Introduction
Endometrial cancer is the most commonly diagnosed
female genital cancer and, in 2012, was ranked the fourth most
common cancer in women worldwide, after breast, lung and colorectal
cancer (1). In China, an estimated
63,400 new cases and 21,800 mortalities were reported for
endometrial cancer in 2015 (2).
Uterine corpus endometrial carcinoma (UCEC) is a common type of
endometrial cancer. The incidence of UCEC increases with age and it
is most frequently diagnosed in women aged between 45 and 65 years
(3). Previous studies have sought to
identify tumor biomarkers and have identified multiple UCEC
biomarkers, including activated leukocyte cell adhesion molecule
(4), sperm-associated antigen 9
(5), L1 cell adhesion molecule
(6), progestogen-associated
endometrial protein (7), heat shock
protein family A (Hsp70) member 5 (8)
and CD151 molecule (Raph blood group) (9). To enhance our understanding of UCEC and
explore more effective and targeted therapies, the present study
aimed to identify gene co-expression networks, hub genes and small
molecule drugs associated with the development of UCEC.
The Cancer Genome Atlas (TCGA) is a comprehensive
genomic database that holds data for >20 types of cancer
obtained from thousands of patients. Data in TCGA includes
whole-genome measurements of multiple genomic features, including
DNA copy numbers, DNA methylation, and gene and microRNA
expression, thereby assisting researchers in assessing cancer
mechanisms at multiple molecular and regulatory levels (10). Furthermore, TGGA data is open access
and available to all researchers in individual work settings.
In the present study, UCEC RNA sequencing data were
collected and analyzed using bioinformatics tools. Differentially
expressed genes (DEGs) were identified and a gene co-expression
network was constructed, enabling the hub genes to be identified.
Relevant small molecule drugs were also assessed. The results of
the present study may provide novel insights into the pathogenesis
of UCEC and thereby expand perspectives on the treatment of this
type of tumor.
Materials and methods
Gene expression data
The RNA expression data (level 3) of UCEC patients
and normal controls without UCEC were downloaded from the TCGA data
portal up until December 2016 (http://cancergenome.nih.gov/) (11). A total of 552 UCEC samples and 35
normal control samples were included in the dataset. The exclusion
criteria were as follows: A histological diagnosis not of UCEC; and
samples without complete data for analysis.
DEG screening
The DEGs between UCEC and normal control tissues
were screened using edgeR software (v..5; http://www.bioconductor.org/packages/release/bioc/html/edgeR.html).
A false discovery rate <0.01 and |log2(fold
change)|>1 were set as the cut-off values to identify the
DEGs.
Cluster analysis
Bidirectional hierarchical clustering according to
the expression of the DEGs was performed using the pheatmap package
(v.1.0.8; https://CRAN.R-project.org/package=pheatmap) in R, as
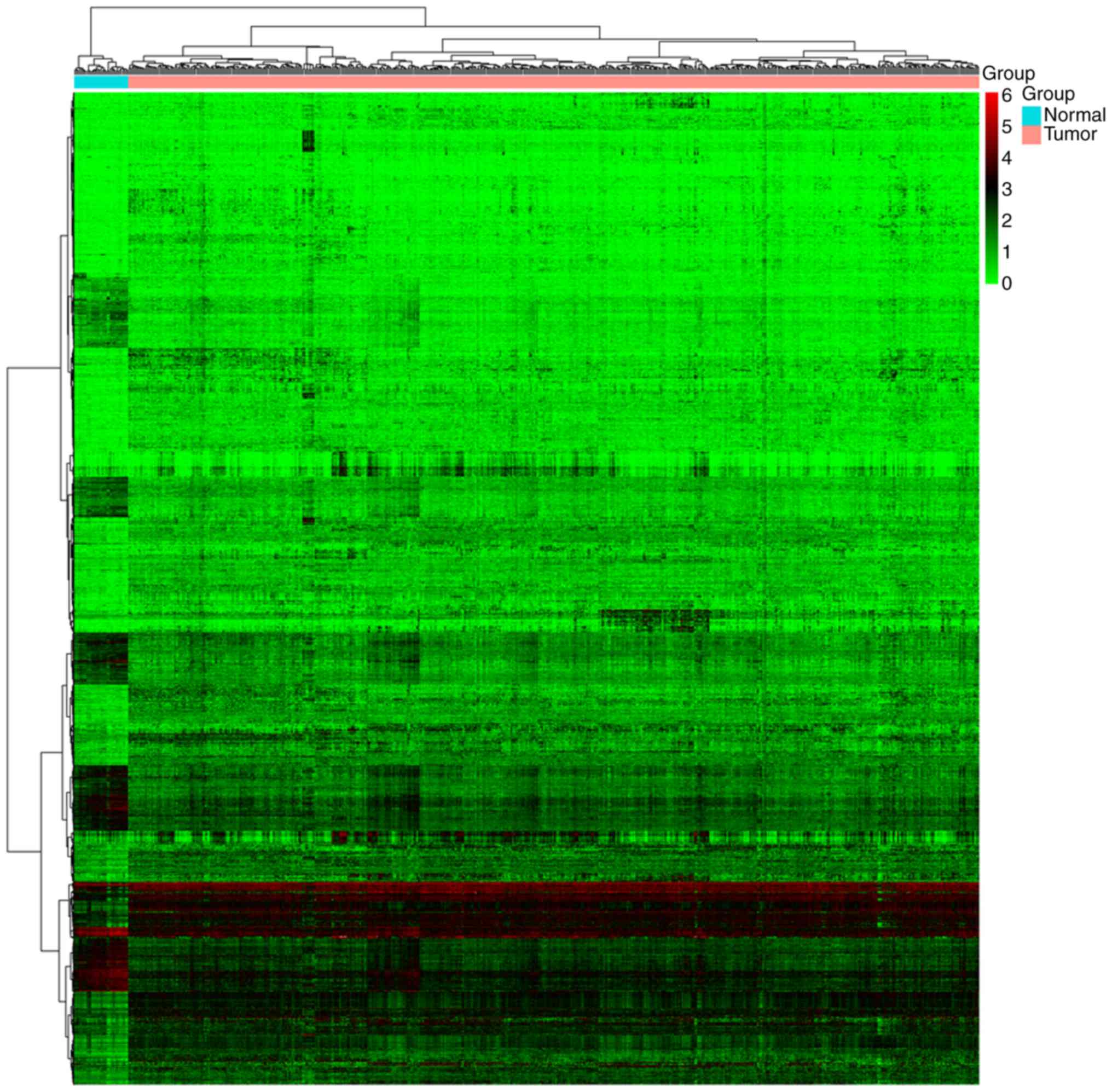
previously described (12). A heat
map was used to represent the data (Fig.
