Introduction
Hepatocellular carcinoma (HCC) is one of the most
common primary malignancies of the liver, the third leading cause
of cancer-associated mortalities after stomach and lung cancer, and
the sixth most common neoplasm worldwide (1,2). Current
treatments for HCC include chemotherapy, radiotherapy and surgical
intervention. However, drug resistance is a growing concern as a
result of the increased application of chemotherapeutics (3). Determining the molecular mechanisms
underlying drug resistance in HCC cells is therefore
imperative.
MicroRNAs (miRNAs/miRs) are small, non-coding RNAs
that modulate post-transcriptional gene expression by binding to
the 3′-untranslated region (3′-UTR) of target mRNAs, in addition to
being involved in the regulation of essential cellular processes,
including proliferation, diversification, metastasis and apoptosis,
particularly in cancer development and progression (4–6). Aberrant
miRNA expression has demonstrated potential as a prognostic or
diagnostic marker in various types of cancer, including breast,
non-small cell lung and colon cancer (7–10). The
expression levels of miRNAs reflect the developmental lineage and
differentiation state of tumors (11). Studies have demonstrated that miR-185
suppresses cell growth and epithelial-mesenchymal transition
progression by targeting Six2, providing a novel target for the
molecular treatment of liver malignancies (12). It has been demonstrated that a loss of
miR-211 expression, and consequently uncontrolled secreted protein,
acidic and rich in cysteine overexpression, may drive progression
of HCC (13).
Previous studies have indicated that the levels of
miR-9 are downregulated in certain types of cancer, such as ovarian
cancer, gastric cancer and neuroblastoma (14–17);
however, it has also been revealed that the expression of miR-9 is
upregulated in colorectal, breast, lung and laryngeal squamous cell
cancer types (18–22). Other studies have indicated that miR-9
may serve a major role in tumor progression and tumorigenesis
(23). The eukaryotic translation
initiation factor 5A-2 (eIF-5A-2), located at 3q26, is one of the
most frequently observed chromosomal alterations in ovarian
carcinoma (24). Overexpression of
eIF-5A-2 has also been observed in other solid tumors and has been
defined as an adverse prognostic marker in HCC (25), ovarian carcinoma (26), bladder carcinoma (27) and non-small cell lung cancer (28), and as a metastasis-promoting factor in
HCC (29) and colorectal carcinoma
(30). In the present study, an miRNA
target prediction website, TargetScan (www.targetscan.org), was used to identify potential
targets of miR-9. The 3′UTR of eIF-5A-2 was identified as a
potential target, and examination of the sequence of the 3′UTR of
eIF-5A-2 identified multiple potential sites for interaction with
miR-9. The mechanisms underlying the role of miR-9 in the
regulation of HCC cells have not been characterized. We
hypothesized that miR-9 may affect the expression of eIF-5A-2 and,
furthermore, may regulate the sensitivity of epithelial HCC cells
to cetuximab.
The present study investigated the effects of
miR-9/eIF-5A-2-regulated cetuximab sensitivity in HCC cell lines
and monitored the expression of miR-9 and eIF-5A-2 mRNA. We aimed
to investigate whether or not miR-9, through regulating the
expression of eIF-5A-2, may affect the sensitivity of HCC cells
with an epithelial phenotype to cetuximab.
Materials and methods
Cell culture
Human HCC cell lines (Hep3B, Huh7, SNU387 and
SNU449) were obtained from American Type Culture Collection (ATCC;
Manassas, VA, USA). Huh7 cells (epithelial phenotype HCC) were
maintained in Dulbecco's modified Eagle's medium containing 10%
fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) and 1% penicillin/streptomycin (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany). SNU387 and SNU449 cells
(mesenchymal phenotype HCC) were cultured in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
and 1% penicillin/streptomycin. Hep3B cells (epithelial phenotype
HCC) were cultured in minimum essential medium (MEM; Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS and 1%
penicillin/streptomycin. All cells were maintained under 5%
CO2 at 37°C in a humidified incubator. Cetuximab and the
miR-9 mimic and inhibitor were all purchased from Guangzhou RiboBio
Co., Ltd. (Guangzhou, China). The eIF-5A-2 siRNA and negative
control siRNA were designed by and purchased from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA). Furthermore, TargetScan
(www.targetscan.org) was to predict the
associated miRNAs for eIF-5A-2. Briefly, we opened TargetScan was
opened and the human species chosen. The gene symbol eIF-5A-2 was
searched to obtain the predicted associated miRNAs.
siRNA transfections
HCC cell lines were transfected with eIF-5A-2 siRNA
(100 nM; Santa Cruz Biotechnology, Inc.; forward,
5′-TATGCAGTGCTCGGCCTTG-3′ and reverse,
5′-TTGGAACATCCATGTTGTGAGTAGA-3′) or a negative control siRNA using
Lipofectamine 2000 (Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocols. The transfection medium (Opti-MEM;
Gibco; Thermo Fisher Scientific, Inc.) was replaced with complete
medium (Gibco; Thermo Fisher Scientific, Inc.) 6 h after
transfection, and the cells were incubated at 37°C for 24 h. All
treatments were initiated 24 h after transfection.
