Introduction
Colorectal cancer (CRC) is one of the most common
types of malignancy in Western countries, and is the cause of a
high number of cancer-associated mortalities every year (1). In the past decades, the prevalence of
CRC in China has increased rapidly, which may be associated with
westernized lifestyle, longer lifespan and a poor CRC screening
system (2,3). Previous studies have shown that the
process of CRC formation refers to a gradual activation of
oncogenes and inactivation of anti-oncogenes (4). However, the mechanisms of CRC
tumorigenesis and tumor progression are still not illustrated
clearly. Consequently, identifying more credible biomarkers may
help with CRC prognosis estimation and provide novel potential
targets for therapy.
Platelet-derived growth factor (PDGF)-D belongs to
the PDGF family, whose members are known as a mesenchymal growth
factors that promote epithelial-stromal communication (5). Platelet-derived growth factor-D (PDGF-D)
exerts its biological functions by specifically binding to its
cognate receptor, platelet-derived growth factor receptor-β
(PDGFR-β), leading to rapid phosphorylation of PDGFR-β and
consequent activation of numerous downstream signaling pathways
(6). Previous studies have shown that
PDGF-D is involved in carcinogenesis, particularly regulating the
course of epithelial-to-mesenchymal transition, which facilitates
tumor proliferation and metastasis (7–9).
Furthermore, PDGF-D has been reported to be associated with the
mechanistic target of rapamycin (mTOR), Notch, nuclear factor-κB
(NF-κB), B-cell lymphoma-2 (Bcl-2) and CXC motif chemokine receptor
4 (CXCR4) signaling pathways (10).
Overexpression of PDGF-D was also observed in various human tumors,
including pancreatic, prostate, gastric and breast cancer,
predicting that PDGF-D is involved in cancer development and
progression (7–9,11).
As there is little clear evidence about the
expression and function of PDGF-D in CRC thus far, it will be
necessary to explore the significant role of PDGF-D in CRC. In the
present study, PDGF-D messenger RNA (mRNA) level and protein
expression were examined in CRC tissues, paired normal tissues and
cell lines. The effects of PDGF-D on CRC behaviors, including
migration, invasion and proliferation, were then surveyed. By
upregulating and downregulating the expression level of PDGF-D in
colon cell lines, the detailed functions of PDGF-D were studied,
with the aim of clarifying its potential mechanism.
Materials and methods
Ethics statement and patient tissue
samples
The present study was approved by the Ethics
Committee of Tongji Medical College, Huazhong University of Science
and Technology (Wuhan, China). A total of 45 CRC tissue samples
were collected from patients with CRC who had received surgical
treatment in the Union Hospital affiliated to Huazhong University
of Science and Technology between September 2014 and August 2015.
The present study included 19 and 26 male and female patients,
respectively (mean age, 52.82 years; age range, 25–87 years). All
samples were obtained with written informed consent from the
patients. Part of these specimens were immersed in 5%
paraformaldehyde solution at 4°C for immunohistochemistry (IHC),
and the remaining were immediately frozen and stored at −80°C.
IHC
CRC specimens were fixed in 5% paraformaldehyde
solution, as described previously (12). For calculating the protein level of
PDGF-D, the general procedure was designed as follows: i) Record
the intensity of dyeing in stained tissues (negative, 0; weak
positive, 1; moderate positive, 2; and strong positive, 3); ii)
calculate the quantity of stained cancer cells (0, <10%; 1,
<25%; 2, <50%; and 3, ≥50%); iii) multiply the two indexes to
calculate the staining index (SI); and iv) the final expression is
considered negative when SI <3 and positive when SI ≥4.
Cell culture and lentivirus
transfection
Human colon cancer SW480 and SW620 cells were
cultured in L15 (HyClone; GE Healthcare Life Sciences, Little
Chalfont, UK), HCT116 cells were cultured in McCoy's 5A (HyClone;
GE Healthcare Life Sciences), DLD-1 cells were cultured in
RPMI-1640 (Wuhan Goodbio Technology, Wuhan, China) and LoVo cells
were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) complete medium with 10% fetal bovine serum
(ScienCell Research Laboratories, Inc., Carlsbad, CA, USA), and
were grown in 25-cm2 polystyrene tissue culture flasks
(Corning Incorporated, Corning, NY, USA) at 37°C in an atmosphere
containing 5% CO2 for between 48 and 72 h. Lentivirus
transfection was used to up- and downregulate the PDGF-D expression
in cell lines. The specific procedure has been described previously
(13). The short hairpin
(sh)PDGF-D-lentivirus and complementary DNA
(cDNA)-PDGF-D-lentivirus were purchased from Shanghai GeneChem.,
Shanghai, China.
