Introduction
Hepatocellular carcinoma (HCC) accounts for ~80% of
all liver cancer cases, and is one of the most common causes of
cancer-associated mortality worldwide (1,2). The
prevalence rate of HCC in China is particularly high, but continues
to increase in numerous western countries (1,3). Despite
HCC being was one of the first cancers to be linked
epidemiologically to a definite risk factor, the underlying
mechanisms of HCC pathogenesis remain unclear (4). The risk factors for HCC vary by
location. In China, Hepatitis B or Hepatitis C virus infections are
the main risk factors (5). The
survival rate of patients with HCC has been extended due to
progress in liver transplantation and other treatments; however,
the insensitivity of chemotherapeutic drugs, cancer recurrence and
metastasis continue to contribute to a poor prognosis (6). Thus, the identification of therapeutic
targets and translation of molecular studies of HCC into clinical
practice is urgently required.
MicroRNAs (miRNAs) are a class of small non-coding
RNAs, which are ~22 nucleotides in length (7–9). A number
of miRNAs have been revealed to be involved in the pathogenesis of
HCC (10–26). The roles of miR-1271 in numerous types
of cancers have previously been investigated. For example, in
gastric cancer, miR-1271 inhibited cell proliferation, invasion and
epithelial-mesenchymal transition (EMT) by targeting forkhead box
Q1 (FOXQ1) (27).
Additionally, in oral squamous cell carcinoma, miR-1271 inhibited
cell growth and metastasis by targeting anaplastic lymphoma
receptor tyrosine kinase (28). In
HCC, a previous study demonstrated that miR-96, miR-129-1-3p,
miR-1291, miR-1303 and miR-1271 differentially regulated Glypican-3
(GPC3) expression levels in HCC cells and that the
upregulation of GPC3 was associated with a concomitant
downregulation of its repressor miR-1271 (29). However, the roles served by miR-1271
in HCC remain unclear.
The present study analyzed the expression level of
miR-1271 in HCC tissues and determined the in vitro function
of miR-1271. The aim of the study was to provide useful evidence
and suggestions for additional investigations.
Patients and methods
Patients
A total of 22 HCC tissue specimens were collected
from the Institute of Liver Disease (The Fourth Hospital of Huaian
City, Huaian, China). Tissue samples were immediately frozen in
liquid nitrogen following isolation. Written informed consent was
obtained from each patient prior to enrollment in the present
study. The present study was approved by the Ethics Committee of
The Fourth Hospital of Huaian. The specimens were processed using a
hematoxylin and eosin staining protocol (30). Senior pathologists of The Fourth
Hospital of Huaian (Huaian, China) evaluated the histological
features of the specimens using LED microscopy at magnifications,
×100 and ×400 (DM300 Microscope, Leica Microsystems GmbH, Wetzlar,
Germany).
Cell culture and antibodies
HCC Huh7 and HepG2 cells were purchased from the
Cell Store of Shanghai Jiaotong University (Shanghai, China). The
Huh7 cell line was maintained in RPMI-1640 medium supplemented with
10% fetal bovine serum (FBS; PAA Laboratories; GE Healthcare Life
Sciences, Chalfont, UK). The human HCC HepG2 cell line was
maintained in Dulbecco's modified Eagle's medium supplemented with
10% FBS (PAA Laboratories; GE Healthcare Life Sciences). Antibodies
specific to FOXQ1 (Anti-FOXQ1 antibody; cat. no., ab51340, working
concentration: 2.5 µg/ml) and β-actin (Anti-β-Actin antibody; cat.
no., ab8226, working concentration: 0.05 µg/ml) were supplied by
Abcam (Cambridge, UK).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), was transcribed to cDNA using an EasyScript First-Strand cDNA
Synthesis SuperMix kit (TransGen Biotech, Inc., Beijing, China)
according to the manufacturer's protocol. The qPCR was carried out
to detect the expression of miR-1271 and FOXQ1 mRNA by using
SYBR-Green qPCR SuperMix-UDG with ROX (Invitrogen; Thermo Fisher
Scientific, Inc.). The mRNA and miRNA primers were synthesized by
Shanghai Genepharma Co. Ltd. (Shanghai, China). U6 and GAPDH were
used as the references for detecting the expression of miR-1271 or
FOXQ1, respectively. The primers used were as follows:
miR-1271-forward (F), 5′-CAGCACTTGGCACCTAGCA-3′, miR-1271-reverse
(R), 5′-TATGGTTGTTCTCCTCTCTGTCTC-3′; FOXQ1-F,
5′-GTGATTTCTTGCTATTGACCGATG-3′, FOXQ1-R,
5′-GCCCAAGGAGACCACAGTTAGA-3′; U6-F, 5′-AGAGCCTGTGGTGTCCG-3′, U6-R,
5′-CATCTTCAAAGCACTTCCCT-3′; and GAPDH-F,
5′-CATCACCATCTTCCAGGAGCG-3′, GAPDH-R, 5′-TGACCTTGCCCACAGCCTTG-3′.
Relative expression levels were calculated by the 2−ΔΔCt
method (31–33). This experiment was repeated at least
three times.
miR mimics and
miR-antisense-oligonucleotides (AOS)
miR-1271 mimics, miR-1271 antisense oligonucleotides
and the negative control (NC) were purchased from Qiagen, Inc.