Cell viability assay
HCC cell lines or siRNA-transfected HCC cell lines
were seeded onto 96-well plates at a density of 5×103
cells/well. The medium was replaced with the corresponding
serum-free medium for 24 h to synchronize the cells, prior to the
culture medium being replaced with complete medium containing the
drug at the indicated concentrations (Cetuximab: Hep3B, 2,229
µg/ml; Huh7, 2,564 µg/ml; SNU387, 3,182 µg/ml; SNU449, 2,450 µg/ml)
for 48 h at 37°C. Following treatment, 10 µl Cell Counting kit-8
solution was added and the cells were incubated at 37°C for a
further 3 h. Subsequently, absorbance at 450 nm was measured using
an MRX II microplate reader (Dynex Technologies, Inc., Chantilly,
VA, USA). Relative cell viability was calculated as a percentage of
the untreated controls.
Western blot analysis
To extract proteins after 48 h of treatment, cell
lysis buffer (Cell Signaling Technology, Inc., Danvers, MA, USA)
containing protease inhibitors (Sigma-Aldrich; Merck KGaA) was
added to harvested HCC cells in an ice bath. Cells were centrifuged
at 12,000 × g for 5 min at 4°C following the lysis. The supernatant
was collected and the protein concentration was measured with a
bicinchoninic acid protein assay kit (Sigma-Aldrich; Merck KGaA).
The protein samples (40 µg/lane) were separated by 10% SDS-PAGE and
transferred to polyvinylidene difluoride membranes (EMD Millipore,
Billerica, MA, USA). Membranes were blocked with Tris-buffered
saline and 0.1% Tween 20 (TBST) containing 5% bovine serum albumin
at 37°C for 2 h, and incubated overnight at 4°C with the eIF-5A-2
primary antibody (Abcam, Cambridge, MA, USA; ab150439; diluted
1:1,000 in TBST). The membranes were washed 3 times with TBST and
were incubated with a horseradish peroxidase-conjugated secondary
antibody (Abcam; ab97051; 1:2,000) for 2 h at room temperature. The
protein bands were detected using an enhanced chemiluminescence kit
(GE Healthcare Life Sciences, Little Chalfont, UK). Bands were
quantified by densitometry using Image Lab version 5.0 (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) and GAPDH (Cell Signaling
Technology, Inc.; cat no. 5174S; diluted 1:2,000 in TBST) was used
as an internal control.
5-ethynyl-2′-deoxyuridine (EdU)
incorporation assay
HCC cell lines were seeded onto 96-well plates at a
density of 5×103 cells/well in their respective growth
medium. The medium was replaced with the corresponding serum-free
medium to synchronize the cells. After 24 h, the serum-free medium
was replaced with growth medium containing drugs at the
aforementioned concentrations (Cetuximab: Hep3B, 2,229 µg/ml; Huh7,
2,564 µg/ml; SNU387, 3,182 µg/ml; SNU449, 2,450 µg/ml) for 48 h.
Cell proliferation was assessed through an EdU assay using the
Click-iTEdU Imaging kit (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) according to the manufacturer's protocols,
and cells were counterstained with 100 µl Hoechst 3342 at room
temperature for 30 min in the dark.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from Hep3B, Huh7, SNU387 and
SUN449 cells using TRIzol reagent (Invitrogen; Carlsbad, CA, USA)
and reverse transcribed into cDNA using Prime Script reagent RT kit
(Takara Biotechnology, Co., Ltd., Dalian, China) and the following
thermocycling conditions: 42°C for 2 min, 15 min at 37°C and 5 sec
at 85°C. Expression of eIF-5A-2 mRNA was normalized to U6 and
relative quantification was performed using the comparative
2−ΔΔCt method (31). SYBR
Green PCR (Takara Biotechnology, Co., Ltd., Dalian, China) was
performed at 95°C for 30 sec, followed by 40 cycles of denaturation
at 95°C for 5 sec and annealing at 60°C for 30 sec. All reactions
were performed in triplicate. The primers were as follows: eIF-5A-2
forward, 5′-TATGCAGTGCTCGGCCTTG-3′, and reverse,
5′-TTGGAACATCCATGTTGTGAGTAGA-3′; and miR-9,
5′-TCTTTGGTTATCTAGCTGTATGA-3′. Negative control: Sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′.
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical analysis was performed using GraphPad Prism 5 software
(GraphPad Software, Inc., La Jolla, CA, USA). Comparisons between 2
independent groups were performed using Student's t-test P<0.05
was considered to indicate a statistically significant
difference.
Results
miR-9 regulates the sensitivity of HCC
cells with an epithelial phenotype to cetuximab
To assess whether miR-9 acted in a synergistic or
antagonistic manner with cetuximab, HCC cell lines were treated
with cetuximab alone, or cetuximab with an miR-9 mimic or inhibitor
for 48 h. A CCK-8 assay was then used to assess the cytotoxicity of
cetuximab. As demonstrated in Fig. 1A and
B, compared with cetuximab treatment alone, the addition of a
miR-9 mimic significantly increased the sensitivity to cetuximab,
while the addition of a miR-9 inhibitor significantly decreased the
sensitivity of epithelial phenotype HCC cells (Hep3B and Huh7). In
the mesenchymal phenotype HCC SNU387 and SNU449 cells, no
significant differences were observed among the groups (Fig. 1C and D). EdU incorporation assays were
also performed to determine the effects on proliferation of each
treatment condition. The results revealed that the proliferation of
HCC cells (Fig. 1E and F) was
significantly reduced following treatment with cetuximab plus the
miR-9 mimic and significantly increased following treatment with
cetuximab plus the miR-9 inhibitor in the Hep3B and Huh7 cells. No
significant differences were observed between the 2 groups in the
mesenchymal phenotype HCC cells (Fig. 1G
and H). Taken together, these results demonstrate that miR-9
may enhance the sensitivity of epithelial phenotype HCC cells to
cetuximab.
