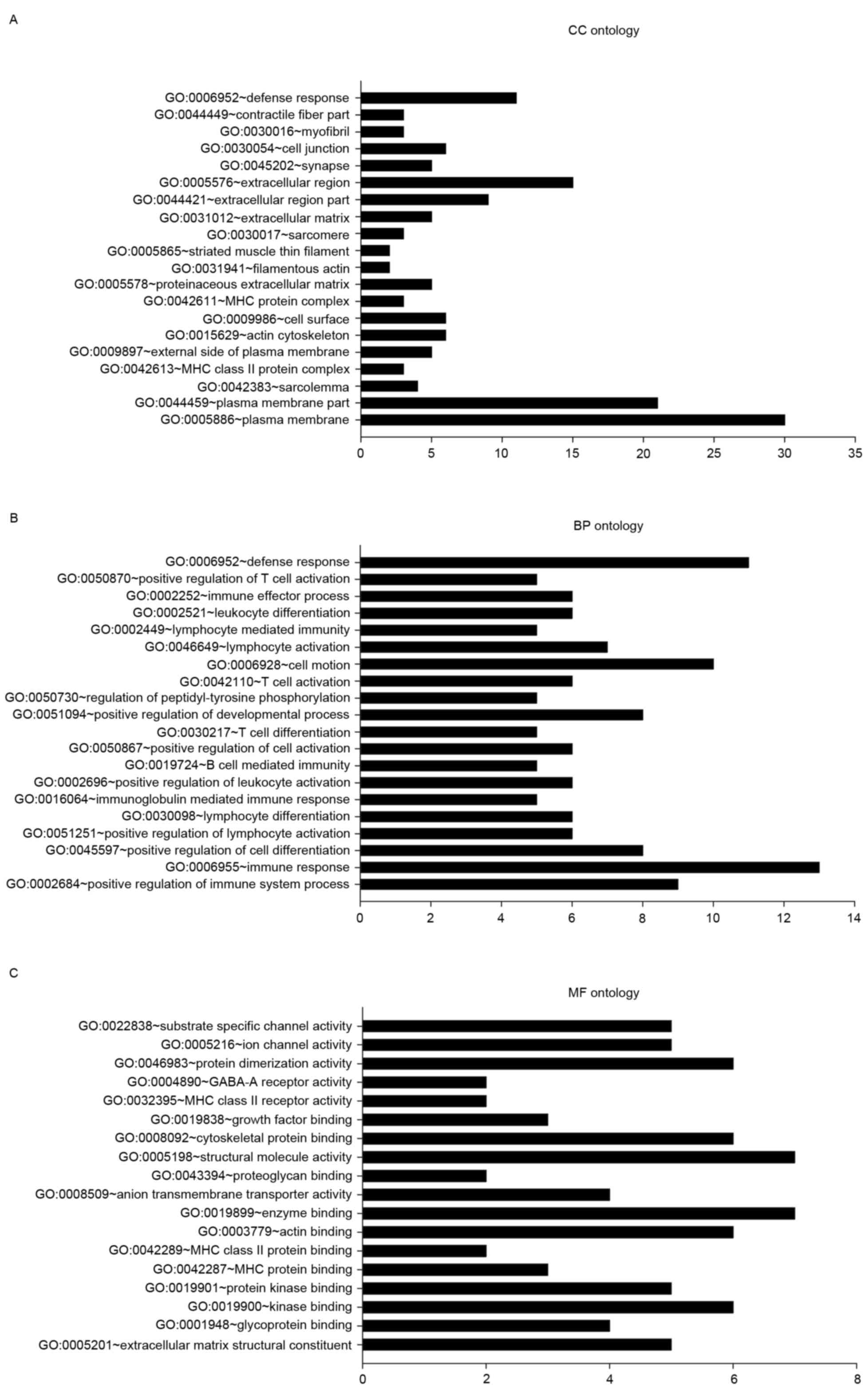
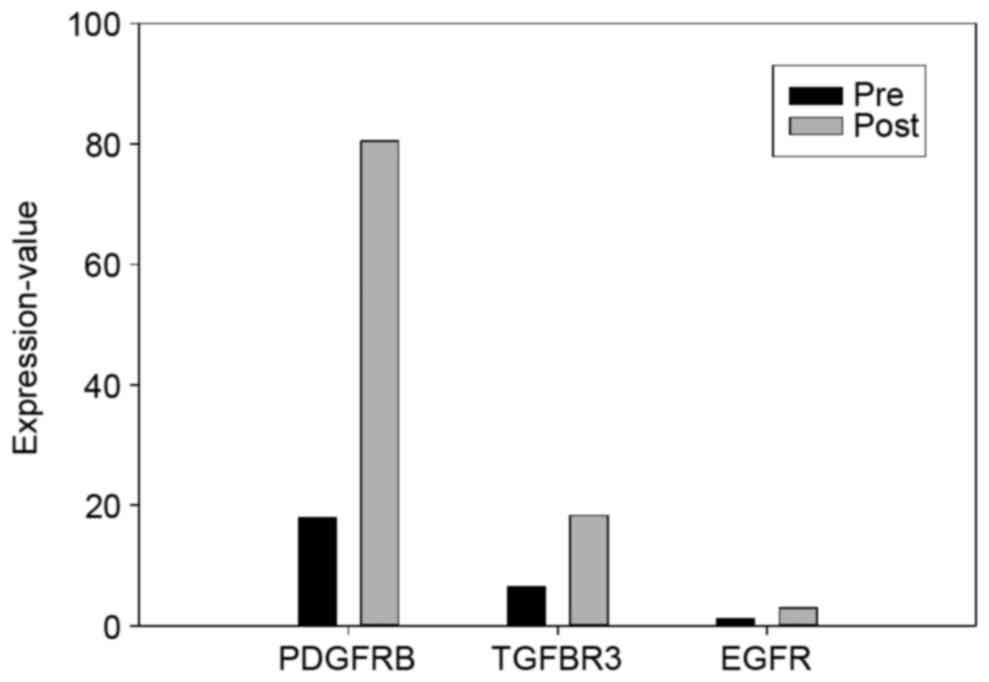
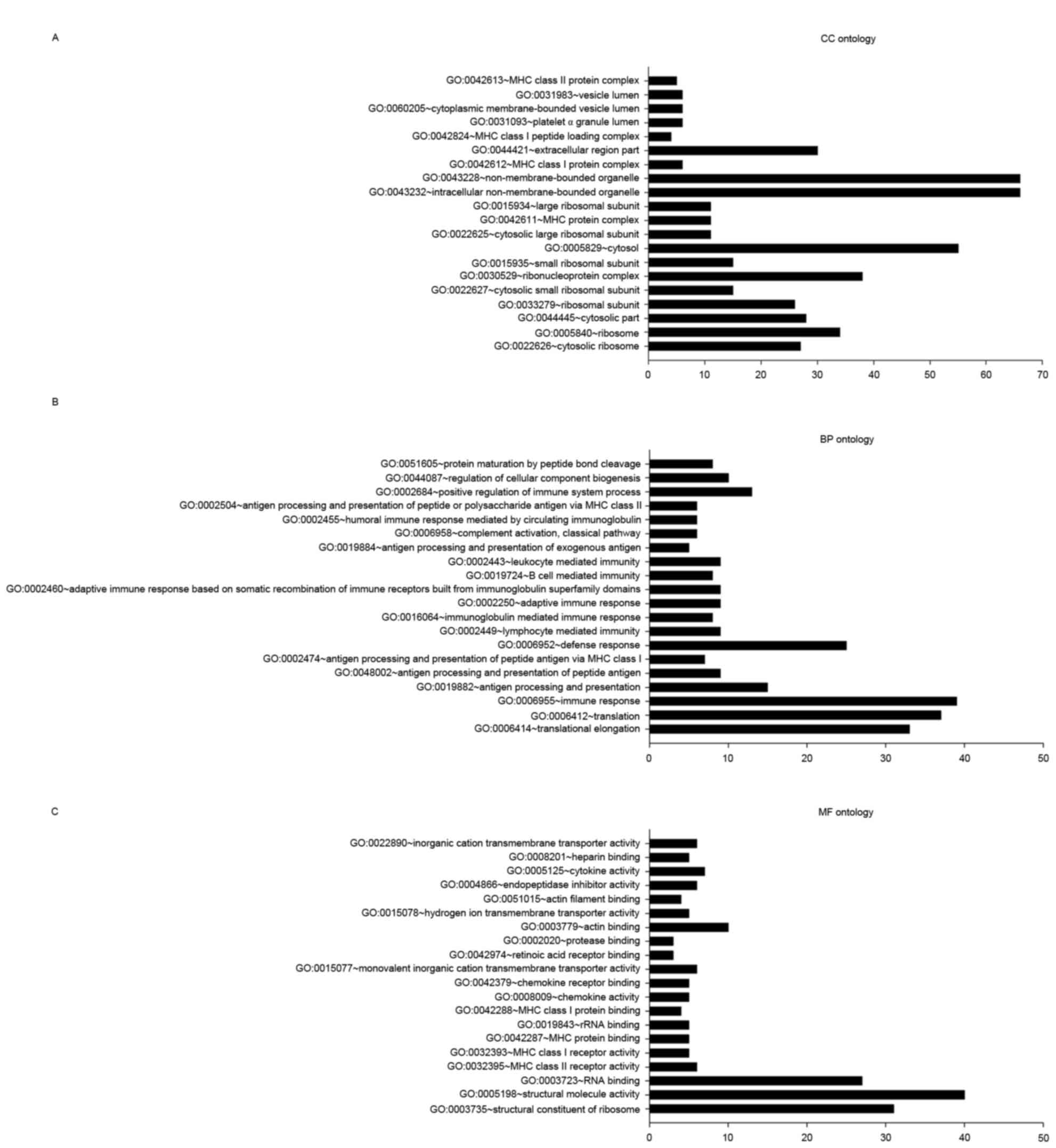
|
1
|
Chapman PB, Hauschild A, Robert C, Haanen
JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M, et
al: Improved survival with vemurafenib in melanoma with BRAF V600E
mutation. N Engl J Med. 364:2507–2516. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Clary BM, Brady MS, Lewis JJ and Coit DG:
Sentinel lymph node biopsy in the management of patients with
primary cutaneous melanoma: Review of a large single-institutional
experience with an emphasis on recurrence. Ann Surg. 233:250–258.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Flaherty KT, Hodi FS and Fisher DE: From
genes to drugs: Targeted strategies for melanoma. Nat Rev Cancer.
12:349–361. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hartsough EJ, Basile KJ and Aplin AE:
