Introduction
Lung cancer is the leading cause of
cancer-associated mortality globally and one of the five top causes
of years of potential life lost in East Asia, in 2013. Non-small
cell lung cancer (NSCLC) accounts for 80% of all lung cancers.
However, the incidence rates and death rates have declined over the
last 20 years (1,2). Due to the advances made in early
diagnosis and treatment, the outcome for patients has improved, and
this has motivated further research concerning novel targets for
the diagnosis and treatment of lung cancer. The disordered
metabolism of tumor cells is involved in rapid cell proliferation,
maintenance of redox homeostasis and epigenetics (3,4). Interest
in this topic has waxed and waned since the observations of Warburg
(5). In previous years, multiple
oncogenes and tumor suppressors have been linked to the regulation
of cancer cell metabolism, making this particular topic one of the
most intense areas of cancer research (6).
Proline is one of the most abundant amino acids in
the cellular microenvironment. Proline metabolism and synthesis are
associated with the tricarboxylic acid cycle, urea cycle and
pentose phosphate pathway. Thus, proline metabolism and synthesis
are critically important for tumor cells (7,8). Proline
dehydrogenase/proline oxidase (PRODH/POX) catalyzes the first step
in proline catabolism, and its function in tumors has attracted
attention since it was reported to be a P53-induced gene in tumor
cell lines (9,10). PRODH/POX is downregulated in multiple
types of human tumor, particularly those of the kidney, stomach,
colon and rectum. It functions as a mitochondrial tumor suppressor
primarily through inhibiting cell proliferation, inducing apoptosis
and suppressing hypoxia-inducible factor 1 signaling (11). The function of proline synthesis in
cancer, however, remains to be comprehensively understood.
Pyrroline-5-carboxylate reductase (PYCR) catalyzes the last step of
proline synthesis, and three isozymes are encoded by three human
genes. A mutation in PYCR1 was described in patients with
autosomal-recessive cutis laxa type 2, indicating a critical
function for proline in normal development (12). An association between PYCR1 gene
expression and breast cancer growth has been reported by a previous
study (13). These studies indicate
that PYCR1 may participate in the process of tumorigenesis.
The aim of the present study was to investigate the
function of PYCR1 in lung cancer. PYCR1 expression was detected and
compared in lung cancer and its adjacent normal lung tissue.
Furthermore, the influence of PYCR1 knockdown on cell
proliferation, the cell cycle and apoptosis was investigated in
order to explore the function of PYCR1 in the tumorigenesis of lung
cancer.
Materials and methods
Patients and tissue specimens
Paired NSCLC samples and adjacent normal tissues
were obtained from 28 patients who underwent primary surgical
resection to treat NSCLC in the Department of Thoracic Surgery,
Jinling Hospital, Nanjing University School of Medicine (Nanjing,
China) between July 2014 and September 2014. The specimens were
collected during surgery and were kept frozen in liquid nitrogen
until RNA and protein extraction. In addition, 62 patients with
NSCLC with clinicopathological data and follow-up information
(61/62) who underwent surgical resection between June 2007 and
November 2008 were enrolled, and their paraffin-embedded specimens
were collected from the Department of Pathology, Jinling Hospital,
for immunohistochemical staining. Follow-up lasted until February
2015, with a median follow-up period of 84.5 months for living
patients (range, 75–94 months). None of these patients received
chemotherapy or radiotherapy prior to surgery. The present study
was reviewed and approved by the Institutional Review Board of
Jinling Hospital, and all patients signed an informed consent form
prior to undergoing surgery.
Oncomine® Platform
Bioinformatics
The gene search function of Oncomine®
Platform (www.oncomine.org) was used to analyze
the mRNA expression of PYCR1 in NSCLC tissues relative to their
normal controls (14). Gene lists based on fold change were obtained
from six NSCLC datasets which were named by the first author and
numbers of patient as follows: Selamat (116), Su (66), Stearman
(39), Hou (156), Beer (96) and Bhattacharjee (203).
Cell culture
All NSCLC cell lines (A549, SPC-A1, H1703, H1299,
PC9, H1915, SK-MES-1) were obtained from the Institute of
Biochemistry and Cell Biology of the Chinese Academy of Sciences
(Shanghai, China). All cells were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) except
for SPC-A1, which was cultured in Dulbecco's modified Eagle's
medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.), supplemented
with 10% heat-inactivated fetal bovine serum (Hyclone; GE
Healthcare, Chicago, IL, USA), 100 U/ml penicillin and 100 mg/ml
streptomycin at 37.8°C in a humidified atmosphere containing 5%
CO2.
Reverse-transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from frozen tissues or
cultured cells using TRIzol (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), according to the manufacturer's protocol.
Total RNA was reverse transcribed to cDNA using a
PrimeScript™ RT Master Mix (Perfect Real Time) cDNA
synthesis kit (Takara Biotechnology Co., Ltd., Dalian, China),
according to the manufacturer's protocol. Amplifications were
performed in a quantitative real-time PCR machine (Agilent
Technologies, Inc., Santa Clara, CA, USA) using SYBR Premix Ex
Taq (Takara Biotechnology Co., Ltd.). The PCR cycling
conditions were conducted as follows: 95°C for 30 sec followed by
40 cycles of 95°C for 5 sec and 60°C for 31 sec. The primer
sequences used were as follows: Human β-actin forward,
5′-AGCGAGCATCCCCCAAAGTT-3′; and reverse, 5′-GGGCACGAAGGCTCATCATT-3′
(15); human PYCR1 forward,
5′-ACACCCCACAACAAGGAGAC-3′; and reverse, 5′-CTGGAGTGTTGGTCATGCAG-3′
(16). All samples were loaded in
triplicates. Relative mRNA expression levels were compared via the
2−ΔΔCq method or log-transformed (17). The data were analyzed using MxPro qPCR
software v1.2 (Agilent Technologies, Inc.).
Western blot analysis
Total protein was extracted from frozen tissues or
cells using RIPA lysis buffer (Beyotime Institute of Biotechnology,
Haimen, China) containing a protease inhibitor cocktail (Roche,
Basel, Switzerland), at 4°C for 20 min, and centrifuged at 14,000 ×
g, for 10 min at 4°C. A total of 50 mg of protein (5 mg/µl sourced
from the tissues and 2.5 mg/µl sourced from the cells) were
separated on 12% SDS-PAGE and blotted onto polyvinylidene fluoride
Immobilon-P membranes (EMD Millipore, Billerica, MA, USA).