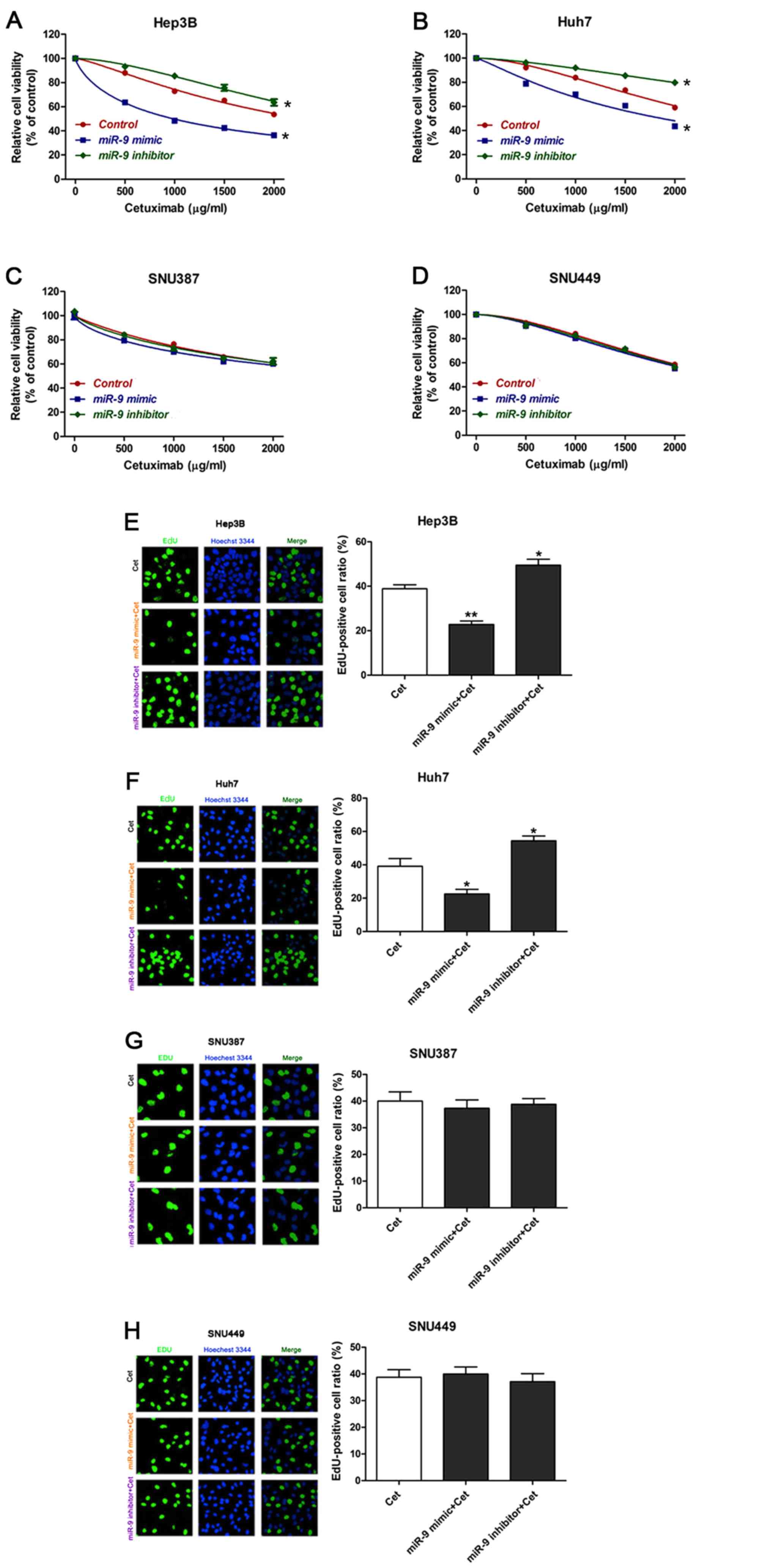 | Figure 1.Overexpression of miR-9 enhances
sensitivity of epithelial HCC cells to cetuximab. (A-D) Viability
of HCC cells treated with cetuximab in the presence of a miR-9
mimic or inhibitor. (E-H) Photomicrographs and bar charts depicting
EdU staining and relative EdU-positive ratios, respectively, of HCC
cells following treatment with cetuximab, with cetuximab plus a
miR-9 mimic or with cetuximab plus miR-9 inhibitor for 48 h.
*P<0.05, **P<0.01 vs. control, Hep3B: PmiR-9
mimic+Cet=0.0025, PmiR-9 inhibitor+Cet=0.0295,
Huh7: PmiR-9 mimic+Cet=0.0356, PmiR-9
inhibitor+Cet=0.0480. All experiments were performed ≥3
times. miR-9, microRNA-9; HCC, hepatocellular carcinoma; EdU,
5-ethynyl-2′-deoxyuridine; Cet, cetuximab. |
miR-9 regulates the expression of
eIF-5A-2
Bioinformatics analysis using Targetscan software
suggested that eIF-5A-2 was a target gene of miR-9 (Fig. 2A). RT-qPCR was used to determine the
miR-9 and eIF-5A-2 mRNA expression in HCC cell lines (Fig. 2B). Mesenchymal phenotype SNU387 and
SNU449 cell lines were revealed to exhibit higher miR-9 expression
than the epithelial phenotype Hep3B and Huh7 cell lines. The
expression of eIF-5A-2 was negatively associated with the
expression of miR-9 in the HCC cells. RT-qPCR was then performed to
evaluate the effects of miR-9 on the expression of eIF-5A-2 mRNA.