1).
Gene co-expression network
construction
Correlations among the DEGs were calculated using
the R/EBcoexpress package in R (version 3.5; http://www.bioconductor.org/packages/release/bioc/html/EBcoexpress.html),
as previously described (13). Genes
with a correlation coefficient >0.9 were included in the gene
co-expression network, which was visualized using Cytoscape
software (version 3.5.1; http://www.cytoscape.org/).
Functional enrichment analysis
A Gene Ontology (GO) analysis (14) was performed on the DEGs in the gene
co-expression network using the Database for Annotation,
Visualization and Integration Discovery (http://david.abcc.ncifcrf.gov/) (15). P<0.05 was set as the cut-off
criteria. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment analysis (16) was
performed using KOBAS 2.0 software (http://kobas.cbi.pku.edu.cn/) (17), with P<0.05 set as the
threshold.
Module analysis
Modules were identified using the MCODE plug-in in
Cytoscape with the cut-off criteria of degree ≥2 and k-core ≥3.
Each module was functionally annotated using the Cytoscape plug-in
BiNGO based on hypergeometric distribution (adjusted P-value
<0.01).
Screening of relevant small molecule
drugs
ConnectivityMap (cMap) (18) is a public database containing
>7,000 expression profiles representing 1,309 compounds
(www.broad.mit.edu/cmap/). Relevant small
molecule drugs were predicted using the cMap tool and those with a
|score|>0.6 were included.
Results
DEGs among UCEC and control