Following blocking with 5% milk solution for 2 h at room
temperature, the membranes were incubated with primary antibodies
against PYCR1 (cat. no. ab150347; dilution, 1:1,000; Abcam,
Cambridge, MA, USA), β-tubulin (cat. no. 2128; dilution, 1:1,000),
cyclin D1 (cat. no. 2978; dilution, 1:1,000), B-cell lymphoma-2
(Bcl-2; cat. no. 2870; dilution, 1:1,000), B-cell lymphoma-extra
large (Bcl-xl; cat. no. 2764; dilution, 1:1,000) and BCL2
associated X, apoptosis regulator (Bax; cat. no. 5023; dilution,
1:1,000), all purchased from Cell Signaling Technology, Inc.,
Danvers, MA, USA, overnight at 4°C. The following day, membranes
were probed with horseradish peroxidase (HRP)-conjugated
anti-rabbit IgG secondary antibody (dilution, 1:10,000; cat. no.
7074; Cell Signaling Technology, Inc., Danvers, MA, USA), for 2 h
at room temperature. The bands were detected using an Immobilon
Western enhanced chemiluminescence HRP substrate kit (cat. no.
WBKLS0500; EMD Millipore).
Immunohistochemistry (IHC)
Sections of formalin-fixed and paraffin-embedded
specimens (5-µm thick) were deparaffinized with xylene and
rehydrated in a descending ethanol series, 100, 95, 70, 50 and 30%.
Heat-induced antigen retrieval was performed by submerging slides
in 0.01 mol/l sodium citrate buffer (pH 6.0), and boiling in a
microwave oven for 10 min. The sections were treated with 10%
normal goat serum (cat. no. ZLI-9021; ZSJQ-BIO, Beijing, China) to
block non-specific binding, for 30 min at room temperature.
Subsequently, the sections were incubated at 4°C overnight with the
aforementioned primary rabbit anti-PYCR1 antibody (1:200; Abcam).
The following day, slides were incubated with HRP-conjugated
anti-rabbit IgG secondary antibody (cat no. PV-9001; ZSJQ-BIO;
dilution, 1:50), at room temperature for 30 min. Finally, the
sections were stained using 3,3′-diaminobenzidine for 10 sec-1 min
at room temperature, and counterstained with 0.5% hematoxylin for 1
sec at room temperature. Subsequently, slides were dehydrated, and
mounted.
IHC staining was independently scored by two
pathologists from the Department of Pathology, Jinling Hospital,
without any prior knowledge of patient characteristics, and any
discrepancy was solved by consensus review. Immunostaining was
assessed in five fields of view for each sample, under a light
microscope (Carl Zeiss AG, Oberkochen, Germany), at ×200
magnification. The presence of brown staining within cells was
considered to be positive staining for PYCR1. Scores representing
the percentage of positive cells were as follows: 0, ≤5%; 1, 6–20%;
2, 21–50% and 3, ≥50%. Staining intensity was scored as follows: 0,
negative staining; 1, weak staining; 2, moderate staining; and 3,
strong staining. The product of the two scores was used as the
total staining score. In the following analysis, the expression
levels were divided into two groups based on the final scores: Low
expression (<3.5) and high expression (≥3).
Cell transfection
A549 (2×105 per well) and H1703
(1.5×105 per well) cells were cultured for 1 day until
they reached 70–90% confluence. Cells were transfected with
PYCR1-specific small interfering (si)RNA or control siRNA using the
Lipofectamine 2000 transfection reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
sequences of the siRNAs used were as follows: Control siRNA sense,
5′-UUCUCCGAACGUGUCACGUTT-3′; and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′; siRNA-PYCR1 sense,
5′-GCCACAGUUUCUGCUCUCATT-3′; and antisense,
5′-UGAGAGCAGAAACUGUGGCTT-3′.
Cell proliferation assays
Cell proliferation was measured using the CCK-8 cell
proliferation kit (Dojindo Molecular Technologies, Inc., Kumamoto,
Japan) and 5-ethynyl-2′deoxyuridine (EdU) assay kit (Guangzhou
Ribobio Co., Ltd., Guangzhou, China), respectively. For the CCK-8
assay, siRNA-PYCR1 or control siRNA-transfected cells were cultured
in a 96-well plate at a density of 3,000 cells per well. CCK-8
solution (20 µl) was added to each well after 0, 24, 48, 72 and 96
h. Cells were incubated at 37°C for 2 h, and the absorbance of
samples was recorded at 450 nm using an epoch microplate
spectrophotometer (BioTek Instruments, Inc., Winooski, VT, USA).
All the experiments were performed in triplicate.
For the EdU incorporation assay, dissociated cells
were exposed to 50 mM EdU (Guangzhou RiboBio Co., Ltd.) for 2 h at
37°C. Following fixation with 4% formaldehyde for 15 min and
permeabilization with 0.5% Triton X-100 for 10 min at room
temperature, the cells were incubated with 1X Apollo reaction
cocktail (Guangzhou RiboBio Co., Ltd.), 100 µl/well for 30 min.
Then, the cells were stained with Hoechst 33342 for 30 min at room
temperature and visualized in 3 fields of view/well under a
fluorescence microscope (Carl Zeiss). The EdU incorporation rate
was expressed as the ratio of EdU-positive cells to total Hoechst
33342-positive cells.
Clone formation assay
SiRNA-PYCR1 or control siRNA-transfected cells were
seeded into a 6-well culture plate at a density of 1,500 cells per
dish. Cells were maintained in DMEM containing 10% FBS for 2 weeks
and stained with crystal violet for 20 min at room temperature, for
colony counting. The visible colonies were manually counted with
the naked eye. All experiments were done in triplicate.
Flow cytometry analysis for cell cycle
and apoptosis
Cells, cultured at a density of
1×105/assay, were trypsinized into single cell
suspensions and fixed with 70% ethanol for 30 min on ice. Then, the
cells were stained with propidium iodide (PI) using the CycleTEST
TM PLUS DNA reagent kit (BD Biosciences, San Jose, CA, USA) and
analyzed using a FACS Calibur flow cytometer (BD Biosciences)
equipped with BD CellQuest software v6.1 (BD Biosciences). For
apoptosis analysis, cultured cells were harvested by trypsinization
and were double stained with Annexin V-fluorescein
isothiocyanate/PI apoptosis detection (BD Biosciences) according to
the manufacturer's protocol, and analyzed with the aforementioned
flow cytometer and software.