eIF-5A-2 mRNA expression was markedly decreased when HCC cells were
transfected with the miR-9 mimic, compared with that in cells
transfected with the negative control mimic. By contrast,
transfection with an miR-9 inhibitor enhanced the expression of
eIF-5A-2 mRNA (Fig. 2C). RT-qPCR was
used to determine the expression of miR-9 following treatment with
an miR-9 mimic or inhibitor, the results demonstrated that miR-9
expression was increased after transfection with miR-9 mimic and
was decreased after transfected with miR-9 inhibitor compared with
the control (Fig. 2D). Western blot
analysis demonstrated downregulation of eIF-5A-2 in miR-9
mimic-transfected HCC cells and upregulation of eIF-5A-2 in miR-9
inhibitor-transfected HCC cells (Fig.
2E).
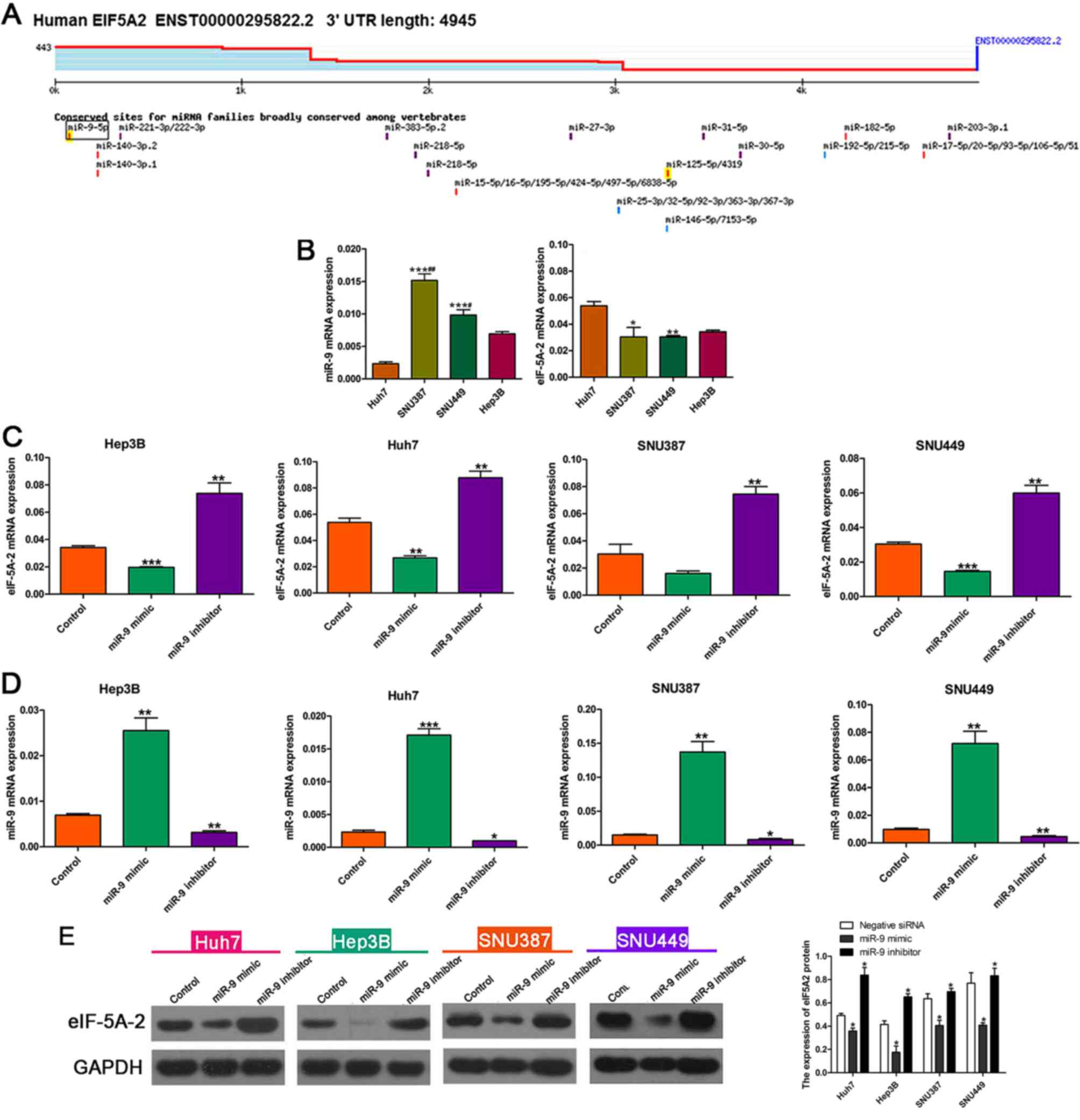 | Figure 2.miR-9 modulates expression of
eIF-5A-2 in HCC cell lines. (A) miR-9 target site in eIF-5A-2
predicted by TargetScan. (B) Expression of miR-9 and eIF-5A-2 mRNA
in HCC cells as determined by RT-qPCR. *P<0.05, **P<0.01,
***P<0.001 vs. Huh7; #P<0.05,
##P<0.01 vs. Hep3B. (C) eIF-5A-2 expression in HCC
cells following treatment with a miR-9 mimic or inhibitor as
determined by RT-qPCR. **P<0.01, ***P<0.001 vs. the control.