(Valencia, CA, USA). miR-ASOs, miR mimics and the respective NC
were transfected into cells at a concentration of 50 nM using
Lipofectamine® 2000 transfection reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) at 37°C for 30 min, according to
the manufacturer's protocol. After transfection, the cells were
maintained at 37°C for 48 h for additional experiments.
MTT assays
After 24 h of transfection of miR-1271 mimics and
anti-miR-940, MDA-MB-231 and BT-549 cells (5×103/well)
were seeded into 96-well plates. Then, MTT experiments were
performed as described previously (34).
Apoptosis assay for flow
cytometry
Huh7 and HepG2 cells were seeded at a density of
0.3×106 overnight, then these cells were harvested and
washed with PBS three times. The Fc receptor was blocked by 3%
fetal bovine albumin at room temperature for 30 min. Cells were
stained with Anti-Annexin V antibody (1:500, cat. no., ab63556;
Abcam) at room temperature for 20 min.
Bioinformatics methods
The miR-1271 targets predicted by algorithms were
acquired from the Target Scan Human database (http://www.targetscan.org/vert_61/) (35–38).
FOXQ1 3′untranslated region (UTR)
reporter analysis
The reporter genes analysis was performed by Chengdu
Technology & Market Co., Ltd. (Chengdu, China). Briefly, the
3′UTR of FOXQ1 was amplified and cloned into the downstream
FOXQ1 3′UTR reporter plasmids (pRL-FOXQ1), using the
following primers: FOXQ1-3′UTR-HF,
5′-AATTCTAGGCGATCGCTCGAGGACTACTGTTTGGGGTTTCTGG-3′; FOXQ1-3′UTR-HR,
5′-GCGGCCGCTCTAGGTTTAAACACACTTGCTTTCAAGGCAGTGG-3′. The thermocycler
conditions were as follows: 95°C for 3 min, then 95°C for 30 sec,
53°C for 30 sec and 72°C for 30 sec for 28 cycles, followed by 72°C
for 2 min. Mutants of FOXQ1 3′UTR were generated using the
Site-Directed Mutagenesis kit (Shanghai Shengong Biotechnology
Company, Shanghai, China). For the luciferase reporter assay, the
cells were co-transfected with miR-1271 mimics, control and
reporter plasmid or the mutant 3′UTR, together with the controls
using Lipofectamine® 2000 at 37°C for 30 min. At 48 h
following transfection, cells were analyzed using the Dual
Luciferase reporter assay system (Promega Corporation, Madison, WI,
USA) as described previously (39).
Western blot analysis
Cells (5×106 cells) were lysed using 50
µl M-PER protein extraction reagent (Pierce; Thermo Fisher
Scientific, Inc.) supplemented with 10 µl protease inhibitor
cocktail (EMD Millipore, Billerica, MA, USA) on ice for 45 min
(40). Protein quantification was
evaluated using a BCA assay and absorbance at 280 nm (Pierce;
Thermo Fisher Scientific, Inc.). Protein (20 µg) was separated
using 10% SDS-PAGE gels, then electrophoretically transferred onto
nitrocellulose membranes (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The membranes were blocked with 5% milk/TBS Tween-20 at 4°C
overnight, and then incubated with antibodies specific to FOXQ1
(Anti-FOXQ1 antibody; cat. no., ab51340, working concentration, 2.5
µg/ml) at 4°C overnight, followed by incubation with a horseradish
peroxidase-conjugated secondary antibody (goat anti-rabbit IgG
H&L, cat. no., ab6721, 1:1,000) at room temperature for 2 h
(Abcam, Cambridge, United Kingdom). The membranes were washed with
TBS three times and then images were visualized by
chemiluminescence and LabWorks Image Acquisition and Analysis
Software 2 (UVP LLC, Upland, CA, USA).
Statistical analysis
Statistical analyses were conducted using SPSS
version 18 (SPSS, Inc., Chicago, IL, USA). Data are presented as
the mean ± standard deviation from three experimental repeats.
