and pathway enrichment
analysis of genes in KRIB cells. |
Table II.
Gene ontology and pathway enrichment
analysis of genes in KRIB cells.
| Category
identity | Name | Count | P-value |
|---|
| Upregulated
genes |
|
|
|
|
GO:0009653 | Anatomical
structure morphogenesis | 106 |
3.88×10−6 |
|
GO:0001944 | Vasculature
development | 38 |
9.85×10−6 |
|
GO:0010646 | Regulation of cell
communication | 108 |
1.24×10−5 |
|
GO:0071294 | Cellular response
to zinc ion |
5 |
1.50×10−5 |
|
GO:0023051 | Regulation of
signaling | 107 |
1.88×10−5 |
|
Pathway:4510 | Focal adhesion | 16 |
7.36×10−4 |
|
Pathway:480 | Glutathione
metabolism |
7 |
1.05×10−3 |
|
Pathway:4360 | Axon guidance | 11 |
3.03×10−3 |
|
Pathway:3320 | PPAR signaling
pathway |
7 |
7.33×10−3 |
|
Pathway:4520 | Adherens
junction |
7 |
9.19×10−3 |
| Downregulated
genes |
|
|
|
|
GO:0000902 | Cell
morphogenesis | 104 | 0.00 |
|
GO:0000904 | Cell morphogenesis
involved in differentiation | 86 | 0.00 |
|
GO:0006928 | Cellular component
movement | 132 | 0.00 |
|
GO:0007275 | Multicellular
organismal development | 299 | 0.00 |
|
GO:0007399 | Nervous system