(D) Expression levels of miR-9 following treatment with a miR-9
mimic or inhibitor as determined by RT-qPCR. *P<0.05,
**P<0.01, ***P<0.001 vs. the control (E) Western blot
analysis was used to detect eIF-5A-2 expression in the various HCC
cell lines in the presence of the miR-9 mimic, inhibitor and
control. *P<0.05 vs. negative siRNA. miR-9, microRNA-9;
eIF-5A-2; eukaryotic translation initiation factor 5A-2; HCC,
hepatocellular carcinoma; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; miRNA,
microRNA; UTR, untranslated region. |
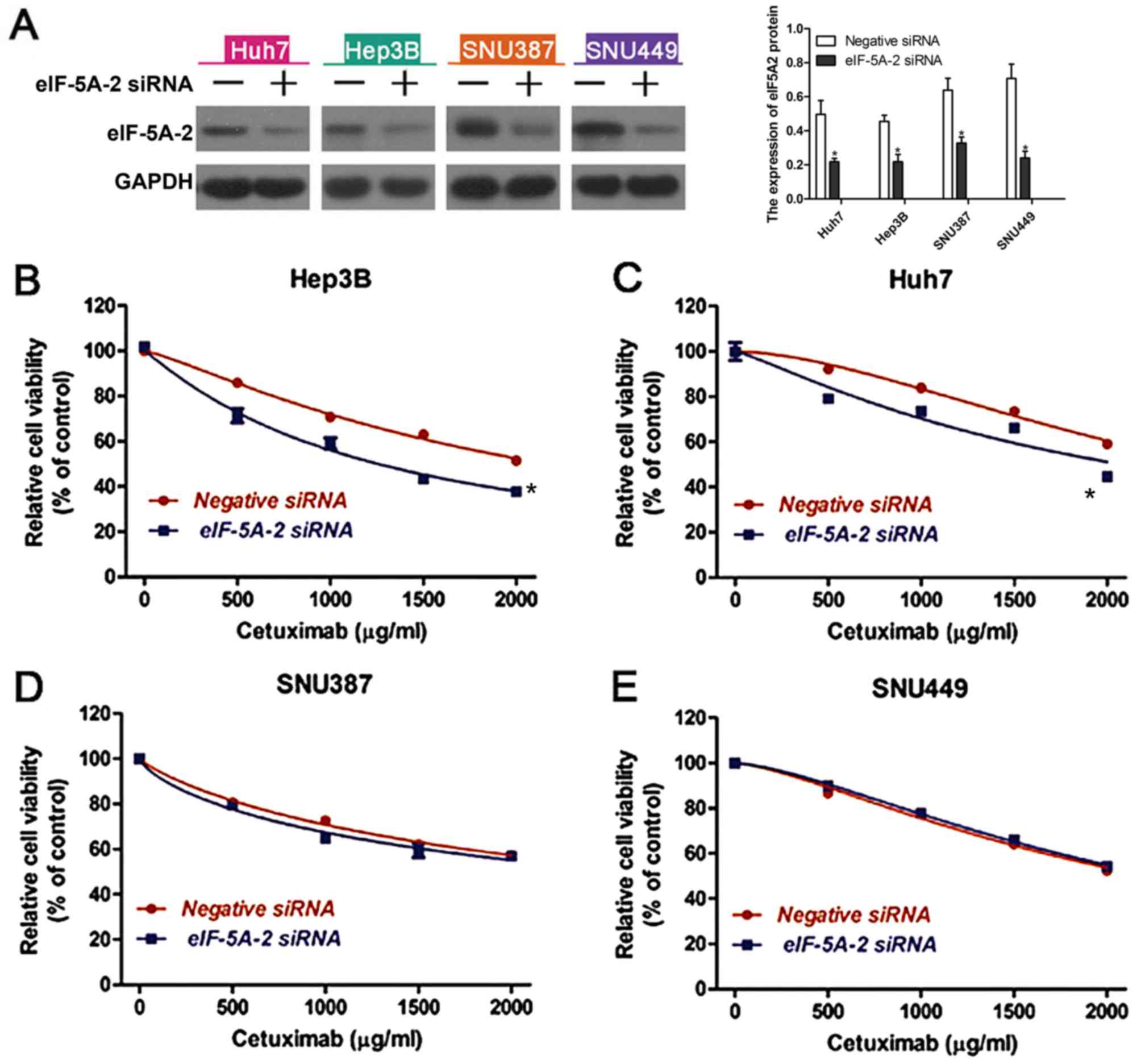
Knockdown of eIF-5A-2 enhances
cetuximab sensitivity in epithelial phenotype HCC cells
To determine whether altered expression of eIF-5A-2
may affect the sensitivity of HCC cells to cetuximab, eIF-5A-2
siRNA or a negative control siRNA were transfected into the HCC
cell lines. Western blot analysis was used to examine the knockdown
efficiency (Fig. 3A). A CCK-8 assay
was then used to measure cell viability following treatment with
cetuximab. It was revealed that knockdown of eIF-5A-2 significantly
improved cetuximab-induced growth inhibition in epithelial HCC
cells (Fig. 3B and C), but had no
significant effect on mesenchymal HCC cells (Fig. 3D and E). These data suggest that
eIF-5A-2 is involved in the sensitivity of epithelial HCC cells to
cetuximab.
miR-9 regulates the sensitivity of
epithelial phenotype HCC cells to cetuximab through eIF-5A-2
In order to determine whether or not miR-9 enhanced
the sensitivity of epithelial HCC cells to cetuximab through
regulation of eIF-5A-2, HCC cells were transfected with eIF-5A-2
siRNA and the viability of HCC cell lines treated with cetuximab
alone, with cetuximab plus a miR-9 mimic or with cetuximab plus a
miR-9 inhibitor, for 48 h was measured. The results demonstrated
that silencing of eIF-5A-2 resulted in no significant differences
in the viability of cells treated with cetuximab with or without a
miR-9 mimic or an inhibitor (Fig.