Two-tailed Student's t-test was used to analyze the difference
between two groups. One-way ANOVA was used to analyze the
difference between three groups. Multiple comparison between the
groups was performed using the Student Newman Keuls method. The
Wilcoxon matched-pairs signed rank test was used to determine the
difference between the expression level of miR-1271 in HCC tissues
and the matched controls. Kaplan-Meier analysis was used to
evaluate the overall survival rate of patients with HCC according
to the expression level of miR-1271 in HCC tissues. The correlation
analysis was performed by two-tailed Pearson's correlation
coefficient analysis. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-1271 is frequently downregulated
in HCC tissues
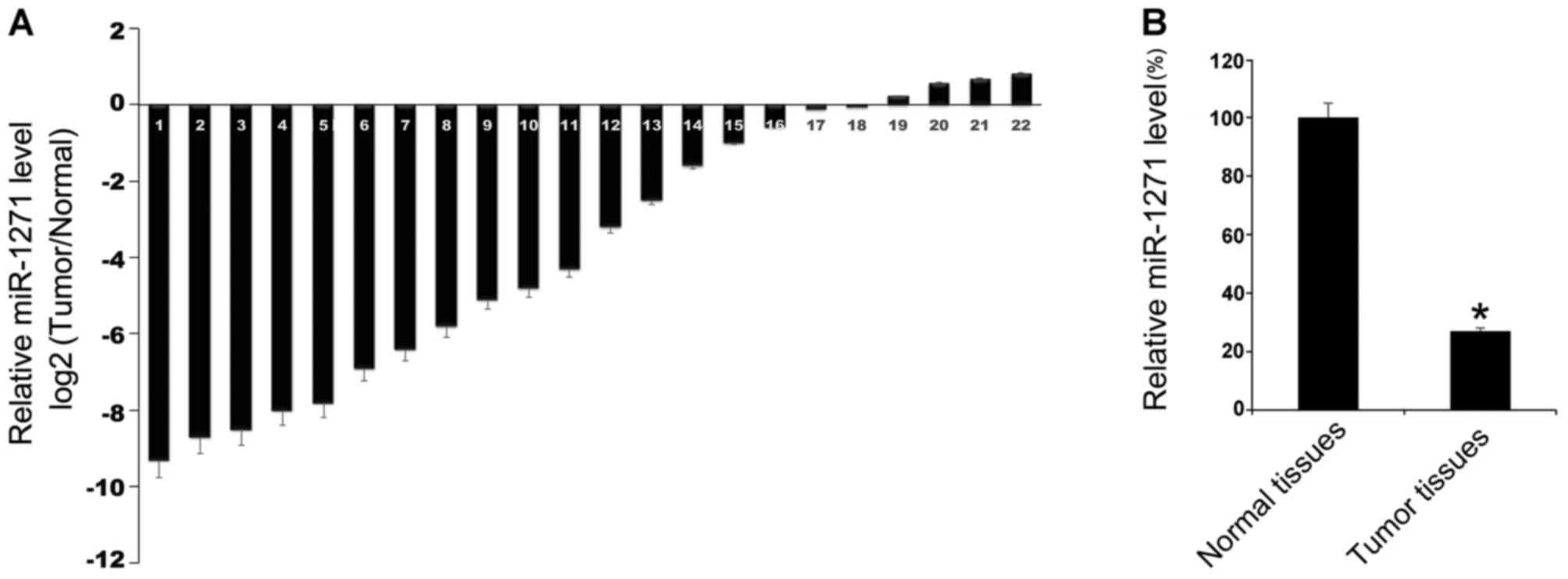
Initially, miR-1271 expression levels in tumor
tissues and the matched adjacent normal tissues of 22 patients with
HCC were evaluated by RT-qPCR. The results indicated that in 22
patients with HCC, compared with the matched non-tumorous tissues,
the majority of HCC tissues exhibited lower miR-1271 expression
levels compared with the corresponding tumor-adjacent normal
tissues (Fig. 1A). Furthermore, the
22 HCC tissues demonstrated a lower mean expression level of
miR-1271 compared with in normal tissues (Fig. 1B). These results indicated that
miR-1271 may serve an important role in HCC.
Overexpression of miR-1271 inhibited
cell growth and promoted apoptosis of cells
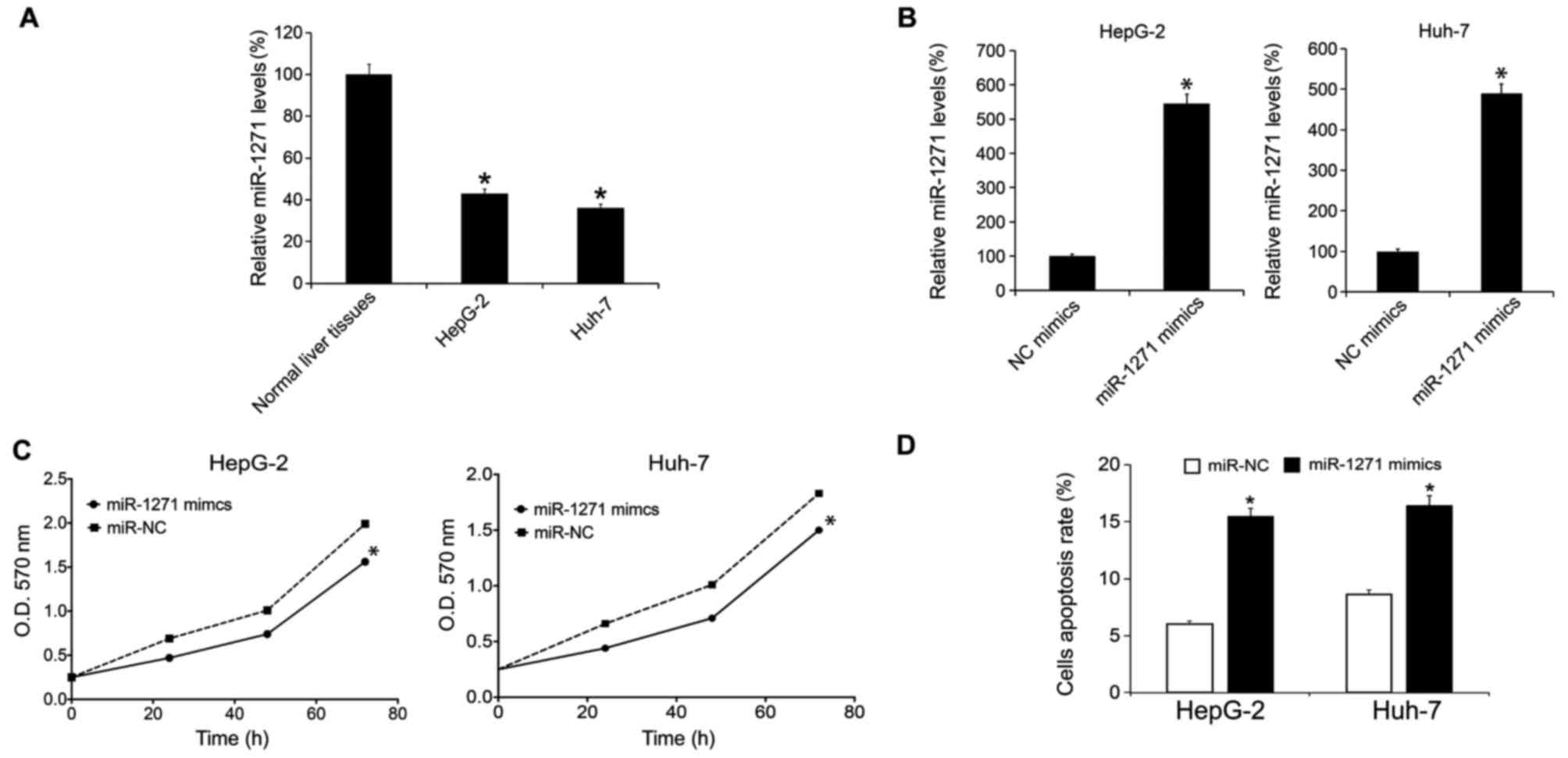
The miR-1271 expression levels in HCC cell lines,
HepG-2 and Huh-7, were analyzed by RT-qPCR. The results indicated