development | 153 | 0.00 |
|
Pathway:4514 | Cell adhesion
molecules | 19 |
1.14×10−5 |
|
Pathway:5144 | Malaria | 11 |
1.74×10−5 |
|
Pathway:5146 | Amoebiasis | 16 |
2.83×10−5 |
|
Pathway:5200 | Pathways in
cancer | 32 |
4.49×10−5 |
|
Pathway:4512 | ECM-receptor
interaction | 13 |
1.40×10−4 |
PPI network and subnetwork
A total of 856 pairs of interacting proteins and 495
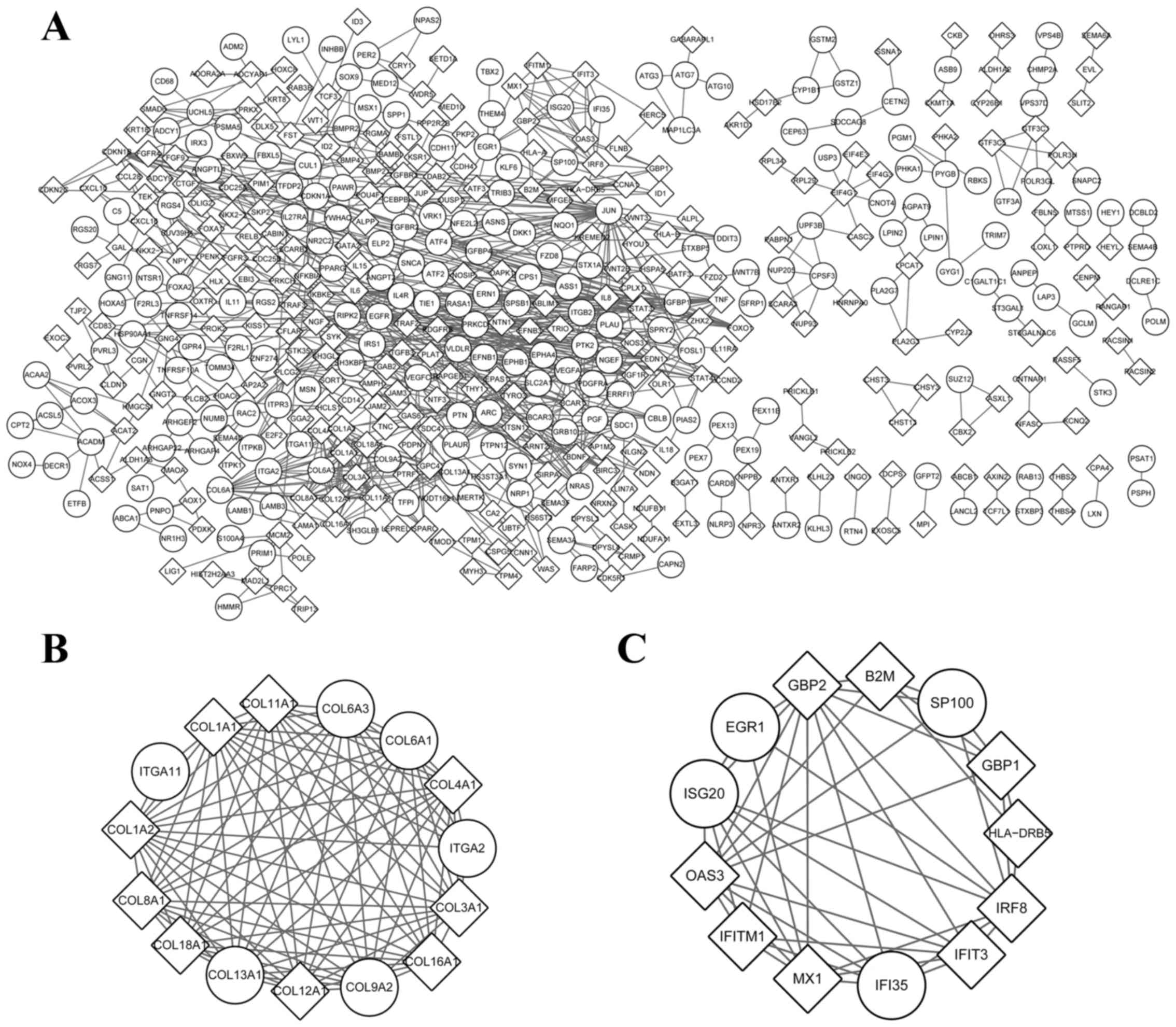
nodes were noted in the PPI network of KHOS (Fig. 1A). The top 6 nodes with a greater
degree of connectivity compared with the others were jun
proto-oncogene (JUN; degree=33), vascular endothelial growth factor
A (VEGFA; degree=29), epidermal growth factor receptor (EGFR;
degree=23), cyclin-dependent kinase inhibitor 1A (degree=19),
interleukin 6 (IL-6; degree=19) and signal transducer and activator
of transcription 3 (STAT3; degree=17). Furthermore, two modules
with P<0.05 were obtained (Fig. 1B and
C). A total of 9 genes, including 7 collagen molecules and 2
integrin family members involved in module 1 were significantly
enriched in terms of ECM-receptor interaction and focal
adhesion.