4A-D). Therefore, we hypothesized that miR-9 increased the
sensitivity of these cells to cetuximab through the regulation of
eIF-5A-2.
Discussion
miRNAs are a family of small interfering RNAs that
modulate gene expression in a sequence-specific manner (32). miRNAs have been revealed to serve
critical roles in cell growth, differentiation, apoptosis and drug
resistance (32). Several studies
have demonstrated that upregulation of miR-9 expression is
associated with enhanced tumor cell invasion and a poor prognosis
(33,34). These studies support a role for miR-9
as an oncogene in these tumor types. Notably, miR-9 has also been
reported as a tumor suppressor in various human cancer types. These
studies have demonstrated that miR-9 may inhibit cancer cell
proliferation. Taken together, these data indicate that miR-9 may
act either as an oncogene or as a tumor suppressor. In the present
study, the effects of miR-9 on HCC cells were examined and miR-9
overexpression was revealed to inhibit HCC cell proliferation. A
CCK-8 assay was used to determine the effect of miR-9 on cell
viability following treatment with cetuximab. An miR-9 mimic
significantly increased the sensitivity of epithelial phenotype HCC
cells to cetuximab, while an miR-9 inhibitor significantly
decreased cetuximab sensitivity in these cells.
eIF-5A-2 serves important roles in cell
proliferation, metastasis and apoptosis, and is considered as a
novel oncogene (28). Accumulating
evidence has demonstrated upregulated expression of eIF-5A-2 in a
number of cancer types, such as bladder cancer, hepatocellular
carcinoma and colon cancer (35–37).
Inhibition of eIF-5A-2 may decrease invasion and metastasis, and
enhance the therapeutic efficacy of drugs in HCC cells (38,39).
Cetuximab, an anti-epidermal growth factor receptor (EGFR)
monoclonal antibody that targets the extracellular domain of EGFR,
is used to treat several cancer types (40); for example, it is used for the
treatment of patients with colorectal cancer and squamous cell
carcinoma of the head and neck (41,42). It
has been demonstrated that combination treatment with the eIF-5A-2
inhibitor, GC7, enhances cetuximab sensitivity by inhibiting
eIF-5A-2 in non-small cell lung cancer (43). Studies identified activated EGFR as a
potential determinant that promoted the resistance of HCC cells to
sorafenib, while inhibition of EGFR with cetuximab increased the
efficacy of sorafenib (44).
Furthermore, a novel signal transducer and activator of
transcription 3 inhibitor, NSC 74859, enhanced the
anti-proliferative activity of cetuximab in HCC (45). However, single use of cetuximab was
only slightly effective in clinical trials and drug resistance may
be an issue. It is therefore imperative to increase the
cytotoxicity of cetuximab in the treatment of HCC. In the present
study, Huh7 cells were the most resistant to cetuximab among all
the HCC cell lines. Notably, the eIF-5A-2 mRNA expression in the
Huh7 cells was higher than that in the other HCC cell lines. The
siRNA-mediated silencing of eIF-5A-2 resulted in enhanced
sensitivity of the HCC cells to cetuximab. We also confirmed that
inhibition of eIF-5A-2 could enhance the cytotoxicity of cetuximab
in HCC. These discoveries indicate that eIF-5A-2 may be involved in
cetuximab resistance in HCC cell lines. In the present study, miR-9
overexpression was revealed to downregulate the expression of
eIF-5A-2 mRNA, while an miR-9 inhibitor increased eIF-5A-2 mRNA
expression. These findings indicate that miR-9 may regulate the
expression of eIF-5A-2. Additionally, the effects of miR-9 on
cetuximab sensitivity were revealed to be eliminated by knockdown
of eIF-5A-2 with siRNA.
In summary, the present study demonstrated that
overexpression of miR-9 downregulated the expression of eIF-5A-2
and enhanced the sensitivity of epithelial phenotype HCC cells to
cetuximab. Taken together, these results indicate that the use of
miR-9 may serve as a potential therapeutic approach for HCC in the
future.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81273260), the Natural
Science Foundation of Henan Province (grant no. 162300410274) and
Zhejiang Provincial Natural Science Foundation of China (grant no.
LQ13H160006).