that there were lower expression levels of miR-1271 in HepG-2 and
Huh-7 cells compared with normal liver tissue (Fig. 2A). The present study subsequently
overexpressed miR-1271 by performing miR-1271 mimic transfection.
Following 48 h, the miR-1271 expression levels in HepG-2 and Huh-7
cells were evaluated by RT-qPCR. It was revealed that miR-1271
mimic transfection upregulated miR-1271 levels in HepG-2 and Huh-7
cells (Fig. 2B). Following the
transfection, cellular proliferation rate was determined by MTT
analysis. The results suggested that transfection of miR-1271
mimics inhibited HepG-2 and Huh-7 cell growth (Fig. 2C). A total of 48 h subsequent to
transfection, cells were prepared for apoptosis analysis and it was
demonstrated that transfection of miR-1271 mimics induced an
increase in the levels of cellular apoptosis (Fig. 2D).
Downregulation of miR-1271 promoted
cell growth and inhibited apoptosis
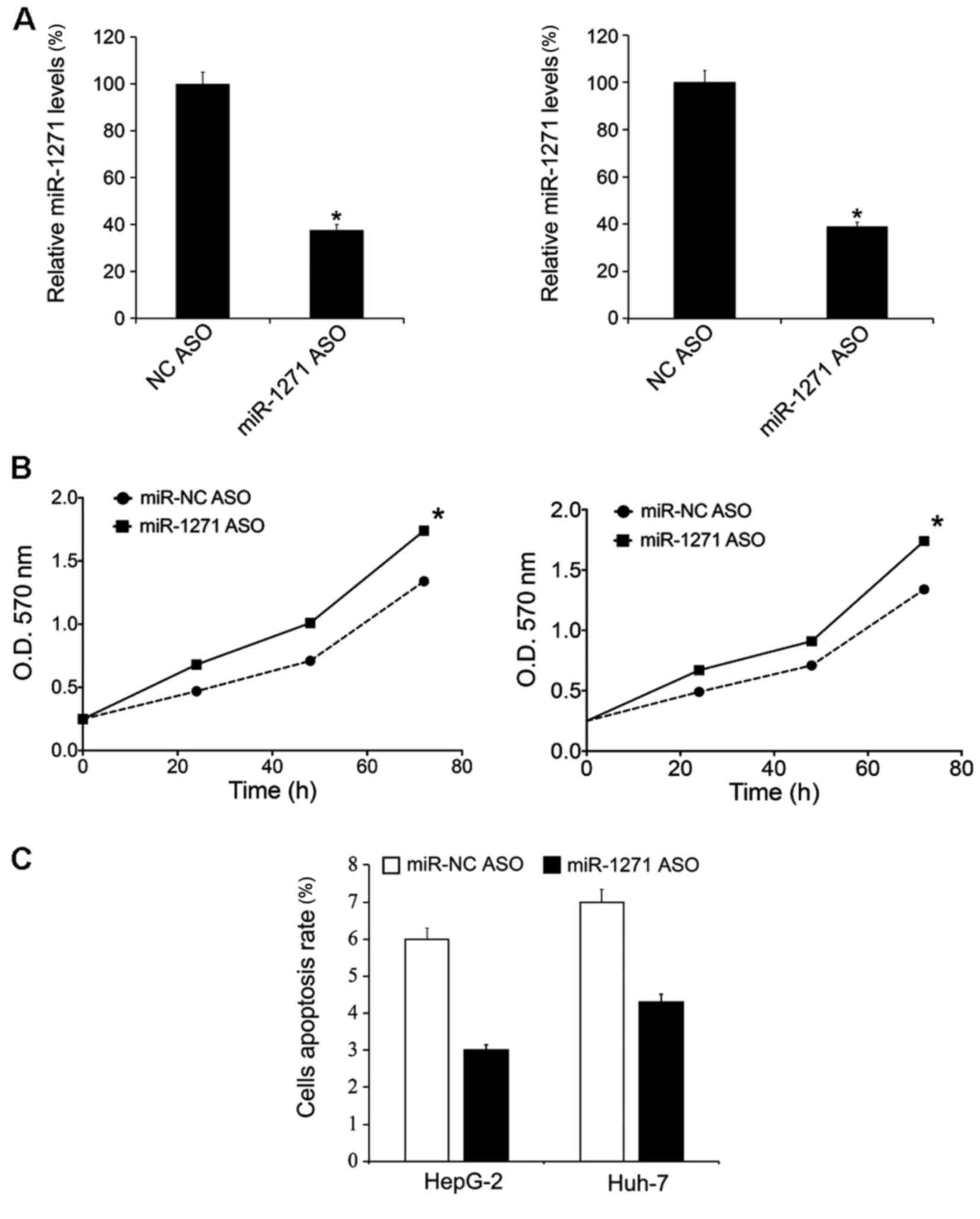
The present study downregulated miR-1271 expression
levels by transfection of miR-1271 ASO. Following 48 h, the
miR-1271 expression levels in HepG-2 and Huh-7 cells were
determined by RT-qPCR and it was revealed that miR-1271 ASO
transfection downregulated the miR-1271 levels in HepG-2 and Huh-7
cells (Fig. 3A). Following the
transfection, cellular proliferation rate was evaluated by MTT
analysis. The present study demonstrated that transfection of
miR-1271 mimics promoted HepG-2 and Huh-7 cell growth (Fig. 3B). Following a 48 h transfection,
cells were prepared for apoptosis analysis and the results revealed