samples
The differential expression analysis identified
3,742 DEGs, comprising 2,580 upregulated and 1,162 downregulated
genes. The results of bidirectional hierarchical clustering of the
3,742 DEGs in the 587 samples (552 UCEC and 35 normal control
samples) that were screened were provided (Fig. 1). The UCEC and control samples
differed in gene expression pattern, suggesting that DEGs could
distinguish between the two types of sample.
GO annotations of DEGs
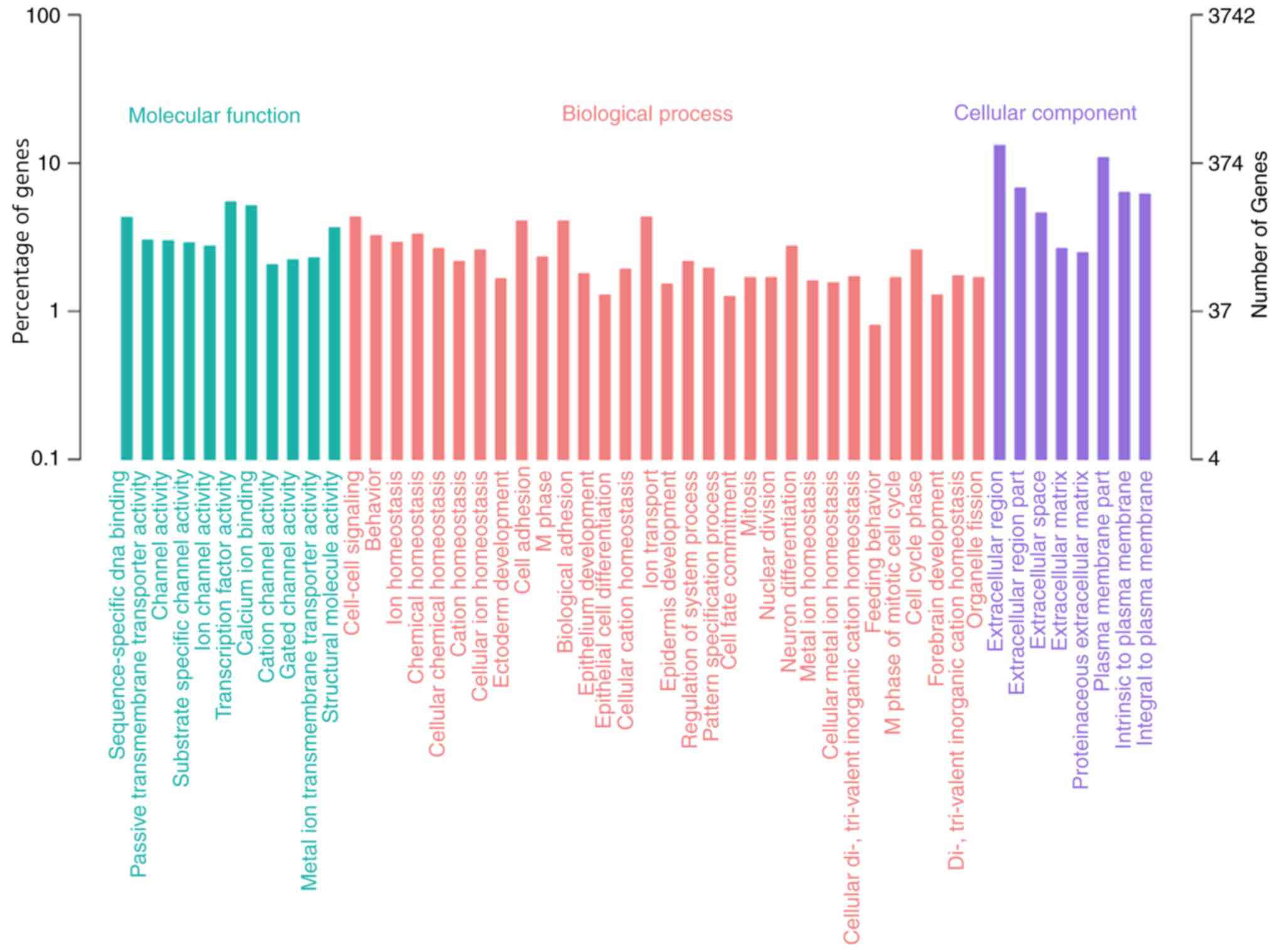
The GO annotations of the DEGs were provided
(Fig. 2). Cancer-associated
processes, including cell adhesion, ion transport and biological
adhesion, were among the significantly associated terms.
Gene co-expression network
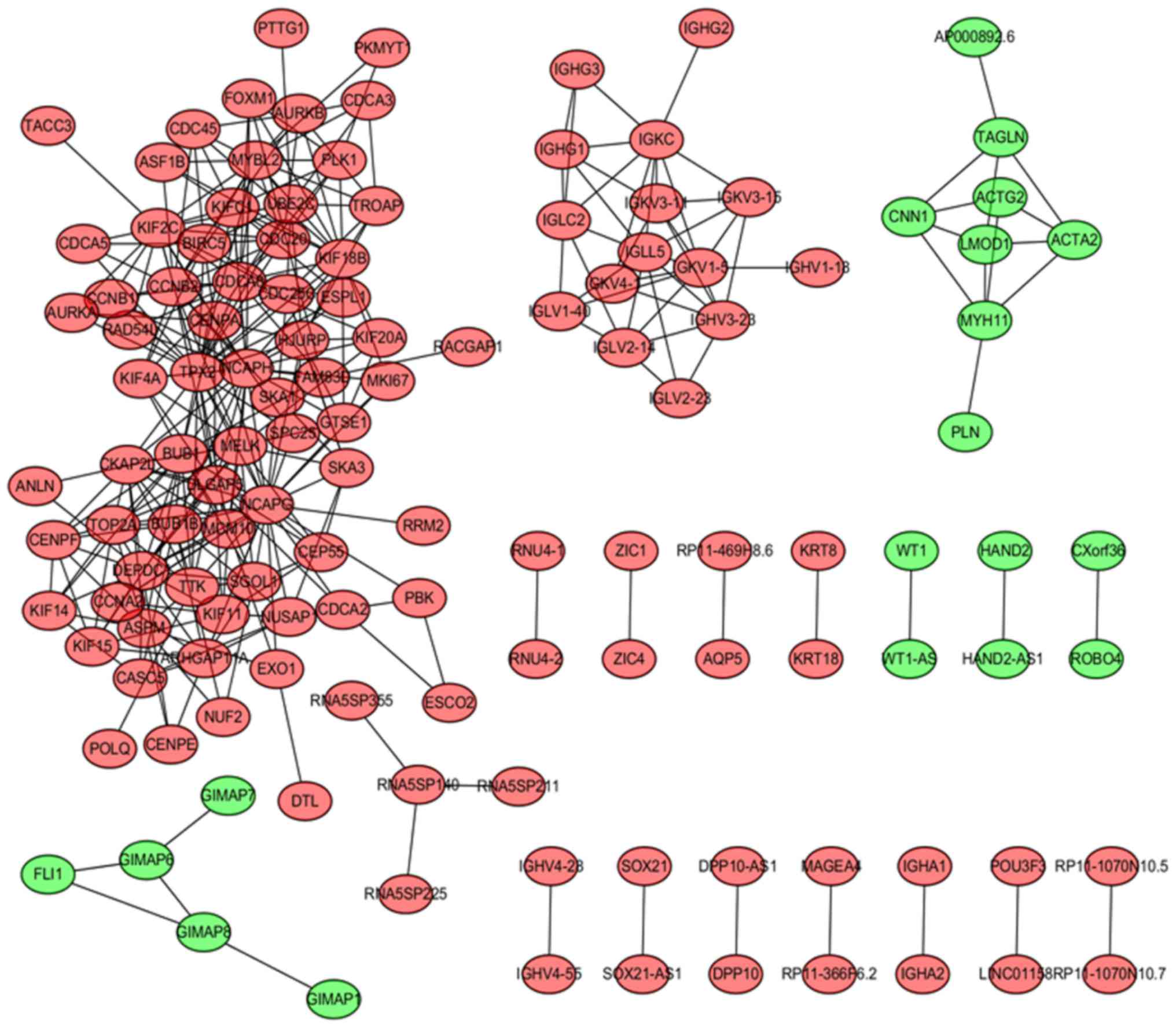
Correlated genes with a correlation coefficient
>0.9 were included in the gene co-expression network (Fig. 3). A total of 129 DEGs (nodes) and 368
edges (lines between nodes) were included, divided into 110
upregulated and 19 downregulated genes. The sizes of the nodes were
proportional to the number of genes in the gene set, and a number
of hub genes (nodes with degrees ≥8) were highlighted, including
cell division cycle 20 (CDC20), cyclin B2, non-SMC condensin I
complex subunit H, BUB1 mitotic checkpoint serine/threonine kinase,
cell division cycle-associated 8, maternal embryonic leucine zipper
kinase, MYB proto-oncogene like 2, TPX2, microtubule nucleation
factor and non-SMC condensin I complex subunit G.