Statistical analysis
Statistical evaluation was performed using SPSS 19.0
software (IBM Corp., Armonk, NY, USA). Two-group comparisons were
performed using the Student's t-test. χ2 and Fisher's
exact tests were used to analyze the associations between PYCR1
expression and clinicopathological features. Kaplan-Meier survival
analyses and Cox's proportional hazards models were utilized to
determine the association between PYCR1 expression and survival.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Elevated PYCR1 expression in NSCLC
tissues and cell lines
To investigate the differences between PYCR1
transcript levels between NSCLC tumor tissues and normal tissues,
28 paired samples of fresh frozen normal tissues and tumor tissues
were randomly selected, and the mRNA expression levels examined
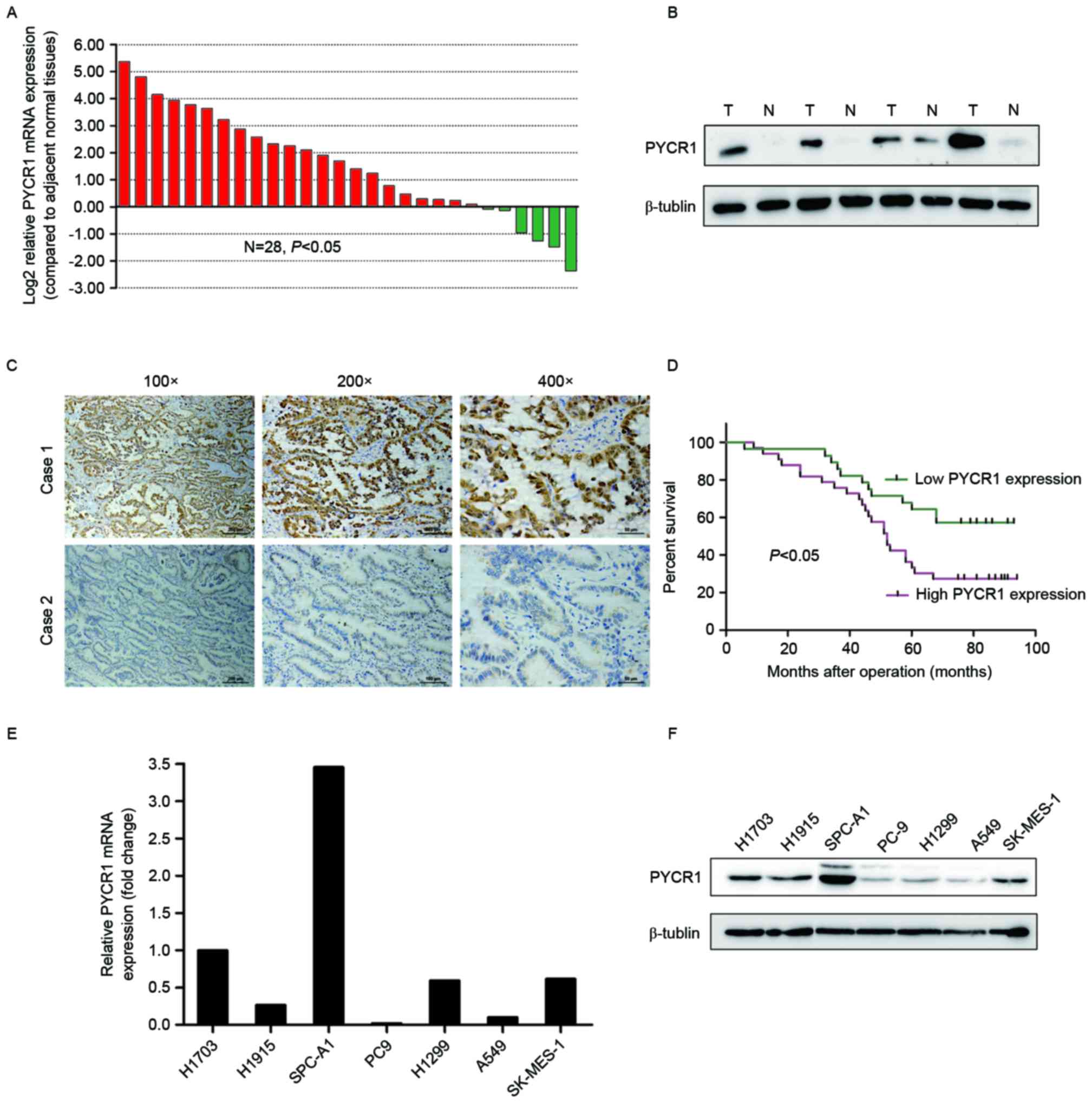
using RT-qPCR. The results revealed that 16/28 patients (57.1%)
demonstrated higher PYCR1 mRNA expression levels in NSCLC specimens
compared with normal tissue specimens (≥2-fold change; Fig. 1A). Overexpression of PYCR1 in lung
cancers was further supported by data analysis from the Oncomine
platform, which revealed higher PYCR1 mRNA expression levels in
lung adenocarcinoma tissues, large cell lung carcinoma tissues and
squamous cell lung carcinoma tissues than in normal lung tissues.
In 4 paired specimens, PYCR1 was overexpressed in all lung tumor
samples compared with normal tissues, as detected by western blot
(Fig. 1B).
To further study whether PYCR1 protein levels were
associated with clinicopathological features and prognosis of
patients with NSCLC, 62 paraffin-embedded archived NSCLC tissues
were examined using immunohistochemical staining. Representative
examples of tumor tissues with either strong or weak staining of
PYCR1 are presented in Fig. 1C. The
associations between PYCR1 cytoplasmic staining in tumor cells and
the clinicopathological features of the 62 patients with NSCLC were
further analyzed (Table I). There was
no significant association between PYCR1 expression and any
clinicopathological characteristic. However, there was a trend
towards higher PYCR1 protein expression in tumor specimens with a
larger size (>4 cm). Clinicopathological characteristics and
PYCR1 protein expression were also analyzed using Cox's univariate
and multivariate hazard regression models to evaluate whether high
PYCR1 expression was an independent risk factor of poor prognosis
(Table II). Kaplan-Meier analysis
demonstrated that poor overall survival was associated with lymph
node metastasis (P=0.010), tumor-node-metastasis (TNM) stage
(P=0.010) and PYCR1 expression (Fig.
1D; P=0.01). For multivariate survival analysis, TNM stage was
associated with overall patient survival [hazard ratio (HR): 1.926,
95% confidence interval (CI): 1.149–3.227, P=0.013]. High PYCR1
protein expression was also revealed to be an independent risk
factor for poor prognosis (HR: 2.48, 95% CI: 1.164–5.286,
P=0.019).
 | Table I.Associations between PYCR1 expression
and various clinicopathologic features of 62 patients with non
small cell lung cancer. |
Table I.