Beneficial effects of RAF inhibitor in mutant BRAF splice
variant-expressing melanoma. Mol Cancer Res. 12:795–802. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Heidorn SJ, Milagre C, Whittaker S, Nourry
A, Niculescu-Duvas I, Dhomen N, Hussain J, Reis-Filho JS, Springer
CJ, Pritchard C and Marais R: Kinase-dead BRAF and oncogenic RAS
cooperate to drive tumor progression through CRAF. Cell.
140:209–221. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vial E, Sahai E and Marshall CJ: ERK-MAPK
signaling coordinately regulates activity of Rac1 and RhoA for
tumor cell motility. Cancer Cell. 4:67–79. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gautschi O, Pauli C, Strobel K, Hirschmann
A, Printzen G, Aebi S and Diebold J: A patient with BRAF V600E lung
adenocarcinoma responding to vemurafenib. J Thorac Oncol.
7:e23–e24. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Das Thakur M, Salangsang F, Landman AS,
Sellers WR, Pryer NK, Levesque MP, Dummer R, McMahon M and Stuart
DD: Modelling vemurafenib resistance in melanoma reveals a strategy
to forestall drug resistance. Nature. 494:251–255. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kalluri R and Zeisberg M: Fibroblasts in
cancer. Nat Rev Cancer. 6:392–401. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McMillin DW, Delmore J, Weisberg E, Negri
JM, Geer DC, Klippel S, Mitsiades N, Schlossman RL, Munshi NC, Kung
AL, et al: Tumor cell-specific bioluminescence platform to identify
stroma-induced changes to anticancer drug activity. Nat Med.
16:483–489. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lovly CM, Dahlman KB, Fohn LE, Su Z,
Dias-Santagata D, Hicks DJ, Hucks D, Berry E, Terry C, Duke M, et
al: Routine multiplex mutational profiling of melanomas enables
enrollment in genotype-driven therapeutic trials. PLoS One.