that transfection of miR-1271 mimics inhibited apoptosis (Fig. 3C).
miR-1271 targets FOXQ1 in HCC
cells
In order to investigate mechanism underlying
miR-1271 in HCC, the present study predicted the potential target
genes of miR-1271 using bioinformatics algorithms. Numerous genes
were predicted (data not shown), including FOXQ1.
FOXQ1 is a member of the forkhead transcription factor
family (41). A previous study
revealed that FOXQ1 is a modulator of TWIST-1
mediated metastatic phenotypes and a biomarker of metastasis
(42). FOXQ1 served a key role
in regulating EMT and aggressiveness of human cancer (43). Therefore, FOXQ1 was selected
for additional investigation. The binding sites with miR-1271 and
the mutant sequence were listed (Fig.
4A). Following the construction of luciferase reporter plasmids
with 3′UTR of FOXQ1 or mutant, miR-1271 mimics and reporter
plasmids were co-transfected into HepG-2 cells: This revealed that
overexpression of miR-1271 reduced the luciferase activity of wild
type 3′UTR reporter; however, in the mutated 3′UTR reporter,
miR-1271 demonstrated less effect (Fig.
4B). Next, HepG-2 cells were transfected with miR-1271 mimics.
After 48 h, western blot analysis was performed to determine the
expression level of FOXQ1 protein, and it was identified that the
transfection of miR-1271 mimics inhibited the FOXQ1 protein
expression in HepG-2 cells (Fig. 4C).
Finally, the FOXQ1 mRNA expression levels in 11 HCC tissues
were examined by PCR. It was demonstrated that the FOXQ1
mRNA expression levels and miR-1271 expression levels were
negatively correlated (Fig. 4D).