Associations between PYCR1 expression
and various clinicopathologic features of 62 patients with non
small cell lung cancer.
|
|
| PYCR1
expression |
|---|
|
|
|
|
|---|
| Variable | No. of cases | High | Low |
P-valuea |
|---|
| Age at diagnosis,
years |
|
|
|
|
|
<60 | 24 | 13 | 11 | 0.933 |
|
≥60 | 38 | 21 | 17 |
|
| Sex |
|
|
|
|
|
Male | 52 | 30 | 22 | 0.737 |
|
Female | 11 | 6 | 5 |
|
| Histological
type |
|
|
|
|
| AC | 27 | 17 | 10 | 0.365 |
|
SCC | 25 | 11 | 14 |
|
|
Otherb | 10 | 6 | 4 |
|
| Tumor
differentiation |
|
|
|
|
| Well,
moderate | 37 | 22 | 15 | 0.374 |
|
Poor | 25 | 12 | 13 |
|
| Tumor size |
|
|
|
|
| ≤4
cm | 35 | 16 | 19 | 0.100 |
| >4
cm | 27 | 18 | 9 |
|
| Lymph node
metastasis |
|
|
|
|
|
Absent | 37 | 19 | 18 | 0.502 |
|
Present | 25 | 15 | 10 |
|
| TNM stage |
|
|
|
|
| I | 20 | 9 | 11 | 0.560 |
| II | 25 | 15 | 10 |
|
|
III | 17 | 10 | 7 |
|
 | Table II.Univariate and multivariate analysis
of prognostic factors in 61 patients with non small cell lung
cancer. |
Table II.
Univariate and multivariate analysis
of prognostic factors in 61 patients with non small cell lung
cancer.
|
| Univariate
analysisa | Multivariate
analysisb |
|---|
|
|
|
|
|---|
| Variable | No. of cases | Mean survival
(months) | P-value | HR (95% CI) | P-value |
|---|
| Age at diagnosis,
years |
|
| 0.794 |
|
|
|
<60 | 24 | 64.04 |
|
|
|
|
≥60 | 37 | 62.78 |
|
|
|
| Sex |
|
| 0.129 |
|
|
|
Male | 51 | 65.44 |
|
|
|
|
Female | 10 | 51.50 |
|
|
|
| Histological
type |
|
| 0.447 |
|
|
| AC | 27 | 65.60 |
|
|
|
|
SCC | 24 | 63.63 |
|
|
|
|
Otherc | 10 | 54.70 |
|
|
|
| Tumor
differentiation |
|
| 0.445 |
|
|
| Well,
moderate | 36 | 60.72 |
|
|
|
|
Poor | 25 | 66.96 |
|
|
|
| Tumor size |
|
| 0.672 |
|
|
| ≤4
cm | 35 | 62.94 |
|
|
|
| >4
cm | 26 | 62.46 |
|
|
|
| Lymph node
metastasis |
|
| 0.010 | 1.060
(0.461–2.439) | 0.890 |
|
Absent | 37 | 70.61 |
|
|
|
|
Present | 24 | 52.52 |
|
|
|
| TNM stage |
|
| 0.010 | 1.926
(1.149–3.227) | 0.013 |
| I | 20 | 74.80 |
|
|
|
| II | 24 | 59.17 |
|
|
|
|
III | 17 | 53.82 |
|
|
|
| PYCR1
expression |
|
| 0.011 | 2.480
(1.164–5.286) | 0.019 |
|
High | 33 | 55.49 |
|
|
|
|
Low | 28 | 72.25 |
|
|
|
In addition, the mRNA and protein levels of PYCR1
were examined in seven NSCLC cell lines by qPCR and western
blotting, respectively (Fig. 1E and
F). The results demonstrated that PYCR1 expression level is