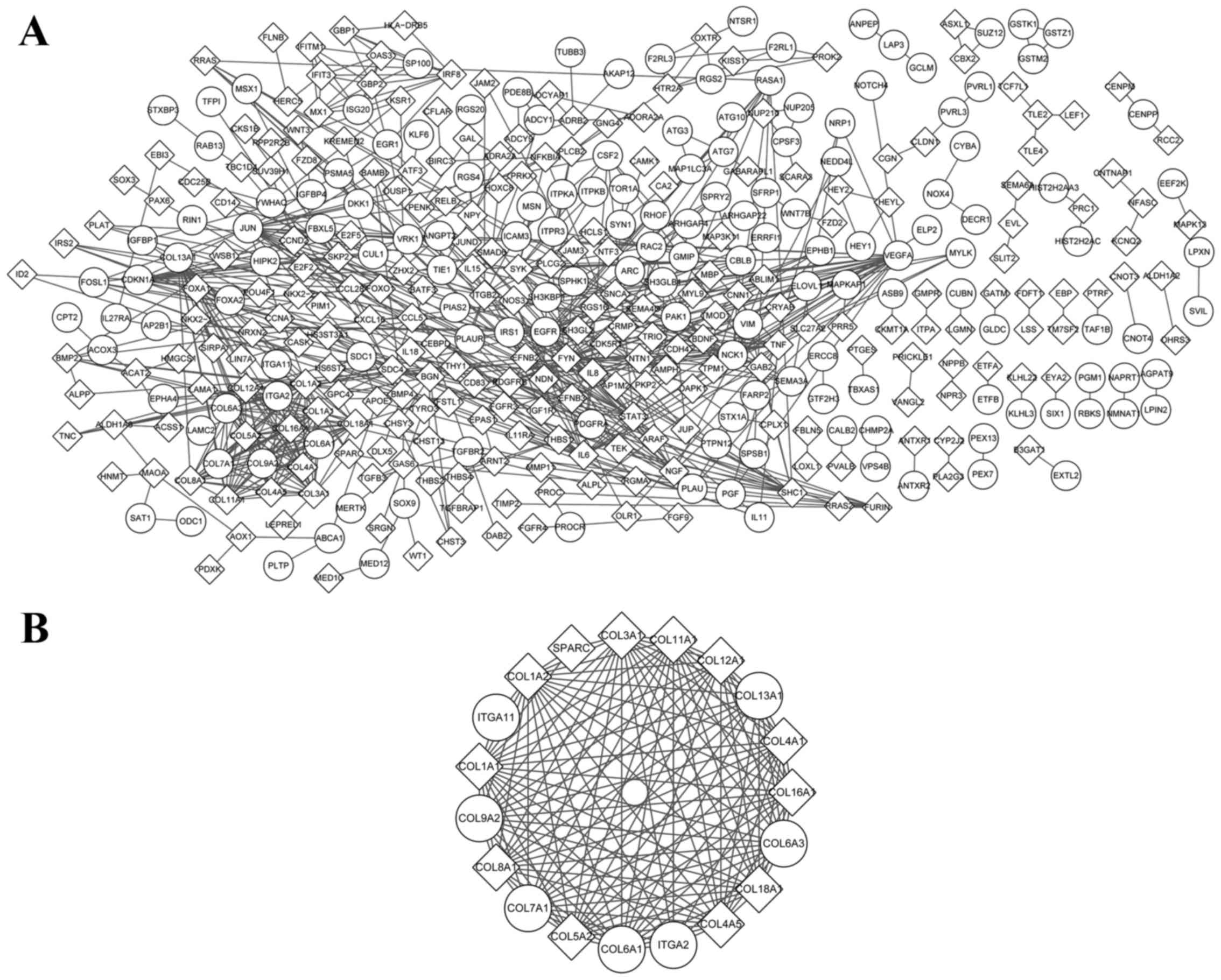
There were 719 interacting pairs and 397 nodes of
the PPI network of KRIB (Fig. 2).
VEGFA (degree=29), Fyn oncogene related to Src, FGR, Yes
(degree=25), JUN (degree=23), EGFR (degree=22) and IL-6
(degree=19), were the top 5 hub nodes. In addition, one module was
screened (Fig. 2), and the
significant pathways enriched by its genes were ECM-receptor
interaction and focal adhesion.
KEGG pathway enrichment and
transcriptional regulatory network analysis of overlapping
DEGs
A total of 421 upregulated and 595 downregulated
overlapping genes were obtained. Pathway of focal adhesion,
glutathione metabolism and endocytosis were enriched by their
upregulated genes, whereas pathways in cancer, focal adhesion and
cell adhesion molecules were enriched by downregulated genes. In
addition, the mitogen-activated protein kinase (MAPK) and
transforming growth factor-β (TGF-β) signaling pathways were
enriched by overlapping genes with decreased expression levels
(Table III).
 | Table III.Kyoto Encyclopedia of genes and
genomes pathway of overlapping genes. |
Table III.