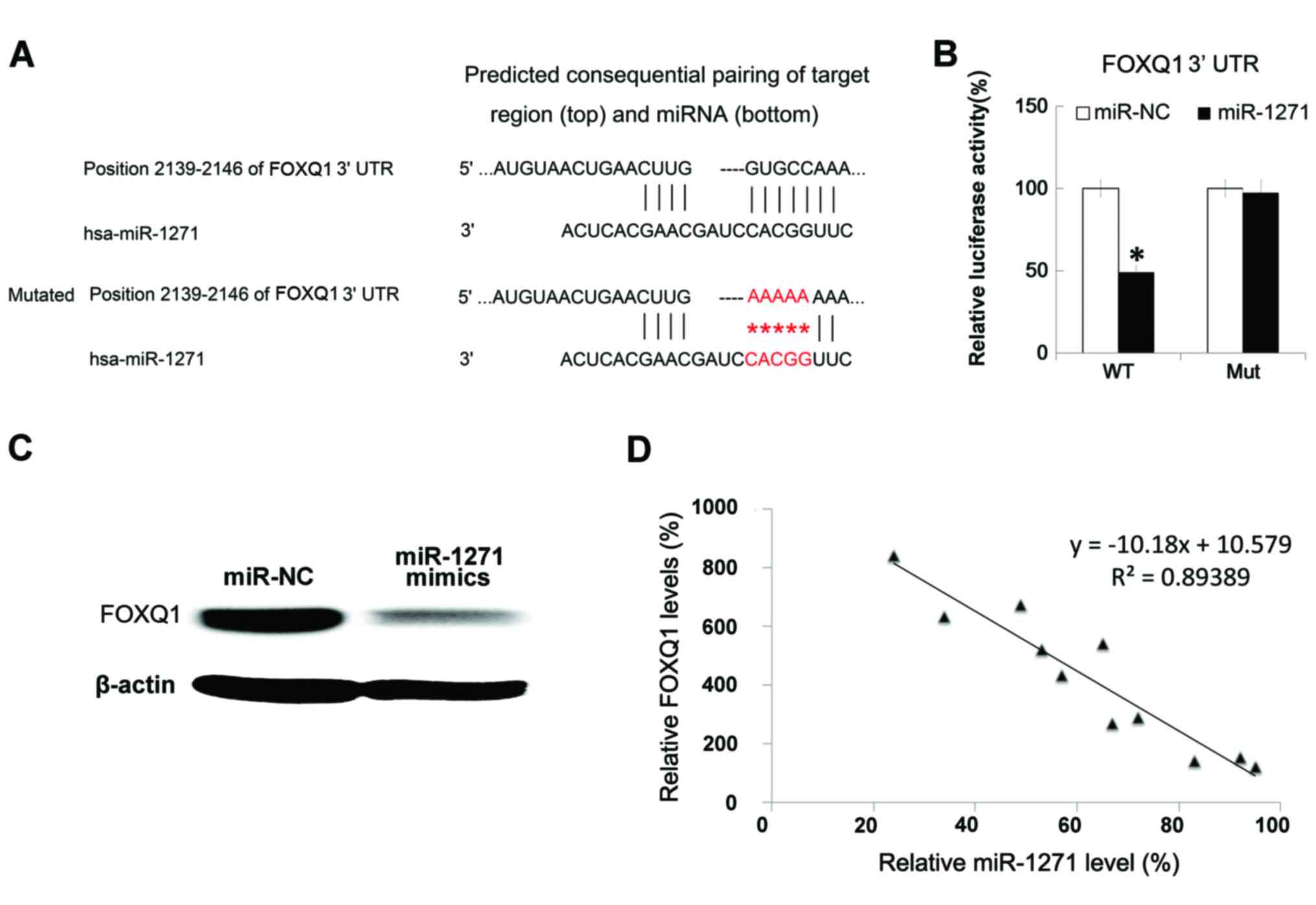 | Figure 4.FOXQ1 was targeted by miR-1271. (A)
TargetScanHuman (www.targetscan.org) demonstrated that FOXQ1 was a
direct target of miR-1271. (B) HepG-2 cells were co-transfected
with miR-1271 mimics, the control and reporter plasmid or the Mut
3′UTR, together with the controls. (C) HepG-2 cells were
transfected with miR-1271 mimics. Subsequent to 48 h, western blot
analysis was performed to determine the expression level of FOXQ1
protein. (D) FOXQ1 mRNA expression levels in 11 HCC tissues were
examined by reverse transcription-quantitative polymerase chain
reaction. The correlation between FOXQ1 mRNA and miR-1271
expression levels were determined by two-tailed Pearson's
correlation coefficient analysis. All data are presented as the
mean ± standard deviation of three separate experiments.
*P<0.05. FOXQ1, Forkhead box Q1; miR, microRNA; 3′UTR,
3′untraslated region; HCC, hepatocellular carcinoma; NC, negative
control; WT, wild type; Mut, mutant. |
Discussion
The present study determined the function of
miR-1271 by alteration of miR-1271 expression levels in HCC cells.
HCC tissues revealed a lower expression level of miR-1271.
Overexpression of miR-1271 inhibited cell growth and promoted
apoptosis, and downregulation of miR-1271 promoted cell growth and
inhibited apoptosis. miR-1271 targeted FOXQ1. The
downregulation of miR-1271 in HCC has been demonstrated in a
previous study (29). The results of
the present study revealed a clear role of miR-1271 in HCC.
The present study demonstrated that FOXQ1 was
targeted by miR-1271. It has previously been revealed that
FOXQ1 is overexpressed in various types of human cancer,
including colorectal (44), breast
(45) and lung cancer (45) and HCC (46,47).
Notably, high FOXQ1 expression levels were independent
prognostic factors of HCC (46). As
FOXQ1 was targeted by miR-1271, the present study suggested
that miR-1271 may be a prognostic factor of HCC.
In addition, FOXQ1 has been reported to be a
target of TGF-β signaling in breast cancer (48) and a novel target of the Wnt-β-catenin
signaling pathway in colorectal cancer (49). miR-124 suppressed tumor growth and
metastasis by targeting FOXQ1 in nasopharyngeal cancer
(50). An additional previous study
suggested that there was a double-negative feedback loop between
miR-422a and its targeted gene, FOXQ1, in HCC (51). To the best of our knowledge, the
present study demonstrated for the first time that FOXQ1 was
directly downregulated by miR-1271 in HCC.