Co-expression network functional
enrichment analysis
GO enrichment analysis identified 20 significantly
overrepresented terms for gene function in the co-expression
network (Table I), including M phase,
nuclear division, mitosis and the M phase of the mitotic cell
cycle. KEGG pathway enrichment analysis indicated that pathways
such as those of the cell cycle, oocyte meiosis,
progesterone-mediated oocyte maturation and tumor protein p53
signaling were significantly enriched among genes in the
co-expression network (Table
II).
 | Table I.GO terms significantly
overrepresented in the genes from the gene co-expression
network. |
Table I.
GO terms significantly
overrepresented in the genes from the gene co-expression
network.
| GO category | GO term | Count | P-value | FDR |
|---|
| GO:0000279 | M phase | 45 |
1.27×10−46 |
1.93×10−43 |
| GO:0000280 | Nuclear
division | 40 |
5.98×10−46 |
9.07×10−43 |
| GO:0007067 | Mitosis | 40 |
5.98×10−46 |
9.07×10−43 |
| GO:0000087 | M phase of the
mitotic cell cycle | 40 |
1.27×10−45 |
1.93×10−42 |
| GO:0048285 | Organelle
fission | 40 |
3.20×10−45 |
4.85×10−42 |
| GO:0022403 | Cell cycle
phase | 46 |
1.33×10−43 |
2.02×10−40 |
| GO:0000278 | Mitotic cell
cycle | 43 |
3.22×10−41 |
4.89×10−38 |
| GO:0007049 | Cell cycle | 53 |
2.40×10−40 |
3.63×10−37 |
| GO:0022402 | Cell cycle
process | 48 |
3.05×10−40 |
4.62×10−37 |
| GO:0051301 | Cell division | 35 |
4.17×10−33 |
6.33×10−30 |
| GO:0005819 | Spindle | 24 |
8.32×10−27 |
9.68×10−24 |
| GO:0015630 | Microtubule
cytoskeleton | 35 |
2.86×10−26 |
3.33×10−23 |
| GO:0007059 | Chromosome
segregation | 18 |
3.26×10−21 |
4.94×10−18 |
| GO:0044430 | Cytoskeletal
part | 37 |
1.25×10−20 |
1.45×10−17 |
| GO:0005856 | Cytoskeleton | 41 |
5.11×10−19 |
5.94×10−16 |
| GO:0043228 |
Non-membrane-bounded organelle | 52 |
5.37×10−18 |
6.24×10−15 |
| GO:0043232 | Intracellular
non-membrane-bounded organelle | 52 |
5.37×10−18 |
6.24×10−15 |
| GO:0000775 | Chromosome,
centromeric region | 17 |
2.29×10−17 |
2.67×10−14 |
| GO:0000793 | Condensed
chromosome | 17 |
4.39×10−17 |
5.10×10−14 |
| GO:0000779 | Condensed
chromosome, centromeric region | 14 |
8.86×10−17 |
1.33×10−13 |
 | Table II.Kyoto Encyclopedia of Genes and
Genomes pathways significantly overrepresented in the genes from
the gene co-expression network. |
Table II.