7:e353092012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chaft JE, Oxnard GR, Sima CS, Kris MG,
Miller VA and Riely GJ: Disease flare after tyrosine kinase
inhibitor discontinuation in patients with EGFR-mutant lung cancer
and acquired resistance to erlotinib or gefitinib: Implications for
clinical trial design. Clin Cancer Res. 17:6298–6303. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Corcoran RB, Ebi H, Turke AB, Coffee EM,
Nishino M, Cogdill AP, Brown RD, Pelle P Della, Dias-Santagata D,
Hung KE, et al: EGFR-mediated re-activation of MAPK signaling
contributes to insensitivity of BRAF mutant colorectal cancers to
RAF inhibition with vemurafenib. Cancer Discov. 2:227–235. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Prahallad A, Sun C, Huang S, Di
Nicolantonio F, Salazar R, Zecchin D, Beijersbergen RL, Bardelli A
and Bernards R: Unresponsiveness of colon cancer to BRAF(V600E)
inhibition through feedback activation of EGFR. Nature.
483:100–103. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun C, Wang L, Huang S, Heynen GJ,
Prahallad A, Robert C, Haanen J, Blank C, Wesseling J, Willems SM,
et al: Reversible and adaptive resistance to BRAF(V600E) inhibition
in melanoma. Nature. 508:118–122. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Trapnell C, Roberts A, Goff L, Pertea G,
Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL and Pachter L:
Differential gene and transcript expression analysis of RNA-seq
experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Robinson MD, McCarthy DJ and Smyth GK:
edgeR: A Bioconductor package for differential expression analysis
of digital gene expression data. Bioinformatics. 26:139–140. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Arcangeli A, Crociani O, Lastraioli E,
Masi A, Pillozzi S and Becchetti A: Targeting ion channels in
cancer: A novel frontier in antineoplastic therapy. Curr Med Chem.
16:66–93. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cheli VT, González DA Santiago, Spreuer V
and Paez PM: Voltage-gated Ca2+ entry promotes oligodendrocyte
progenitor cell maturation and myelination in vitro. Exp Neurol.
265:69–83. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tsavaler L, Shapero MH, Morkowski S and
Laus R: Trp-p8, a novel prostate-specific gene, is up-regulated in
prostate cancer and other malignancies and shares high homology
with transient receptor potential calcium channel proteins. Cancer
Res. 61:3760–3769. 2001.PubMed/NCBI
|
|
22
|
Imming P, Sinning C and Meyer A: Drugs,
their targets and the nature and number of drug targets. Nat Rev
Drug Discov. 5:821–834. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kunzelmann K: Ion channels and cancer. J
Membr Biol. 205:159–173. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen X, Ji ZL and Chen YZ: TTD:
Therapeutic target database. Nucleic Acids Res. 30:412–415. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gurnett CA, Veile R, Zempel J, Blackburn
L, Lovett M and Bowcock A: Disruption of sodium bicarbonate
transporter SLC4A10 in a patient with complex partial epilepsy and
mental retardation. Arch Neurol. 65:550–553. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Neri D and Supuran CT: Interfering with pH
regulation in tumours as a therapeutic strategy. Nat Rev Drug
Discov. 10:767–777. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ong GL, Goldenberg DM, Hansen HJ and
Mattes MJ: Cell surface expression and metabolism of major
histocompatibility complex class II invariant chain (CD74) by
diverse cell lines. Immunology. 98:296–302. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kaufman JL, Niesvizky R, Stadtmauer EA,
Chanan-Khan A, Siegel D, Horne H, Wegener WA and Goldenberg DM:
Phase I, multicentre, dose-escalation trial of monotherapy with
milatuzumab (humanized anti-CD74 monoclonal antibody) in relapsed
or refractory multiple myeloma. Br J Haematol. 163:478–486. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Thompson JA, Srivastava MK, Bosch JJ,
Clements VK, Ksander BR and Ostrand-Rosenberg S: The absence of
invariant chain in MHC II cancer vaccines enhances the activation
of tumor-reactive type 1 CD4+ T lymphocytes. Cancer Immunol
Immunother. 57:389–398. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ruggero D and Pandolfi PP: Does the
ribosome translate cancer? Nat Rev Cancer. 3:179–192. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Weening JJ, D'Agati VD, Schwartz MM,
Seshan SV, Alpers CE, Appel GB, Balow JE, Bruijn JA, Cook T,
Ferrario F, et al: The classification of glomerulonephritis in
systemic lupus erythematosus revisited. Kidney Int. 65:521–530.
2004. View Article : Google Scholar : PubMed/NCBI
|