In conclusion, the present study identified miR-1271
as a novel tumor suppressor that inhibits HCC cell growth and
induces cellular apoptosis by targeting FOXQ1 in HCC. These
results indicated that miR-1271 could be potential molecular target
for additional investigation.
Acknowledgements
The present study was supported by the Key Project
of Research and Development of Huai'an (grant no., HAS2014007)
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Venook AP, Papandreou C, Furuse J and de
Guevara LL: The incidence and epidemiology of hepatocellular
carcinoma: A global and regional perspective. Oncologist. 15 Suppl
4:S5–S13. 2010. View Article : Google Scholar
|
|
4
|
Beasley RP: Hepatitis B virus. The major
etiology of hepatocellular carcinoma. Cancer. 61:1942–1956. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song P, Feng X, Zhang K, Song T, Ma K,
Kokudo N, Dong J, Yao L and Tang W: Screening for and surveillance
of high-risk patients with HBV-related chronic liver disease:
Promoting the early detection of hepatocellular carcinoma in China.
Biosci Trends. 7:1–6. 2013.PubMed/NCBI
|
|
6
|
Karaman B, Battal B, Sari S and Verim S:
Hepatocellular carcinoma review: Current treatment, and
evidence-based medicine. World J Gastroenterol. 20:18059–18060.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently down-regulated in human hepatocellular carcinoma. Cancer
Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Budhu A, Jia HL, Forgues M, Liu CG,
Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al:
Identification of metastasis-related microRNAs in hepatocellular
carcinoma. Hepatology. 47:897–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu T, Zhu Y, Xiong Y, Ge YY, Yun JP and
Zhuang SM: MicroRNA-195 suppresses tumorigenicity and regulates
G1/S transition of human hepatocellular carcinoma cells.
Hepatology. 50:113–121. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Su H, Yang JR, Xu T, Huang J, Xu L, Yuan Y
and Zhuang SM: MicroRNA-101, down-regulated in hepatocellular
carcinoma, promotes apoptosis and suppresses tumorigenicity. Cancer
Res. 69:1135–1142. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Braconi C and Patel T: MicroRNA expression
profiling: A molecular tool for defining the phenotype of
hepatocellular tumors. Hepatology. 47:1807–1809. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Murakami Y, Tamori A, Itami S, Tanahashi
T, Toyoda H, Tanaka M, Wu W, Brojigin N, Kaneoka Y, Maeda A, et al:
The expression level of miR-18b in hepatocellular carcinoma is
associated with the grade of malignancy and prognosis. BMC Cancer.
13:992013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gramantieri L, Fornari F, Callegari E,
Sabbioni S, Lanza G, Croce CM, Bolondi L and Negrini M: MicroRNA
involvement in hepatocellular carcinoma. J Cell Mol Med.
12:2189–2204. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang B, Majumder S, Nuovo G, Kutay H,
Volinia S, Patel T, Schmittgen TD, Croce C, Ghoshal K and Jacob ST:
Role of microRNA-155 at early stages of hepatocarcinogenesis
induced by choline-deficient and amino acid-defined diet in C57BL/6
mice. Hepatology. 50:1152–1161. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Coulouarn C, Factor VM, Andersen JB,
Durkin ME and Thorgeirsson SS: Loss of miR-122 expression in liver
cancer correlates with suppression of the hepatic phenotype and
gain of metastatic properties. Oncogene. 28:3526–3536. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ji J, Shi J, Budhu A, Yu Z, Forgues M,
Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al: MicroRNA
expression, survival, and response to interferon in liver cancer. N
Engl J Med. 361:1437–1447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ji J, Yamashita T, Budhu A, Forgues M, Jia
HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH, et al: Identification
of microRNA-181 by genome-wide screening as a critical player in
EpCAM-positive hepatic cancer stem cells. Hepatology. 50:472–480.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ladeiro Y, Couchy G, Balabaud C,
Bioulac-Sage P, Pelletier L, Rebouissou S and Zucman-Rossi J:
MicroRNA profiling in hepatocellular tumors is associated with
clinical features and oncogene/tumor suppressor gene mutations.
Hepatology. 47:1955–1963. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song G, Sharma AD, Roll GR, Ng R, Lee AY,
Blelloch RH, Frandsen NM and Willenbring H: MicroRNAs control
hepatocyte proliferation during liver regeneration. Hepatology.
51:1735–1743. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ura S, Honda M, Yamashita T, Ueda T,
Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K and Kaneko
S: Differential microRNA expression between hepatitis B and
hepatitis C leading disease progression to hepatocellular
carcinoma. Hepatology. 49:1098–1112. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Lee AT, Ma JZ, Wang J, Ren J, Yang
Y, Tantoso E, Li KB, Ooi LL, Tan P and Lee CG: Profiling microRNA
expression in hepatocellular carcinoma reveals microRNA-224
up-regulation and apoptosis inhibitor-5 as a microRNA-224-specific
target. J Biol Chem. 283:13205–13215. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xiang XJ, Deng J, Liu YW, Wan LY, Feng M,
Chen J and Xiong JP: MiR-1271 inhibits cell proliferation, invasion
and EMT in gastric cancer by targeting FOXQ1. Cell Physiol Biochem.
36:1382–1394. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kong D, Zhang G, Ma H and Jiang G:
miR-1271 inhibits OSCC cell growth and metastasis by targeting ALK.