Kyoto Encyclopedia of Genes and
Genomes pathways significantly overrepresented in the genes from
the gene co-expression network.
| Term | Count | P-value |
|---|
| hsa04110: Cell
cycle | 13 |
3.65×10−16 |
| hsa04114: Oocyte
meiosis | 11 |
3.06×10−13 |
| hsa04914:
Progesterone-mediated oocyte maturation | 7 |
3.60×10−8 |
| hsa04115: p53
signaling pathway | 4 |
8.02×10−5 |
| hsa05166: HTLV-I
infection | 5 |
1.41×10−3 |
| hsa04270: Vascular
smooth muscle contraction | 3 |
7.17×10−3 |
| hsa04068: FoxO
signaling pathway | 3 |
9.19×10−3 |
Modules and functions
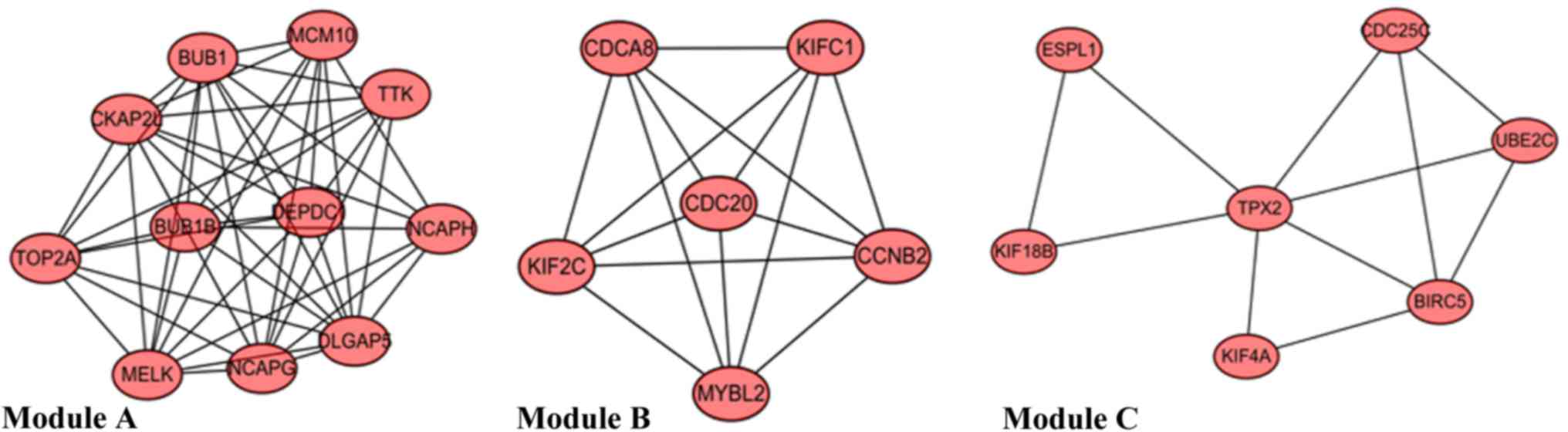
Three modules (designated A-C) were identified from
the gene co-expression network (Fig.
4). Module A included 11 DEGs implicated in the mitotic cell
cycle and cell cycle phases. Module B contained 6 DEGs associated
with nuclear division. Module C comprised 7 DEGs associated with
the M phase of the mitotic cell cycle (Table III).
 | Table III.Functional terms of the three
modules. |
Table III.
Functional terms of the three
modules.
| GO-ID | P-value | FDR | No. of genes |
|---|
| Module A |
|
|
|
|
GO:0000279~M phase |
9.60×10−7 |
1.12×10−3 | 6 |
|
GO:0000278~mitotic cell
cycle |
1.71×10−6 |
2.01×10−3 | 6 |
|
GO:0022403~cell cycle
phase |
2.98×10−6 |
3.50×10−3 | 6 |
|
GO:0007067~mitosis |
8.04×10−6 |
9.43×10−3 | 5 |
|
GO:0000280~nuclear
division |
8.04×10−6 |
9.43×10−3 | 5 |
|
GO:0000087~M phase of mitotic
cell cycle |
8.64×10−6 |
1.01×10−2 | 5 |
|
GO:0048285~organelle
fission |
9.43×10−6 |
1.11×10−2 | 5 |
|
GO:0022402~cell cycle
process |
1.37×10−5 |
1.60×10−2 | 6 |
|
GO:0007059~chromosome
segregation |
1.69×10−5 |
1.98×10−2 | 4 |
|
GO:0000793~condensed
chromosome |
3.41×10−5 |
3.10×10−2 | 4 |
|
GO:0005819~spindle |
5.04×10−5 |
4.59×10−2 | 4 |
|
GO:0005694~chromosome |
5.32×10−5 |
4.83×10−2 | 5 |
| Module B |
|
|
|
|
GO:0000280~nuclear
division |
3.36×10−7 |
3.63×10−4 | 5 |
|
GO:0007067~mitosis |
3.36×10−7 |
3.63×10−4 | 5 |
|
GO:0000087~M phase of mitotic
cell cycle |
3.61×10−7 |
3.90×10−4 | 5 |
|
GO:0048285~organelle
fission |
3.95×10−7 |
4.27×10−4 | 5 |
|
GO:0000279~M phase |
1.69×10−6 |
1.82×10−3 | 5 |
|
GO:0000278~mitotic cell
cycle |
2.69×10−6 |
2.93×10−3 | 5 |
|
GO:0015630~microtubule
cytoskeleton |
3.37×10−6 |
2.94×10−3 | 5 |
|
GO:0022403~cell cycle
phase |
4.22×10−6 |
4.56×10−3 | 5 |
|
GO:0022402~cell cycle
process |
1.46×10−5 |
1.57×10−2 | 5 |
| Module C |
|
|
|
|
GO:0000280~nuclear
division |
3.36×10−7 |
3.63×10−4 | 5 |
|
GO:0007067~mitosis |
3.36×10−7 |
3.63×10−4 | 5 |
|
GO:0000087~M phase of mitotic
cell cycle |
3.61×10−7 |
3.90×10−4 | 5 |
|
GO:0048285~organelle
fission |
3.95×10−7 |
4.27×10−4 | 5 |
|
GO:0000279~M phase |
1.69×10−6 |
1.82×10−3 | 5 |
|
GO:0000278~mitotic cell
cycle |
2.69×10−6 |
2.91×10−3 | 5 |
|
GO:0015630~microtubule
cytoskeleton |
3.37×10−6 |
2.94×10−3 | 5 |
|
GO:0022403~cell cycle
phase |
4.22×10−6 |
4.56×10−3 | 5 |
|
GO:0022402~cell cycle
process |
1.46×10−5 |
1.57×10−2 | 5 |
Relevant small molecule drugs
A total of 7 small molecule drugs were identified,
including esculetin, antazoline and isometheptene. Esculetin had
the highest negative connectivity score compared with the 6 other
small molecule drugs (−0.844; Table
IV).