highly variable in NSCLC cell lines. According to the results, two
NSCLC cell lines with high PYCR1 expression (SPC-A1 and H1703) were
selected to perform further in vitro investigations.
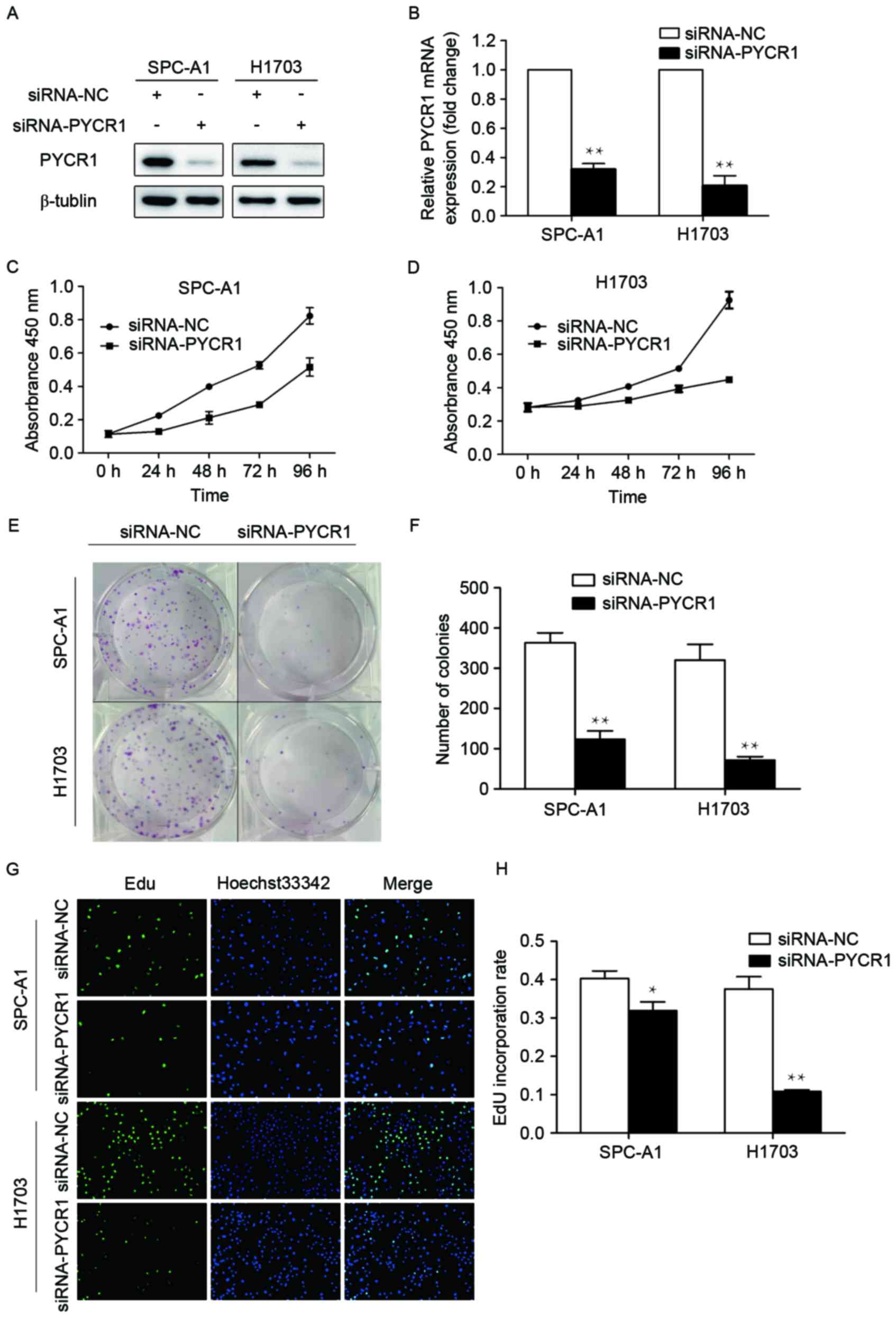
Knockdown of PYCR1 expression in NSCLC
cells suppresses cell proliferation in vitro
As demonstrated by the aforementioned results,
tumors with a larger size tended to have relatively high PYCR1
expression, and overexpression of PYCR1 may be involved in
survival. Therefore, the influence of PYCR1 expression on the
proliferation of human NSCLC cells was studied. SiRNA was used to
specifically knock down PYCR1 expression in SPC-A1 and H1703 cells
(Fig. 2A and B). PYCR1 knockdown
reduced SPC-A1 and H1703 cell proliferation, as assessed using a
CCK-8 cell proliferation kit (Fig. 2C and
D). The colony-forming ability of the population was also
significantly attenuated following knockdown of PYCR1 (Fig. 2E and F). In addition, EdU
incorporation assays were performed to explore the effect of PYCR1
knockdown on DNA replication. Following transfection with
siRNA-PYCR1, the percentage of EdU-positive cells was decreased by
21% in SPC-A1 and 71% in H1703 cells (Fig. 2G and H). These data revealed that
PYCR1 is involved in NSCLC cell proliferation.
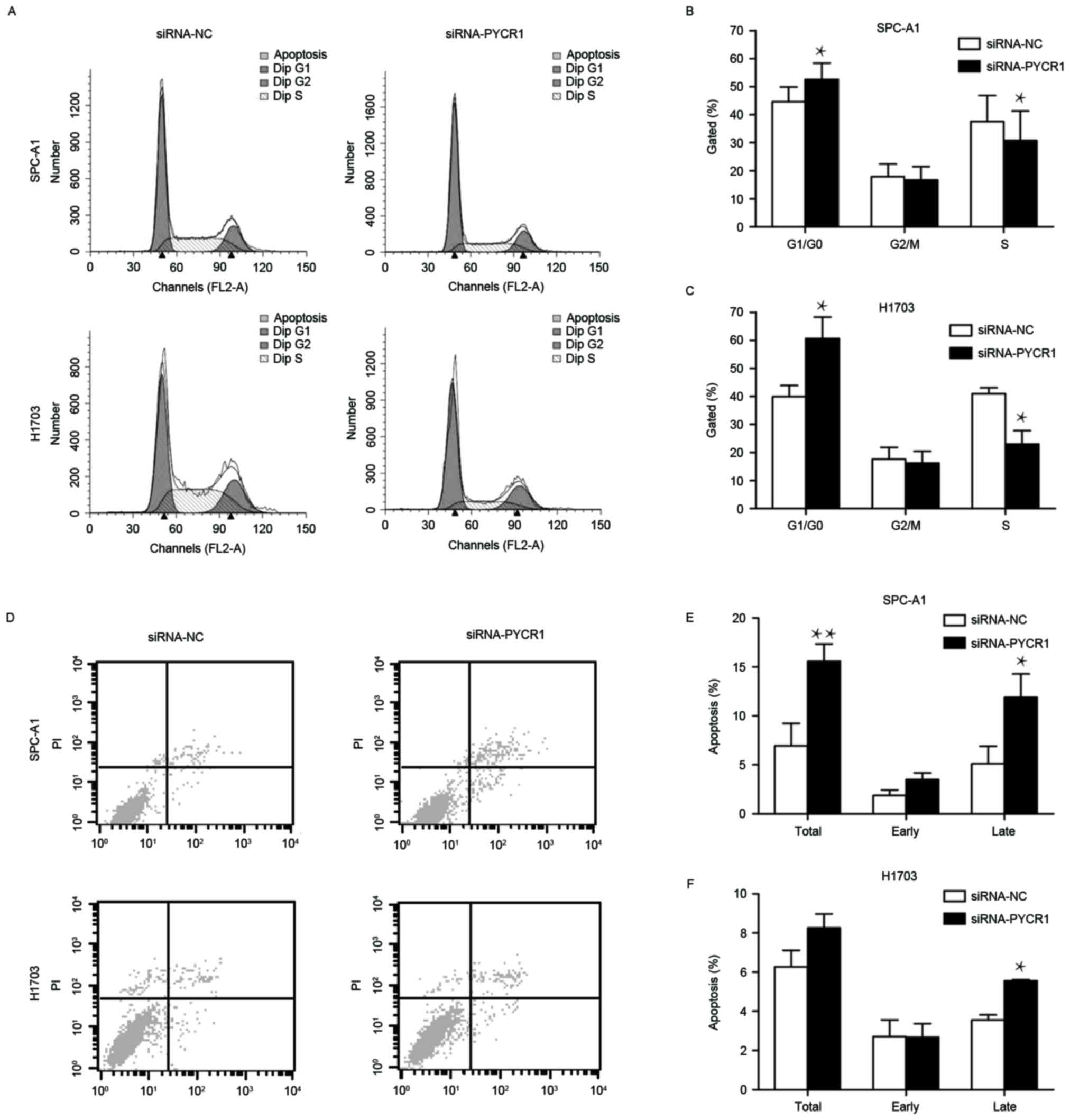
Silencing of PYCR1 induces cell cycle
arrest and apoptosis
To further examine the function of PYCR1 knockdown
in cell cycle and apoptosis, the cell cycle and apoptosis were
analyzed in siRNA-PYCR1 or control siRNA-transfected SPC-A1 and
H1703 cells using flow cytometry. In si-PYCR1-transfected SPC-A1
cells, the proportion of cells in the G1 phase increased by 14.6%
and the proportion of cells in S phase decreased by 21.6% (Fig. 3A and B). The altered proportion of G1
and S phase cells was more apparent in siRNA-PYCR1-transfected
H1703 cells (Fig. 3A and C). PYCR1
knockdown also induced apoptosis of SPC-A1 cells, in particular
late apoptosis (Fig. 3D and E). In
H1703 cells, although there was no significant difference in the
total apoptosis rate between si-PYCR1-transfected cells and control
cells, the late apoptosis rate was increased following PYCR1
silencing (Fig. 3D and F). Taken
together, these results demonstrated that the loss of PYCR1 results
in the arrest of the cell cycle in the G1 phase and the induction
of apoptosis in NSCLC cells.