Kyoto Encyclopedia of genes and
genomes pathway of overlapping genes.
| Category
identity | Name | Count | P-value |
|---|
| Upregulated
overlapping gene |
|
|
|
|
4510 | Focal adhesion | 13 |
2.96×10−3 |
|
480 | Glutathione
metabolism | 5 |
3.11×10−2 |
|
4144 | Endocytosis | 10 |
3.18×10−2 |
|
4360 | Axon guidance | 8 |
3.46×10−2 |
|
4520 | Adherens
junction | 6 |
3.67×10−2 |
| Downregulated
overlapping gene |
|
5200 | Pathways in
cancer | 27 |
2.52×10−4 |
|
4510 | Focal adhesion | 17 |
3.93×10−3 |
|
4514 | Cell adhesion
molecules | 13 |
4.25×10−3 |
|
4512 | ECM-receptor
interaction | 10 |
4.40×10−3 |
|
4010 | MAPK signaling
pathway | 19 |
1.26×10−2 |
|
4350 | TGF-β signaling
pathway | 9 |
1.72×10−2 |
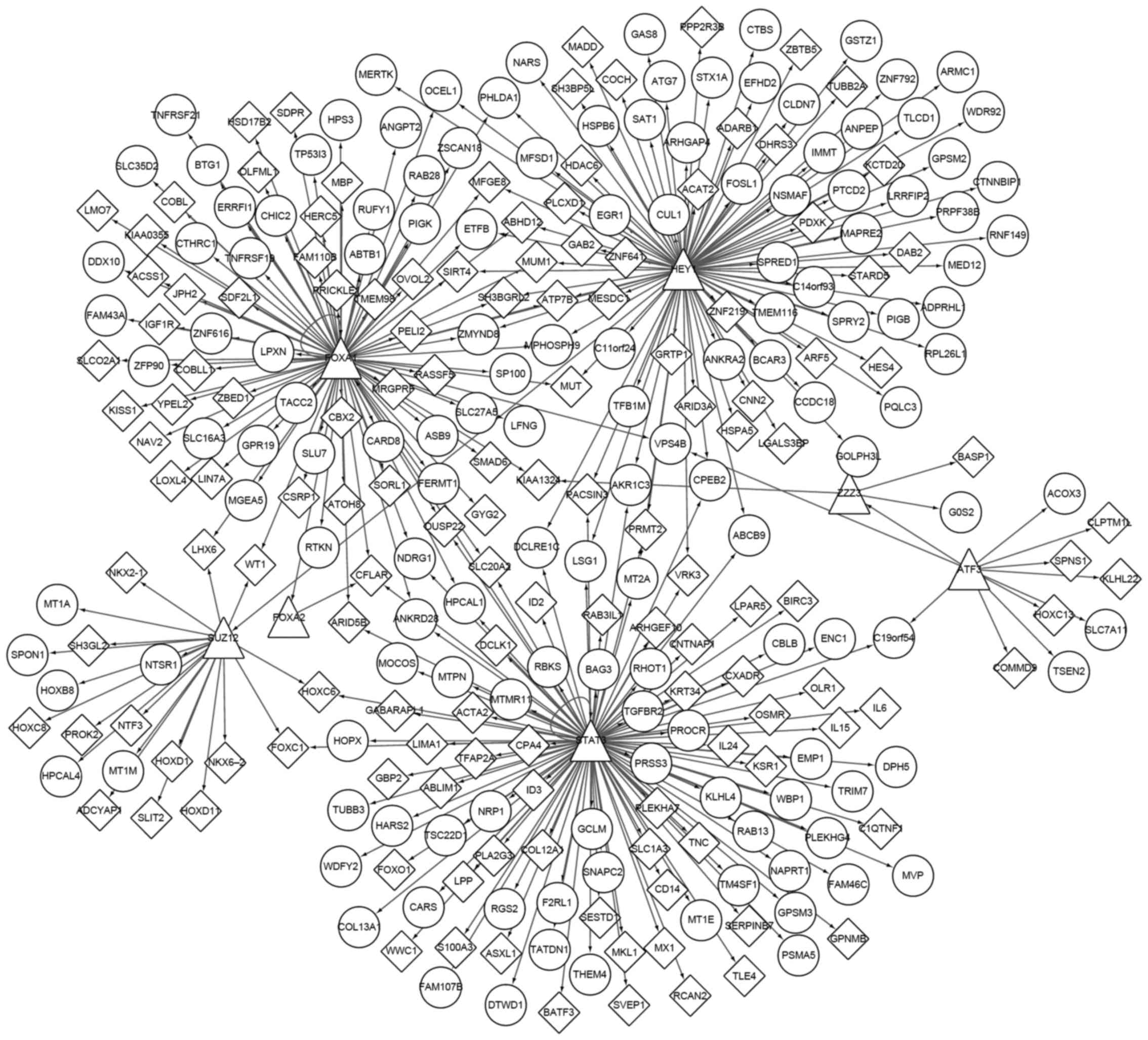
Fig. 3 demonstrates
the transactional regulatory network of overlapping DEGs. A total
of 7 TFs, including activating transcription factor 3, forkhead box
(FOX)A1, STAT3, FOXA2, hes-related family bHLH transcription factor
with YRPW motif 1 (HEY1), SUZ12 polycomb repressive complex 2
subunit and zinc finger ZZ-type containing 3 were identified. The
first three of these were downregulated and the others were
overexpressed.