 | Table IV.Relevant small molecule drugs. |
Table IV.
Relevant small molecule drugs.
| cMap name | Correlation | P-value |
|---|
| Esculetin | −0.844 |
7.55×10−3 |
| Antazoline | −0.729 |
1.07×10−2 |
| Isometheptene | −0.722 |
1.20×10−2 |
| Oxamniquine | 0.717 |
1.31×10−2 |
| Pyrimethamine | −0.642 |
1.49×10−2 |
| Carmustine | −0.795 |
1.75×10−2 |
| Cefapirin | 0.636 |
4.28×10−2 |
Discussion
The results of the present study indicated that
multiple DEGs are associated with UCEC pathophysiology. Matrix
metalloproteinases (MMPs) serve an important function in degrading
the extracellular matrix and basement membrane components (19). Karahan et al identified that
MMP-9 was expressed in a high percentage of primary endometrial
carcinoma tissues and that MMP-9 expression was potentially
associated with parameters of tumor aggressiveness (20). Furthermore, MMP-9 has been revealed to
be overexpressed in UCEC and associated with UCEC progression
(21). Topoisomerase 2α, a key enzyme
in DNA replication, has been implicated as a predictive marker in
treating endometrial cancer with taxane-containing adjuvant
chemotherapy (22). Enhancer of zeste
homolog 2 (EZH2), a critical component of the polycomb repressive
complex 2, is associated with cell proliferation, invasion,
adhesion and metastasis in several cancer types (23), and is overexpressed in numerous types
of cancer, including breast cancer (24), colorectal cancer (25), ovarian cancer (26) and nasopharyngeal cancer (27). A previous study indicated that EZH2
may predict a more aggressive endometrial carcinoma and may
therefore potentially represent a therapeutic target for this type
of cancer (28). Baculoviral
inhibitor of apoptosis (IAP) repeat containing 5 gene (BIRC5), a
member of the IAP gene family, may inhibit the activation of
caspase to negatively regulate apoptosis (29). In endometrial cancer, increased BIRC5
expression has been associated with poor progression-free survival
and may serve as an independent prognostic factor (30).
It is well established that CDC20 is an essential
developmental gene; disrupting CDC20 function in mice induces
mortality in embryos (31). Previous
studies have also suggested that CDC20 functions oncogenically in
human tumorigenesis. CDC20 serves important functions in the cell
cycle (32), cell apoptosis (33) and in targeting downstream substrates
for ubiquitination and subsequent degradation (34). Furthermore, CDC20 has been revealed to
be upregulated and may represent a useful prognostic biomarker in
endometrial cancer (35).
In the present study, esculetin was identified as a
significant small molecule drug in the development of UCEC.
Esculetin (6,7-dihydroxycoumarin), a coumarin derived from natural
plants, inhibits cancer cells from proliferating and induces
multiple types of human cancer cells to apoptose, including breast
(36), colon (37,38) and
gastric cancer (39), malignant
melanoma (40), hepatocellular
carcinoma (41,42) and oral squamous cancer (43,44).