PYCR1 promotes the cell cycle and
inhibits apoptosis through regulating cyclin D1, Bcl-2 and Bcl-xl
expression
In order to investigate the molecular mechanism
underlying the influence of PYCR1 on the cell cycle and apoptosis
in NSCLC, key associated regulators were focused on. The cyclin D
family are a group of closely related G1 cyclins, and of these
cyclin D1 exhibits a more widespread function in human cancers
compared with others (18). Thus, it
was hypothesized that PYCR1 may promote the cell cycle from the G1
phase into the S phase by regulating the expression of cyclin D1.
To test this hypothesis, the expression levels of cyclin D1
following PYCR1 silencing were examined by western blot. The
results revealed a significant decrease of cyclin D1 expression in
siRNA-PYCR1-transfected SPC-A1 and H1703 cells (Fig. 4A and B).
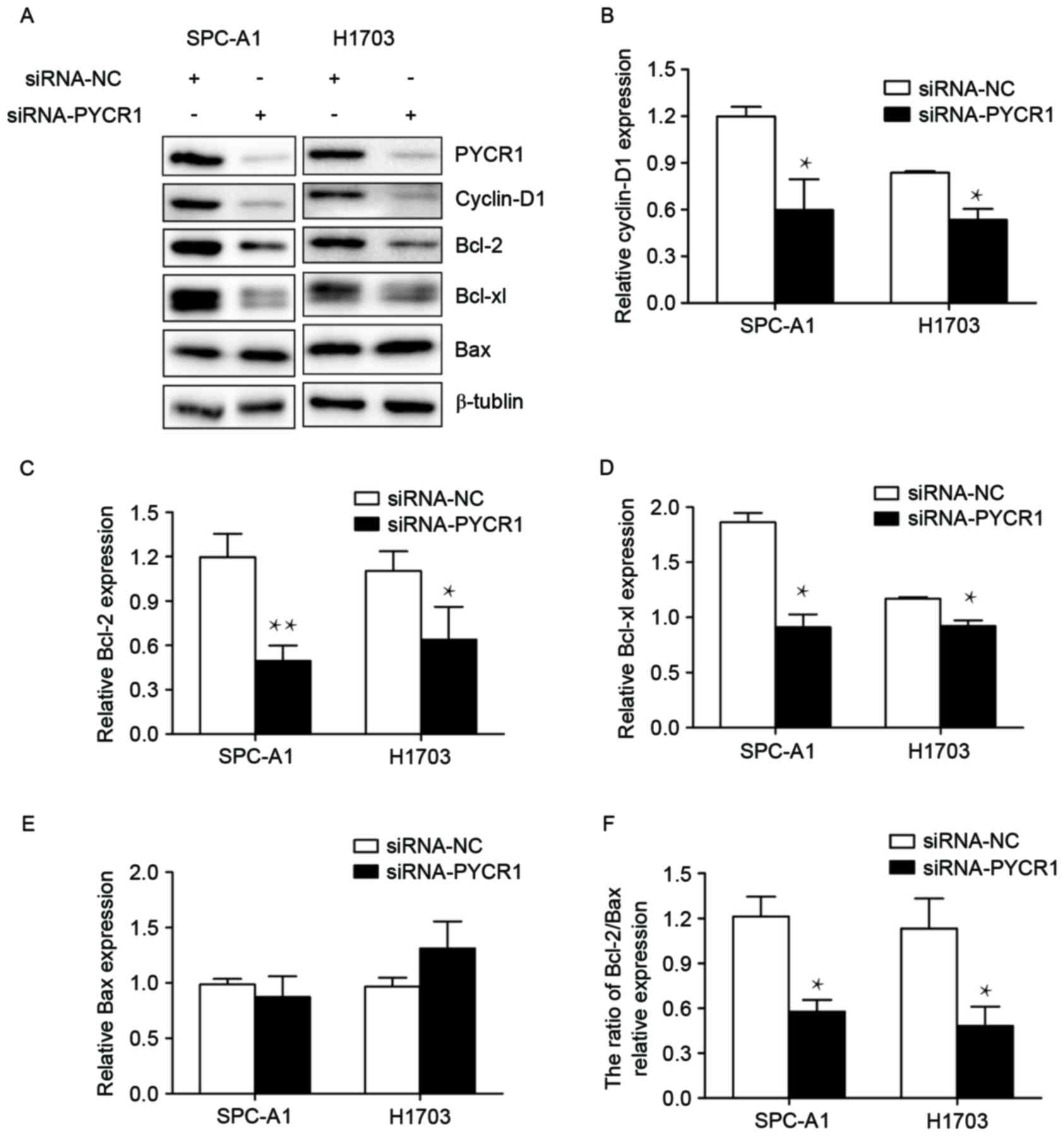 | Figure 4.Cyclin D1, Bcl-2 and Bcl-xl were
downregulated by the knockdown of PYCR1. (A) Protein expression
levels were assessed using western blot analysis following
silencing of PYCR1. Quantification was performed for (B) cyclin D1,
(C) Bcl-2, (D) Bcl-xl, (E) Bax and (F) the Bcl-2/Bax ratio.
β-tublin was used as an internal control. The data are expressed as
the mean ± standard error of the mean of three independent
experiments. Student's t-test was used to assess significance.
*P<0.05 and **P<0.01 vs. siRNA-NC. Bcl-2, B-cell lymphoma-2;
Bcl-xl, B-cell lymphoma-extra large; PYCR1, pyrroline-5-carboxylate
reductase 1; Bax, BCL2 associated X, apoptosis regulator; siRNA,
small interfering RNA; NC, negative control. |
As for apoptosis, the Bcl-2 family is essential to
mitochondrial-controlled apoptosis. As PYCR1 is located on the
surface of mitochondrion (19), it
was hypothesized that PYCR1 may inhibit apoptosis through its
connection to the Bcl-2 family. The results of the present study
demonstrated that, following PYCR1 knockdown, Bcl-2 expression
levels were significantly decreased in SPC-A1 and H1703 cells
(Fig. 4A and C). There was also a
decline in Bcl-xl expression in the two cell lines (Fig. 4A and D). The Bcl-2/Bax ratio was also
decreased, although Bax expression was not affected by the
knockdown of PYCR1 (Fig. 4A, E and
F).