Discussion
Due to the high risk for recurrence and the poor
survival of metastatic OS patients (23), two different metastatic OS cell lines
and one non-metastatic cell lines were used to investigate the
mechanism underlying metastasis. The GO and pathway enrichment
analysis results were similar of these two metastatic types of OS.
Besides, a large number of overlapping genes, including 7 TFs, were
obtained. These genes and biological functions may be important in
OS tumor metastasis.
The most prominent node in the PPI networks of KHOS
and KRIB was VEGFA, which is a growth factor that is activated in
vasculogenesis, angiogenesis and endothelial cell growth (24). It is not only a commonly upregulated
angiogenic factor during tumor growth, but also the most frequent
lymphangiogenic factor contributing to the lymphatic metastasis
(25). A previous study suggested
that VEGFA promoted tumor growth and metastasis via the VEGF-VEGFR1
signaling pathway (26). In addition,
EGFR, which is a member of the EGF family, was another common hub
node with a high degree (27). It was
revealed that EGFR participated in the promotion of cell division,
migration and angiogenesis, and inhibited cell apoptosis (28). Furthermore, these two genes were
enriched in the development and differentiation process, including
anatomical structure morphogenesis and vasculature development.
Yang et al (29) demonstrated
that Twist, which is a master regulator of embryonic morphogenesis,
contributed to metastasis by promoting an epithelial-mesenchymal
transition (29). Therefore, tumor
metastasis is the recapitulation of morphogenetic processes
(30). Morphogenetic factors,
including hepatocyte growth factor/scatter factor have
metastasis-promoting effects on breast carcinoma cells (31). Based on these studies, VEGFA and EGFR
may be factors involved in the morphogenetic processes to promote
tumor metastasis.
The KEGG pathway enrichment analysis results of
module 1 of the KHOS group was similar to that of module 1 of the
KRIB group. ECM-receptor interaction and focal adhesion were the
observed pathways enriched by multiple collagen genes and integrin
genes. These results were in accordance with previous studies
(32,33) and demonstrated the accuracy and
practicability of them. Additionally, functional enrichment
analysis of overlapping genes of these two group would be more
direct to exhibit commonality of metastatic OS.
Focal adhesion was the significant pathway enriched
by upregulated and downregulated overlapping genes. Furthermore,
these downregulated genes remained enriched in signaling pathways
that included the MAPK and TGF-β signaling pathways. The former
pathway serves an important role in the early metastasis of the
tumor (34,35), whereas the latter has positive and
negative effects on carcinogenesis (36), and further investigation for is
required to fully elucidate the molecular mechanism.
Finally, a total of 7 TFs, which regulated multiple
genes, were screened. Among them, FOXA1, STAT3 and HEY1 were more
important in the regulatory network. Candy et al (37) indicated that overexpressed HEY1 was a
negative prognostic factor for colorectal cancer, and it is closely
associated with vascular and perineural invasion and lymph node
metastasis. Therefore, the upregulated HEY1 may be associated with
OS metastasis. In addition, FOXA1 was revealed to be a participant
in embryonic development, mediating tissue-specific gene expression
in differentiated tissues (38). This
gene encoded protein may be another important factor involved in
the similar process of morphogenesis.