Esculetin has also been reported to assist in inducing cancer cells
to apoptose (45,46). In the present study, esculetin had a
higher negative connectivity score compared with the 6 other small
molecule drugs, suggesting that esculetin could inhibit UCEC from
progressing. To the best of our knowledge, no study has yet
reported on the association between esculetin and endometrial
cancer. Verifying whether esculetin represents a novel therapeutic
agent in treating UCEC requires further studies. Overall, the
present study identified multiple key genes in UCEC and clinically
relevant small molecule agents, thereby improving our understanding
of UCEC and expanding perspectives on targeted therapy for this
type of cancer.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81671434).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rutgers JK: Update on pathology, staging
and molecular pathology of endometrial (uterine corpus)
adenocarcinoma. Future Oncol. 11:3207–3218. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Devis L, Moiola CP, Masia N,
Martinez-Garcia E, Santacana M, Stirbat TV, Brochard-Wyart F,
García Á, Alameda F, Cabrera S, et al: Activated leukocyte cell
adhesion molecule (ALCAM) is a marker of recurrence and promotes
cell migration, invasion, and metastasis in early-stage
endometrioid endometrial cancer. J Pathol. 241:475–487. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baser E, Togrul C, Ozgu E, Ayhan S, Caglar
M, Erkaya S and Gungor T: Sperm-associated antigen 9 is a promising
marker for early diagnosis of endometrial cancer. Asian Pac J
Cancer Prev. 14:7635–7638. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Smogeli E, Davidson B, Cvancarova M, Holth
A, Katz B, Risberg B, Kristensen G and Lindemann K: L1CAM as a
prognostic marker in stage I endometrial cancer: A validation
study. BMC Cancer. 16:5962016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lenhard M, Heublein S, Kunert-Keil C,
Vrekoussis T, Lomba I, Ditsch N, Mayr D, Friese K and Jeschke U:
Immunosuppressive Glycodelin A is an independent marker for poor
prognosis in endometrial cancer. BMC Cancer. 13:6162013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Matsuo K, Gray MJ, Yang DY, Srivastava SA,
Tripathi PB, Sonoda LA, Yoo EJ, Dubeau L, Lee AS and Lin YG: The
endoplasmic reticulum stress marker, glucose-regulated protein-78
(GRP78) in visceral adipocytes predicts endometrial cancer
progression and patient survival. Gynecol Oncol. 128:552–559. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Voss MA, Gordon N, Maloney S, Ganesan R,
Ludeman L, McCarthy K, Gornall R, Schaller G, Wei W, Berditchevski
F and Sundar S: Tetraspanin CD151 is a novel prognostic marker in
poor outcome endometrial cancer. Br J Cancer. 104:1611–1618. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu Y, Zhu Y, Müller P, Mitra R and Ji Y:
Characterizing cancer-specific networks by integrating TCGA data.
Cancer Inform. 13 Suppl 2:S125–S131. 2014.
|
|
11
|
Cancer Genome Atlas Research Network, .
Comprehensive genomic characterization defines human glioblastoma
genes and core pathways. Nature. 455:1061–1068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng
Z, Zhu G, Qi J, Ma H, Nian H and Wang Y: RNA-seq analyses of
multiple meristems of soybean: Novel and alternative transcripts,
evolutionary and functional implications. BMC Plant Biol.
14:1692014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dawson JA, Ye S and Kendziorski C:
R/EBcoexpress: An empirical Bayesian framework for discovering
differential co-expression. Bioinformatics. 28:1939–1940. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hulsegge I, Kommadath A and Smits MA:
Globaltest and GOEAST: Two different approaches for Gene Ontology
analysis. BMC Proc. 3 Suppl 4:pp. S102009; View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
16
|
Altermann E and Klaenhammer TR:
PathwayVoyager: Pathway mapping using the Kyoto Encyclopedia of
Genes and Genomes (KEGG) database. BMC Genomics. 6:602005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu J, Mao X, Cai T, Luo J and Wei L: KOBAS
server: A web-based platform for automated annotation and pathway
identification. Nucleic Acids Res. 34(Web Server Issue): W720–W724.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lamb J, Crawford ED, Peck D, Modell JW,
Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, et
al: The Connectivity Map: Using gene-expression signatures to
connect small molecules, genes, and disease. Science.
313:1929–1935. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Knapinska AM, Estrada CA and Fields GB:
The roles of matrix metalloproteinases in pancreatic cancer. Prog
Mol Biol Transl Sci. 148:339–354. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karahan N, Güney M, Baspinar S, Oral B,
Kapucuoglu N and Mungan T: Expression of gelatinase (MMP-2 and
MMP-9) and cyclooxygenase-2 (COX-2) in endometrial carcinoma. Eur J
Gynaecol Oncol. 28:184–188. 2007.PubMed/NCBI
|
|
21
|
Yu F, Jiang Q, Zhou Y, Yang Z, Yu X, Wang
H, Liu Z, Wang L, Fang W and Guo S: Abnormal expression of matrix
metalloproteinase-9 (MMP9) correlates with clinical course in
Chinese patients with endometrial cancer. Dis Markers. 32:321–327.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ito F, Furukawa N and Nakai T: Evaluation
of TOP2A as a predictive marker for endometrial cancer with
taxane-containing adjuvant chemotherapy. Int J Gynecol Cancer.
26:325–330. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shen L, Cui J, Liang S, Pang Y and Liu P:
Update of research on the role of EZH2 in cancer progression. Onco
Targets Ther. 6:321–324. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamaguchi H, Du Y, Nakai K, Ding M, Chang
SS, Hsu JL, Yao J, Wei Y, Nie L, Jiao S, et al: EZH2 contributes to
the response to PARP inhibitors through its PARP-mediated poly-ADP
ribosylation in breast cancer. Oncogene. 2017. View Article : Google Scholar
|
|
25
|
Lian Y, Yan C, Ding J, Xia R, Ma Z, Hui B,
Ji H, Zhou J and Wang K: A novel lncRNA, LL22NC03-N64E9.1,
represses KLF2 transcription through binding with EZH2 in
colorectal cancer. Oncotarget. 8:59435–59445. 2017.PubMed/NCBI
|
|
26
|
Zhou F, Chen J and Wang H: MicroRNA-298
inhibits malignant phenotypes of epithelial ovarian cancer by
regulating the expression of EZH2. Oncol Lett. 12:3926–3932.
2016.PubMed/NCBI
|
|
27
|
Yu L, Lu J, Zhang B, Liu X, Wang L, Li SY,
Peng XH, Xu X, Tian WD and Li XP: miR-26a inhibits invasion and
metastasis of nasopharyngeal cancer by targeting EZH2. Oncol Lett.