References
|
1
|
Faloppi L, Scartozzi M, Maccaroni E, Di
Pietro Paolo M, Berardi R, Del Prete M and Cascinu S: Evolving
strategies for the treatment of hepatocellular carcinoma: From
clinical-guided to molecularly-tailored therapeutic options. Cancer
Treat Rev. 37:169–177. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sapisochin G, de Sevilla EF, Echeverri J
and Charco R: Management of ‘very early’ hepatocellular carcinoma
on cirrhotic patients. World J Hepatol. 6:766–775. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thomas RK: Overcoming drug resistance in
ALK-rearranged lung cancer. N Engl J Med. 370:1250–1251. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Selcuklu SD, Donoghue MT, Rehmet K, de
Souza Gomes M, Fort A, Kovvuru P, Muniyappa MK, Kerin MJ, Enright
AJ and Spillane C: MicroRNA-9 inhibition of cell proliferation and
identification of novel miR-9 targets by transcriptome profiling in
breast cancer cells. J Biol Chem. 287:29516–29528. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Song Y, Li J, Zhu Y, Dai Y, Zeng T, Liu L,
Li J, Wang H, Qin Y, Zeng M, et al: MicroRNA-9 promotes tumor
metastasis via repressing E-cadherin in esophageal squamous cell
carcinoma. Oncotarget. 5:11669–11680. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cui EH, Li HJ, Hua F, Wang B, Mao W, Feng
XR, Li JY and Wang X: Serum microRNA 125b as a diagnostic or
prognostic biomarker for advanced NSCLC patients receiving
cisplatin-based chemotherapy. Acta Pharmacol Sin. 34:309–313. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Perell K, Vincent M, Vainer B, Petersen
BL, Federspiel B, Møller AK, Madsen M, Hansen NR, Friis-Hansen L,
Nielsen FC and Daugaard G: Development and validation of a microRNA
based diagnostic assay for primary tumor site classification of
liver core biopsies. Mol Oncol. 9:68–77. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xin H, Li X, Yang B, Zhang L, Han Z and
Han C: Blood-based multiple-microRNA assay displays a better
diagnostic performance than single-microRNA assay in the diagnosis
of breast tumor. Tumour Biol. 35:12635–12643. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhu SM, Chen CM, Jiang ZY, Yuan B, Ji M,
Wu FH and Jin J: MicroRNA-185 inhibits cell proliferation and
epithelial-mesenchymal transition in hepatocellular carcinoma by
targeting Six2. Eur Rev Med Pharmacol Sci. 20:1712–1719.
2016.PubMed/NCBI
|
|
13
|
Deng B, Qu L, Li J, Fang J, Yang S, Cao Z,
Mei Z and Sun X: MiRNA-211 suppresses cell proliferation, migration
and invasion by targeting SPARC in human hepatocellular carcinoma.
Sci Rep. 6:266792016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Guo LM, Pu Y, Han Z, Liu T, Li YX, Liu M,
Li X and Tang H: MicroRNA-9 inhibits ovarian cancer cell growth
through regulation of NF-kappaB1. FEBS J. 276:5537–5546. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsai KW, Liao YL, Wu CW, Hu LY, Li SC,
Chan WC, Ho MR, Lai CH, Kao HW, Fang WL, et al: Aberrant
hypermethylation of miR-9 genes in gastric cancer. Epigenetics.
6:1189–1197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang H, Qi M, Li S, Qi T, Mei H, Huang K,
Zheng L and Tong Q: microRNA-9 targets matrix metalloproteinase 14
to inhibit invasion, metastasis, and angiogenesis of neuroblastoma
cells. Mol Cancer Ther. 11:1454–1466. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zheng L, Qi T, Yang D, Qi M, Li D, Xiang
X, Huang K and Tong Q: microRNA-9 suppresses the proliferation,
invasion and metastasis of gastric cancer cells through targeting
cyclin D1 and Ets1. PLoS One. 8:e557192013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Muraoka T, Soh J, Toyooka S, Maki Y, Shien
K, Furukawa M, Ueno T, Tanaka N, Yamamoto H, Asano H, et al: Impact
of aberrant methylation of microRNA-9 family members on non-small
cell lung cancers. Mol Clin Oncol. 1:185–189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J, Zhao H, Tang D, Wu J, Yao G and
Zhang Q: Overexpressions of microRNA-9 and microRNA-200c in human
breast cancers are associated with lymph node metastasis. Cancer
Biother Radiopharm. 28:283–288. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu S, Jia S and Xu P: MicroRNA-9 as a
novel prognostic biomarker in human laryngeal squamous cell
carcinoma. Int J Clin Exp Med. 7:5523–5528. 2014.PubMed/NCBI
|
|
21
|
Zhou X, Marian C, Makambi KH, Kosti O,
Kallakury BV, Loffredo CA and Zheng YL: MicroRNA-9 as potential
biomarker for breast cancer local recurrence and tumor estrogen
receptor status. PLoS One. 7:e390112012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu L, Chen H, Zhou D, Li D, Bai R, Zheng
S and Ge W: MicroRNA-9 up-regulation is involved in colorectal
cancer metastasis via promoting cell motility. Med Oncol.
29:1037–1043. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun J, Fang K, Shen H and Qian Y:
MicroRNA-9 is a ponderable index for the prognosis of human
hepatocellular carcinoma. Int J Clin Exp Med. 8:17748–17756.
2015.PubMed/NCBI
|
|
24
|
Clement PM, Johansson HE, Wolff EC and
Park MH: Differential expression of eIF5A-1 and eIF5A-2 in human
cancer cells. FEBS J. 273:1102–1114. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lee NP, Tsang FH, Shek FH, Mao M, Dai H,
Zhang C, Dong S, Guan XY, Poon RT and Luk JM: Prognostic
significance and therapeutic potential of eukaryotic translation
initiation factor 5A (eIF5A) in hepatocellular carcinoma. Int J
Cancer. 127:968–976. 2010.PubMed/NCBI
|
|
26
|
Yang GF, Xie D, Liu JH, Luo JH, Li LJ, Hua
WF, Wu HM, Kung HF, Zeng YX and Guan X: Expression and
amplification of eIF-5A2 in human epithelial ovarian tumors and
overexpression of EIF-5A2 is a new in dependent predictor of
outcome in patients with ovarian carcinoma. Gynecol Oncol.