In conclusion, recapitulation of morphogenetic
processes, a disordered signaling system, and activated cell
adhesion and migration were involved in the facilitated tumor
metastasis. Furthermore, morphogenesis and vasculature development
induced angiogenesis, and tumor growth and adhesion, or even the
lymph node metastasis induced the metastatic OS. The present study
further illustrated the OS tumorigenesis mechanism and displayed
the crucial roles of genes such as VEGFA, EGFR and HEY1. The
practical clinical significance has not yet been clearly described
and requires further investigation.
References
|
1
|
Wu J, Hsieh MY, Wang Y, Roberts E, Vallejo
D, Lee A and Ermel R: Antitumor properties of ouabain in lipid
double emulsion in orthotopic canine osteosarcoma xenografted mouse
model. Cancer Res. 74:54122014. View Article : Google Scholar
|
|
2
|
Roos A, Yustein JT and Donehower L:
Oncogenic role of Runx2 in the development of osteosarcoma. Cancer
Res. 73:5036. 2013. View Article : Google Scholar
|
|
3
|
Pompetti F, Rizzo P, Simon RM, Freidlin B,
Mew DJ, Pass HI, Picci P, Levine AS and Carbone M: Oncogene
alterations in primary, recurrent, and metastatic human bone
tumors. J Cell Biochem. 63:37–50. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Oda Y, Naka T, Takeshita M, Iwamoto Y and
Tsuneyoshi M: Comparison of histological changes and changes in
nm23 and c-MET expression between primary and metastatic sites in
osteosarcoma: A clinicopathologic and immunohistochemical study.
Human Pathol. 31:709–716. 2000. View Article : Google Scholar
|
|
5
|
Liang S, Ren Z, Han X, Yang J, Shan L, Li
L, Wang B, Zhang Q, Mu T, Chen K, et al: PLA2G16 expression in
human osteosarcoma is associated with pulmonary metastasis and poor
prognosis. PLoS One. 10:e01272362015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hughes DP: How the NOTCH pathway
contributes to the ability of osteosarcoma cells to
metastasizePediatric and Adolescent Osteosarcoma. Jaffe N, Bruland
OS and Bielack S: 152. Springer; New York, NY: pp. 479–496. 2010,
View Article : Google Scholar
|
|
7
|
Casar B, Rimann I, Kato H, Shattil SJ,
Quigley JP and Deryugina EI: In vivo cleaved CDCP1 promotes early
tumor dissemination via complexing with activated β1 integrin and
induction of FAK/PI3K/Akt motility signaling. Oncogene. 33:255–268.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim LC, Song L and Haura EB: Src kinases
as therapeutic targets for cancer. Nat Rev Clin Oncol. 6:587–595.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rubin EM, Guo Y, Tu K, Xie J, Zi X and
Hoang BH: Wnt inhibitory factor 1 decreases tumorigenesis and
metastasis in osteosarcoma. Mol Cancer Ther. 9:731–741. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Krishnan K, Khanna C and Helman LJ: The
biology of metastases in pediatric sarcomas. Cancer J. 11:306–313.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Eccles SA and Welch DR: Metastasis: Recent
discoveries and novel treatment strategies. Lancet. 369:1742–1757.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fidler IJ: The pathogenesis of cancer
metastasis: The ‘seed and soil’ hypothesis revisited. Nat Rev
Cancer. 3:453–458. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Diao CY, Guo HB, Ouyang YR, Zhang HC, Liu
LH, Bu J, Wang ZH and Xiao T: Screening for metastatic osteosarcoma
biomarkers with a DNA microarray. Asian Pac J Cancer Prev.
15:1817–1822. 2013. View Article : Google Scholar
|
|
15
|
Smyth GK: Limma: Linear models for
microarray dataBioinformatics and computational biology solutions
using R and Bioconductor. Gentleman R, Carey VJ, Huber W, Irizarry
RA and Dudoit S: Springer; New York: pp. 397–420. 2005, View Article : Google Scholar
|
|
16
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene Ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nepusz T, Yu H and Paccanaro A: Detecting
overlapping protein complexes in protein-protein interaction
networks. Nat Methods. 9:471–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2008. View Article : Google Scholar
|
|
21
|
da Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Meyer LR, Zweig AS, Hinrichs AS, Karolchik
D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al:
The UCSC Genome Browser database: Extensions and updates 2013.