Discussion
Disordered glutamine metabolism is involved in lung
cancer progression, and has been under intense study to search for
promising anticancer therapeutic targets (20). Glutamate is a source of proline
synthesis, and this relationship drew the attention of our group to
the potential function of proline synthesis in lung cancer.
Pyrroline-5-carboxylate (P5C) is an intermediate produced from
glutamate by P5C synthase, or from ornithine by ornithine
aminotransferase in proline synthesis pathways, and this is then
converted to proline through PYCR. Of the three known isozymes of
PYCR, PYCR1 is primarily involved in the conversion of glutamate to
proline in human melanoma cells (21). A DNA microarray analysis of prostate
cancer in 2002 revealed that the expression levels of PYCR1 were
significantly increased in prostate cancer (22). The results of the present study
demonstrated that PYCR1 is overexpressed in human NSCLC, as
detected by RT-qPCR and western blot. Then, by analyzing the
clinical significance of PYCR1 expression, it was revealed that in
NSCLC tumors with dimensions >4 cm, PYCR1 expression was higher
than those with smaller dimensions (66.7% vs. 45.7%). Following
multivariate survival analysis, high PYCR1 expression was revealed
to be an independent risk factor for poor overall survival, as well
as higher TNM stage.
The involvement of PYCR1 in the proliferation and
apoptosis of NSCLC cell lines was also investigated. The results
indicated that silencing PYCR1 decreased cell proliferation,
resulted in cell cycle arrest at the G1 phase and induced
apoptosis. The different medium used for culturing SPC-A1 and H1703
did not alter the effect of PYCR1 on proliferation and apoptosis.
H1703 cells were cultured in RPMI-1640 with 0.1 mM proline, while
SPC-A1 cells were cultured in DMEM without proline. The similar
results in the two cell lines may suggest that the influence of
PYCR1 knockdown was not mitigated even with proline at a
concentration usually used for essential amino acids (0.1 mM).
Possemato et al (13) revealed
that knockdown of PYCR1 reduced tumor formation in breast cancer
cell lines. A previous study also confirmed the results of the
present study: Silencing of aldehyde dehydrogenase 18 family member
A1 (P5CS), PYCR1, 2 and L may decrease tumor cell proliferation in
several cancer cell lines, including one NSCLC cell line, PC9
(23). However, PYCR1 knockdown did
not affect cell cycle and apoptosis in PC9 cells in this study,
which is incompatible with the results of the present study. The
discordance may be because the expression levels of PYCR1 were
decreased in PC9 cells compared with SPC-A1 and H1703 cells, as
demonstrated in Fig. 1E and F. The
variation in PYCR1 expression across different cell lines may cause
differential results in in vitro studies. Repeated studies
should be performed in more NSCLC cell lines to confirm the
involvement of PYCR1 in the cell cycle and apoptosis in this type
of cancer.
The oncogenic transcription factor MYC
proto-oncogene, bHLH transcription factor (MYC) is involved in cell
proliferation, most notably by targeting G1-specific
cyclin-dependent kinases, and it is also associated with apoptosis
control (24,25). MYC stimulates mitochondrial
biogenesis, which may be important in terms of cell cycle
promotion, by helping prepare for cell division (26). MYC promotes proline synthesis from
glutamine to proline through increasing the expression of
glutaminase, P5CS, and PYCR1. It also inhibits the expression of
POX/PRODH and thereby inhibits its function, which is to suppress
cell growth and induce apoptosis (9,27). Thus,
the proline metabolism enzymes PYCR1 and POX/PRODH may contribute,
at least in part, to the effect of MYC on cell growth and
apoptosis.
The signaling pathway through which PYCR1 promotes
cell proliferation and inhibits apoptosis is still unknown. Cyclin
D1 is a key regulator of the G1/S checkpoints and forms a complex
with cyclin dependent kinase (CDK) 4/6 (28). Cyclin D1 is also involved in the
regulation of apoptosis, and whether it induces or inhibits
apoptosis depends on the cell type, its expression level and the
growth conditions (29). Due to its
involvement in controlling the cell cycle and apoptosis, as well as
in lung cancer pathogenesis (30),
cyclin D1 expression was detected in siRNA-transfected SPC-A1 and
H1703 cells and was revealed to be significantly decreased compared
with control cells. In addition, overexpression of cyclin D1
resulted in resistance to cisplatin-mediated apoptosis via the
maintenance of Bcl-2 and Bcl-xl protein levels in an ela-myc
transgene-expressing pancreatic tumor cell line (31). Bcl-2 family members are critical
regulators of the mitochondrial apoptotic pathway through the
balance of and competitive dimerization between the anti-apoptotic
members, including Bcl-2 and Bcl-xl, and pro-apoptotic members,
including Bax and Bcl-2 antagonist/killer 1 (32). High expression of Bcl-2 is associated
with an improved outcome in patients with NSCLC with a non-squamous
histology, which suggests its involvement in NSCLC (33). In the present study, Bcl-2 and Bcl-xl
expression, as well as the ratio of Bcl-2/Bax, were revealed to be
downregulated following PYCR1 silencing. Taken together, these
studies suggested that PYCR1 may promote tumor cell growth by
regulating cyclin D1, Bcl-2 and Bcl-xl. Nevertheless, whether the
downregulation of Bcl-2 and Bcl-xl was directly caused by PYCR1 or
through the low expression of cyclin D1 is worth further
research.
In conclusion, the results of the present study
demonstrated that PYCR1 is overexpressed in NSCLC, and its high
expression is an independent risk factor for poor prognosis. PYCR1
may be a key promoter of tumor cell proliferation. In the future,
these data should be expanded upon using in vivo models to
further elucidate the involvement of PYCR1 in tumorigenesis.