112:314–318. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Luo JH, Hua WF, Rao HL, Liao YJ, Kung HF,
Zeng YX, Guan XY, Chen W and Xie D: Overexpression of EIF-5A2
predict s tumor recurrence and progression in pTa/pT1 urothelial
carcinoma of the bladder. Cancer Sci. 100:896–902. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He LR, Zhao HY, Li BK, Liu YH, Liu MZ,
Guan XY, Bian XW, Zeng YX and Xie D: Overexpression of eIF5A-2 is
an adverse prognostic marker of survival in stage I non-small cell
lung cancer patients. Int J Cancer. 129:143–150. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tang DJ, Dong SS, Ma NF, Xie D, Chen L, Fu
L, Lau SH, Li Y, Li Y and Guan XY: Overexpression of eukaryotic
initiation factor 5A2 enhances cell motility and promotes tumor
metastasis in hepatocellular carcinoma. Hepatology. 51:1255–1263.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhu W, Cai MY, Tong ZT, Dong SS, Mai SJ,
Liao YJ, Bian XW, Lin MC, Kung HF, Zeng YX, et al: Overexpression
of EIF5A2 promotes colorectal carcinoma cell aggressiveness by
upregulating MTA1 through C-myc to induce
epithelial-mesenchymaltransition. Gut. 61:562–575. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sun Z, Han Q, Zhou N, Wang S, Lu S, Bai C
and Zhao RC: MicroRNA-9 enhances migration and invasion through
KLF17 in hepatocellular carcinoma. Mol Oncol. 7:884–894. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ye M, Du YL, Nie YQ, Zhou ZW, Cao J and Li
YF: Overexpression of activated leukocute cell adhesion molecule in
gastric cancer is associated with advanced stages and poor
prognosis and miR-9 deregulation. Mol Med Rep. 11:2004–2012. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xie D, Ma NF, Pan ZZ, Wu HX, Liu YD, Wu
GQ, Kung HF and Guan XY: Overexpression of EIF-5A2 is associated
with metastasis of human colorectal carcinoma. Hum Pathol.
39:80–86. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shek FH, Fatima S and Lee NP: Implications
of the use of eukaryotic translation initiation factor 5A (eIF5A)
for prognosis and treatment of hepatocellular carcinoma. Int J
Hepatol. 2012:7609282012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wei JH, Cao JZ, Zhang D, Liao B, Zhong WM,
Lu J, Zhao HW, Zhang JX, Tong ZT, Fan S, et al: EIF5A2 predicts
outcome in localised invasive bladder cancer and promotes bladder
cancer cell aggressiveness in vitro and in vivo. Br J Cancer.
110:1767–1777. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lou B, Fan J, Wang K, Chen W, Zhou X,
Zhang J, Lin S, Lv F and Chen Y: N1-guanyl-1,7-diaminoheptane (GC7)
enhances the therapeutic efficacy of doxorubicin by inhibiting
activation of eukaryotic translation initiation factor 5A2 (eIF5A2)
and preventing the epithelial-mesenchymal transition in
hepatocellular carcinoma cells. Exp Cell Res. 319:2708–2717. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu RR, Lv YS, Tang YX, Wang YF, Chen XL,
Zheng XX, Xie SZ, Cai Y, Yu J and Zhang XN: Eukaryotic translation
initiation factor 5A2 regulates the migration and invasion of
hepatocellular carcinoma cells via pathways involving reactive
oxygen species. Oncotarget. 7:24348–24360. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Iida M, Brand TM, Starr MM, Huppert EJ,
Luthar N, Bahrar H, Coan JP, Pearson HE, Salgia R and Wheeler DL:
Overcoming acquired resistance to cetuximab by dual targeting HER
family receptors with antibody-based therapy. Mol Cancer.
13:2422014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bonner JA, Harari PM, Giralt J, Azarnia N,
Shin DM, Cohen RB, Jones CU, Sur R, Raben D, Jassem J, et al:
Radiotherapy plus cetuximab for squamous-cell carcinoma of the head
and neck. N Engl J Med. 354:567–578. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cuninham D, Humblet Y, Siena S, Khayat D,
Bleiberg H, Santoro A, Best D, Mueser M, Harstrick A, Verslype C,
et al: Cetuximab monotherapy and cetuximab plus irinotecan in
irinotecan-refractory metastatic colorectal cancer. N Engl J Med.
351:337–345. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang X, Jiang R, Cui EH, Feng WM, Guo HH,
Gu DH, Tang CW, Xue T and Bao Y: N1-guanyl-1,7-diaminoheptane
enhances the chemosensitivity of NSCLC cells to cetuximab through
inhibition of eukaryotic translation initiation factor 5A2
activation. Eur Rev Med Pharmacol Sci. 20:1244–1250.
2016.PubMed/NCBI
|
|
44
|
Ezzoukhry Z, Louandre C, Trécherel E,
Godin C, Chauffert B, Dupont S, Diouf M, Barbare JC, Maziere JC and
Galmiche A: EGFR activation is a potential determinant of primary
resistance of hepatocellular carcinoma cells to sorafenib. Int J
Cancer. 131:2961–2969. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chen W, Shen X, Xia X, Xu G, Ma T, Bai X
and Liang T: NSC 74859-mediated inhibition of STAT3 enhances the
anti-proliferative activity of cetuximab in hepatocellular
carcinoma. Liver Int. 32:70–77. 2012. View Article : Google Scholar : PubMed/NCBI
|