Nucleic Acids Res. 41:(Database issue). D64–D69. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
PosthumaDeBoer J, Witlox MA, Kaspers GJ
and Van Royen BJ: Molecular alterations as target for therapy in
metastatic osteosarcoma: A review of literature. Clin Exp
Metastasis. 28:493–503. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liang D, Chang JR, Chin AJ, Smith A, Kelly
C, Weinberg ES and Ge R: The role of vascular endothelial growth
factor (VEGF) in vasculogenesis, angiogenesis, and hematopoiesis in
zebrafish development. Mech Dev. 108:29–43. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Björndahl MA, Cao R, Burton JB,
Brakenhielm E, Religa P, Galter D, Wu L and Cao Y: Vascular
endothelial growth factor-a promotes peritumoral lymphangiogenesis
and lymphatic metastasis. Cancer Res. 65:9261–9268. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu W, Xu J, Wang M, Wang Q, Bi Y and Han
M: Tumor-derived vascular endothelial growth factor (VEGF)-A
facilitates tumor metastasis through the VEGF-VEGFR1 signaling
pathway. Int J Oncol. 39:1213–1220. 2011.PubMed/NCBI
|
|
27
|
Zimmermann M, Zouhair A, Azria D and
Ozsahin M: The epidermal growth factor receptor (EGFR) in head and
neck cancer: Its role and treatment implications. Radiation Oncol.
1:112006. View Article : Google Scholar
|
|
28
|
Italiano A, Saint-Paul MC, Caroli-Bosc FX,
François E, Bourgeon A, Benchimol D, Gugenheim J and Michiels JF:
Epidermal growth factor receptor (EGFR) status in primary
colorectal tumors correlates with EGFR expression in related
metastatic sites: Biological and clinical implications. Ann Oncol.
16:1503–1507. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang J, Mani SA, Donaher JL, Ramaswamy S,
Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A and
Weinberg RA: Twist, a master regulator of morphogenesis, plays an
essential role in tumor metastasis. Cell. 117:927–939. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Berx G, Raspé E, Christofori G, Thiery JP
and Sleeman JP: Pre-EMTing metastasis? Recapitulation of
morphogenetic processes in cancer. Clin Exp Metastasis. 24:587–597.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meiners S, Brinkmann V, Naundorf H and
Birchmeier W: Role of morphogenetic factors in metastasis of
mammary carcinoma cells. Oncogene. 16:9–20. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lu P, Weaver VM and Werb Z: The
extracellular matrix: A dynamic niche in cancer progression. J Cell
Biol. 196:395–406. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Felding-Habermann B: Integrin adhesion
receptors in tumor metastasis. Clin Exp Metastasis. 20:203–213.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Khanna C, Wan X, Bose S, Cassaday R, Olomu
O, Mendoza A, Yeung C, Gorlick R, Hewitt SM and Helman LJ: The
membrane-cytoskeleton linker ezrin is necessary for osteosarcoma
metastasis. Nat Med. 10:182–186. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wan X, Mendoza A, Khanna C and Helman LJ:
Rapamycin inhibits ezrin-mediated metastatic behavior in a murine
model of osteosarcoma. Cancer Res. 65:2406–2411. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Derynck R, Akhurst RJ and Balmain A:
TGF-beta signaling in tumor suppression and cancer progression. Nat
Genet. 29:117–129. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Candy P, Phillips MR, Redfern AD, Colley
SM, Davidson JA, Stuart LM, Wood BA, Zeps N and Leedman PJ:
Notch-induced transcription factors are predictive of survival and
5-fluorouracil response in colorectal cancer patients. Br J Cancer.
109:1023–1030. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bernardo GM and Keri RA: FOXA1: A
transcription factor with parallel functions in development and
cancer. Biosci Rep. 32:113–130. 2012. View Article : Google Scholar : PubMed/NCBI
|