Acknowledgements
The authors would like to thank Dr Zhongdong Lee
from The Department of Cardiothoracic Surgery, Jinling Hosptial,
for assisting in collecting specimens.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
PYCR1
|
pyrroline-5-carboxylate reductase
1
|
|
PRODH/POX
|
proline dehydrogenase/proline
oxidase
|
|
siRNA
|
small interfering RNA
|
|
Bcl-2
|
B cell lymphoma-2
|
|
TNM
|
tumor-node-metastasis
|
References
|
1
|
GBD 2013 Mortality and Causes of Death
Collaborators: Global, regional, and national age-sex specific
all-cause and cause-specific mortality for 240 causes of death,
1990–2013: A systematic analysis for the Global Burden of Disease
Study 2013. Lancet. 385:117–171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: The metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Johnson C, Warmoes MO, Shen X and Locasale
JW: Epigenetics and cancer metabolism. Cancer Lett. 356:309–314.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Warburg O: On the origin of cancer cells.
Science. 123:309–314. 1956. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cairns RA, Harris IS and Mak TW:
Regulation of cancer cell metabolism. Nat Rev Cancer. 11:85–95.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Phang JM and Liu W: Proline metabolism and
cancer. Front Biosci (Landmark Ed). 17:1835–1845. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Phang JM, Liu W, Hancock CN and Fischer
JW: Proline metabolism and cancer: Emerging links to glutamine and
collagen. Curr Opin Clin Nutr Metab Care. 18:71–77. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maxwell SA and Davis GE: Differential gene
expression in p53-mediated apoptosis-resistant vs.
Apoptosis-sensitive tumor cell lines. Proc Natl Acad Sci USA.
97:pp. 13009–13014. 2000; View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maxwell SA and Rivera A: Proline oxidase
induces apoptosis in tumor cells, and its expression is frequently
absent or reduced in renal carcinomas. J Biol Chem. 278:9784–9789.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu W and Phang JM: Proline dehydrogenase
(oxidase) in cancer. Biofactors. 38:398–406. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guernsey DL, Jiang H, Evans SC, Ferguson
M, Matsuoka M, Nightingale M, Rideout AL, Provost S, Bedard K, Orr
A, et al: Mutation in pyrroline-5-carboxylate reductase 1 gene in
families with cutis laxa type 2. Am J Hum Genet. 85:120–129. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Possemato R, Marks KM, Shaul YD, Pacold
ME, Kim D, Birsoy K, Sethumadhavan S, Woo HK, Jang HG, Jha AK, et
al: Functional genomics reveal that the serine synthesis pathway is
essential in breast cancer. Nature. 476:346–350. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fang Y, Fu D, Tang W, Cai Y, Ma D, Wang H,
Xue R, Liu T, Huang X, Dong L, et al: Ubiquitin C-terminal
Hydrolase 37, a novel predictor for hepatocellular carcinoma
recurrence, promotes cell migration and invasion via interacting
and deubiquitinating PRP19. Biochim Biophys Acta. 1833:559–572.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jariwala U, Prescott J, Jia L, Barski A,
Pregizer S, Cogan JP, Arasheben A, Tilley WD, Scher HI, Gerald WL,
et al: Identification of novel androgen receptor target genes in
prostate cancer. Mol Cancer. 6:392007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Musgrove EA: Cyclins: Roles in mitogenic
signaling and oncogenic transformation. Growth Factors. 24:13–19.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
De Ingeniis J, Kazanov MD, Shatalin K,
Gelfand MS, Osterman AL and Sorci L: Glutamine versus ammonia
utilization in the NAD synthetase family. PLoS One. 7:e391152012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mohamed A, Deng X, Khuri FR and Owonikoko
TK: Altered glutamine metabolism and therapeutic opportunities for
lung cancer. Clin Lung Cancer. 15:7–15. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
De Ingeniis J, Ratnikov B, Richardson AD,
Scott DA, Aza-Blanc P, De SK, Kazanov M, Pellecchia M, Ronai Z,
Osterman AL and Smith JW: Functional specialization in proline
biosynthesis of melanoma. PLoS One. 7:e451902012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ernst T, Hergenhahn M, Kenzelmann M, Cohen
CD, Bonrouhi M, Weninger A, Klären R, Gröne EF, Wiesel M, Güdemann
C, et al: Decrease and gain of gene expression are equally
discriminatory markers for prostate carcinoma: A gene expression
analysis on total and microdissected prostate tissue. Am J Pathol.
160:2169–2180. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu W, Hancock CN, Fischer JW, Harman M
and Phang JM: Proline biosynthesis augments tumor cell growth and
aerobic glycolysis: Involvement of pyridine nucleotides. Sci Rep.
5:172062015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bretones G, Delgado MD and Leon J: Myc and
cell cycle control. Biochim Biophys Acta. 1849:506–516. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
McMahon SB: MYC and the control of
apoptosis. Cold Spring Harb Perspect Med. 4:a0144072014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Morrish F and Hockenbery D: MYC and
mitochondrial biogenesis. Cold Spring Harb Perspect Med. 4:pii:
a0142252014. View Article : Google Scholar
|
|
27
|
Liu W, Le A, Hancock C, Lane AN, Dang CV,
Fan TW and Phang JM: Reprogramming of proline and glutamine
metabolism contributes to the proliferative and metabolic responses
regulated by oncogenic transcription factor c-MYC. Proc Natl Acad
Sci USA. 109:pp. 8983–8988. 2012; View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kato JY, Matsuoka M, Strom DK and Sherr
CJ: Regulation of cyclin D-dependent kinase 4 (cdk4) by
cdk4-activating kinase. Mol Cell Biol. 14:2713–2721. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han EK, Ng SC, Arber N, Begemann M and
Weinstein IB: Roles of cyclin D1 and related genes in growth
inhibition, senescence and apoptosis. Apoptosis. 4:213–219. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gautschi O, Ratschiller D, Gugger M,
Betticher DC and Heighway J: Cyclin D1 in non-small cell lung
cancer: A key driver of malignant transformation. Lung Cancer.
55:1–14. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Biliran H Jr, Wang Y, Banerjee S, Xu H,
Heng H, Thakur A, Bollig A, Sarkar FH and Liao JD: Overexpression
of cyclin D1 promotes tumor cell growth and confers resistance to
cisplatin-mediated apoptosis in an elastase-myc
transgene-expressing pancreatic tumor cell line. Clin Cancer Res.
11:6075–6086. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Adams JM and Cory S: The Bcl-2 protein
family: Arbiters of cell survival. Science. 281:1322–1326. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Anagnostou VK, Lowery FJ, Zolota V,
Tzelepi V, Gopinath A, Liceaga C, Panagopoulos N, Frangia K, Tanoue
L, Boffa D, et al: High expression of BCL-2 predicts favorable
outcome in non-small cell lung cancer patients with non squamous
histology. BMC Cancer. 10:1862010. View Article : Google Scholar : PubMed/NCBI
|