Neoplasma. 62:559–566. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Maurel M, Jalvy S, Ladeiro Y, Combe C,
Vachet L, Sagliocco F, Bioulac-Sage P, Pitard V, Jacquemin-Sablon
H, Zucman-Rossi J, et al: A functional screening identifies five
microRNAs controlling glypican-3: Role of miR-1271 down-regulation
in hepatocellular carcinoma. Hepatology. 57:195–204. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fischer AH, Jacobson KA, Rose J and Zeller
R: Hematoxylin and eosin staining of tissue and cell sections. CSH
Protoc. 2008:pdb.prot49862008.PubMed/NCBI
|
|
31
|
Li D, Liu X, Lin L, Hou J, Li N, Wang C,
Wang P, Zhang Q, Zhang P, Zhou W, et al: MicroRNA-99a inhibits
hepatocellular carcinoma growth and correlates with prognosis of
patients with hepatocellular carcinoma. J Biol Chem.
286:36677–36685. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Song B, Zhang C, Li G, Jin G and Liu C:
MiR-940 inhibited pancreatic ductal adenocarcinoma growth by
targeting MyD88. Cell Physiol Biochem. 35:1167–1177. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mosmann T: Rapid colorimetric assay for
cellular growth and survival: Application to proliferation and
cytotoxicity assays. J Immunol Methods. 65:55–63. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Grimson A, Farh KK, Johnston WK,
Garrett-Engele P, Lim LP and Bartel DP: MicroRNA targeting
specificity in mammals: Determinants beyond seed pairing. Mol Cell.
27:91–105. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Garcia DM, Baek D, Shin C, Bell GW,
Grimson A and Bartel DP: Weak seed-pairing stability and high
target-site abundance decrease the proficiency of lsy-6 and other
microRNAs. Nat Struct Mol Biol. 18:1139–1146. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Grentzmann G, Ingram JA, Kelly PJ,
Gesteland RF and Atkins JF: A dual-luciferase reporter system for
studying recoding signals. RNA. 4:479–486. 1998.PubMed/NCBI
|
|
40
|
Zhang H, Cai X, Wang Y, Tang H, Tong D and
Ji F: microRNA-143, down-regulated in osteosarcoma, promotes
apoptosis and suppresses tumorigenicity by targeting Bcl-2. Oncol
Rep. 24:1363–1369. 2010.PubMed/NCBI
|
|
41
|
Carlsson P and Mahlapuu M: Forkhead
transcription factors: Key players in development and metabolism.
Dev Biol. 250:1–23. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Abba M, Patil N, Rasheed K, Nelson LD,
Mudduluru G, Leupold JH and Allgayer H: Unraveling the role of
FOXQ1 in colorectal cancer metastasis. Mol Cancer Res.
11:1017–1028. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qiao Y, Jiang X, Lee ST, Karuturi RK, Hooi
SC and Yu Q: FOXQ1 regulates epithelial-mesenchymal transition in
human cancers. Cancer Res. 71:3076–3086. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kaneda H, Arao T, Tanaka K, Tamura D,
Aomatsu K, Kudo K, Sakai K, De Velasco MA, Matsumoto K, Fujita Y,
et al: FOXQ1 is overexpressed in colorectal cancer and enhances
tumorigenicity and tumor growth. Cancer Res. 70:2053–2063. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Feng J, Zhang X, Zhu H, Wang X, Ni S and
Huang J: FoxQ1 overexpression influences poor prognosis in
non-small cell lung cancer, associates with the phenomenon of EMT.
PLoS One. 7:e399372012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang W, He S, Ji J, Huang J, Zhang S and
Zhang Y: The prognostic significance of FOXQ1 oncogene
overexpression in human hepatocellular carcinoma. Pathol Res Pract.
209:353–358. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xia L, Huang W, Tian D, Zhang L, Qi X,
Chen Z, Shang X, Nie Y and Wu K: Forkhead box Q1 promotes
hepatocellular carcinoma metastasis by transactivating ZEB2 and
VersicanV1 expression. Hepatology. 59:958–973. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Fan DM, Feng XS, Qi PW and Chen YW:
Forkhead factor FOXQ1 promotes TGF-b1 expression and induces
epithelial-mesenchymal transition. Mol Cell Biochem. 397:179–186.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Christensen J, Bentz S, Sengstag T,
Shastri VP and Anderle P: FOXQ1, a novel target of the Wnt pathway
and a new marker for activation of Wnt signaling in solid tumors.
PLoS One. 8:e600512013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Peng XH, Huang HR, Lu J, Liu X, Zhao FP,
Zhang B, Lin SX, Wang L, Chen HH, Xu X, et al: MiR-124 suppresses
tumor growth and metastasis by targeting Foxq1 in nasopharyngeal
carcinoma. Mol Cancer. 13:1862014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhang J, Yang Y, Yang T, Yuan S, Wang R,
Pan Z, Yang Y, Huang G, Gu F, Jiang B, et al: Double-negative
feedback loop between microRNA-422a and forkhead box (FOX)G1/Q1/E1
regulates hepatocellular carcinoma tumor growth and metastasis.
Hepatology. 61:561–573. 2015. View Article : Google Scholar : PubMed/NCBI
|