5:1223–1228. 2013.PubMed/NCBI
|
|
28
|
Jia N, Li Q, Tao X, Wang J, Hua K and Feng
W: Enhancer of zeste homolog 2 is involved in the proliferation of
endometrial carcinoma. Oncol Lett. 8:2049–2054. 2014.PubMed/NCBI
|
|
29
|
Ma F, Zhang H, Zhai Y, Huang W, Zhao C, Ou
S, Zhou H, Yuan W, Wang Z, Wang H, et al: Functional polymorphism
−31C/G in the promoter of BIRC5 gene and risk of nasopharyngeal
carcinoma among chinese. PLoS One. 6:e167482011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chuwa AH, Sone K, Oda K, Ikeda Y, Fukuda
T, Wada-Hiraike O, Inaba K, Makii C, Takeuchi M, Oki S, et al:
Significance of survivin as a prognostic factor and a therapeutic
target in endometrial cancer. Gynecol Oncol. 141:564–569. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li M, York JP and Zhang P: Loss of Cdc20
causes a securin-dependent metaphase arrest in two-cell mouse
embryos. Mol Cell Biol. 27:3481–3488. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hadjihannas MV, Bernkopf DB, Brückner M
and Behrens J: Cell cycle control of Wnt/b-catenin signalling by
conductin/axin2 through CDC20. EMBO Rep. 13:347–354. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Harley ME, Allan LA, Sanderson HS and
Clarke PR: Phosphorylation of Mcl-1 by CDK1-cyclin B1 initiates its
Cdc20-dependent destruction during mitotic arrest. EMBO J.
29:2407–2420. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xie Q, Wu Q, Mack SC, Yang K, Kim L,
Hubert CG, Flavahan WA, Chu C, Bao S and Rich JN: CDC20 maintains
tumor initiating cells. Oncotarget. 6:13241–13254. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gayyed MF, El-Maqsoud NM, Tawfiek ER, El
Gelany SA and Rahman MF: A comprehensive analysis of CDC20
overexpression in common malignant tumors from multiple organs: Its
correlation with tumor grade and stage. Tumour Biol. 37:749–762.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chang HT, Chou CT, Lin YS, Shieh P, Kuo
DH, Jan CR and Liang WZ: Esculetin, a natural coumarin compound,
evokes Ca(2+) movement and activation of Ca(2+)-associated
mitochondrial apoptotic pathways that involved cell cycle arrest in
ZR-75-1 human breast cancer cells. Tumour Biol. 37:4665–4678. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim AD, Han X, Piao MJ, Hewage SR, Hyun
CL, Cho SJ and Hyun JW: Esculetin induces death of human colon
cancer cells via the reactive oxygen species-mediated mitochondrial
apoptosis pathway. Environ Toxicol Pharmacol. 39:982–989. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim AD, Hewage SR Madduma, Piao MJ, Kang
KA, Cho SJ and Hyun JW: Esculetin induces apoptosis in human colon
cancer cells by inducing endoplasmic reticulum stress. Cell Biochem
Funct. 33:487–494. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pan H, Wang BH, Lv W, Jiang Y and He L:
Esculetin induces apoptosis in human gastric cancer cells through a
cyclophilin D-mediated mitochondrial permeability transition pore
associated with ROS. Chem Biol Interact. 242:51–60. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jeon YJ, Jang JY, Shim JH, Myung PK and
Chae JI: Esculetin, a coumarin derivative, exhibits
anti-proliferative and pro-apoptotic activity in G361 human
malignant melanoma. J Cancer Prev. 20:106–112. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang J, Lu ML, Dai HL, Zhang SP, Wang HX
and Wei N: Esculetin, a coumarin derivative, exerts in vitro and in
vivo antiproliferative activity against hepatocellular carcinoma by
initiating a mitochondrial-dependent apoptosis pathway. Braz J Med
Biol Res. 48:245–253. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kuo HC, Lee HJ, Hu CC, Shun HI and Tseng
TH: Enhancement of esculetin on Taxol-induced apoptosis in human
hepatoma HepG2 cells. Toxicol Appl Pharmacol. 210:55–62. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cho JH, Shin JC, Cho JJ, Choi YH, Shim JH
and Chae JI: Esculetin (6,7-dihydroxycoumarin): A potential cancer
chemopreventive agent through suppression of Sp1 in oral squamous
cancer cells. Int J Oncol. 46:265–271. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kok SH, Yeh CC, Chen ML and Kuo MY:
Esculetin enhances TRAIL-induced apoptosis through DR5 upregulation
in human oral cancer SAS cells. Oral Oncol. 45:1067–1072. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Arora R, Sawney S, Saini V, Steffi C,
Tiwari M and Saluja D: Esculetin induces antiproliferative and
apoptotic response in pancreatic cancer cells by directly binding
to KEAP1. Mol Cancer. 15:642016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jeon YJ, Cho JH, Lee SY, Choi YH, Park H,
Jung S, Shim JH and Chae JI: Esculetin induces apoptosis through
EGFR/PI3K/Akt signaling pathway and nucleophosmin relocalization. J
Cell Biochem. 117:1210–1221. 2016. View Article : Google Scholar : PubMed/NCBI
|