Detection of apoptosis by flow
cytometry
Subsequent to successful transfection, the SW480 and
HCT116 cells were collected to conduct detection of apoptosis by
flow cytometry, and the specific operation steps have been
presented in a previous study (14).
The Annexin V/propidium iodide apoptosis detection kit was used
according to the manufacturer's protocol (Wuhan Antgene
Biotechnology, Wuhan, China).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The detailed procedures and notes of RT-qPCR were
reported in a previous study (15).
In brief, TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) was used to extract total RNA from cells, CRC tissues and
paired normal controls. Subsequently, the RT of total RNAs was
performed with PrimeScript RT Master mix (Takara Bio, Inc., Otsu,
Japan). Finally, the expected nucleic acid amplification products
were detected in the StepOnePlusÔ Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) with SYBR-Green Master
mix (Takara Bio, Inc.). mRNA quantity of each gene was calculated
using the 2−ΔΔCq method (16) and normalized to GAPDH. The primer
sequences of PDGF-D (forward, GTG GAG GAA ATT GTG GCT GT and
reverse, CGT TCA TGG TGA TCC AACTG), and the internal control
(GAPDH forward, GGG GAG CCA AAA GGG TCA TCA TCT and reverse, GAC
GCC TGC TTC ACC ACC TTC TTG) were designed according to a previous
study (17).
Western blot analysis
The specimens and cells were lysed in an appropriate
amount (100 µl) of lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China) with phenylmethylsulfonyl fluoride
for total protein extraction. The protein concentration in the
lysates was then detected by bicinchoninic acid assay. Equal
quantities of total protein (10 µl/lane) were loaded onto SDS-PAGE
(12% gel) and then separated by electrophoresis. The separated
proteins were transferred to a polyvinylidene fluoride membrane,
which was subsequently blocked with 5% skimmed milk and
TBS-Tween-20 for 1 h at room temperature. The membranes were then
incubated with primary antibodies at 4°C overnight. The primary
antibodies used in the study were as follows: Anti-PDGF-D (cat. no.
ab181845; dilution, 1:500; Abcam, Cambridge, MA, USA), anti-PDGFR-β
(cat. no. Esap11645; dilution, 1:500; Wuhan Elabscience
Biotechnology, Wuhan, China), anti-Notch1 (cat. no. SC-6014;
dilution, 1:500; Santa Cruz Biotechnology, Inc., Dallas, TX, USA)
and anti-MMP-9 (cat. no. ab58803; dilution, 1:500; Abcam).
Subsequently, either the horseradish peroxidase (HRP-) labeled goat
anti-rabbit immunoglobulin G (cat. no. GB23303; dilution, 1:1,000;
Wuhan Goodbio Technology) or the HRP-labeled goat anti-mouse IgG
(cat. no. GB23301; dilution, 1:1,000; Wuhan Goodbio Technology)
secondary antibody was applied to the membrane for 1 h at room
temperature. An enhanced chemiluminescence system (cat. no. G2014;
Wuhan Goodbio Technology) and Image J software (version 2; National
Institutes of Health, Bethesda, MD, USA) were used to analyze the
protein expression.
Transwell migration and Matrigel
invasion assays
Cell Transwell assays were performed to test the
invasion and migration of cancer cells. The specific procedure was
previously reported (18). In the
present study, Transwell chambers (24-well, 8-µm pore membranes)
were purchased from Corning Incorporated. Matrigel (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) was used in cell invasion analysis,
but not in the migration assay.
Cell proliferation assays
A Cell Counting kit (CCK)-8 assay (Beyotime
Institute of Biotechnology) was used to detect cell proliferation.
Following inoculation onto a 96-well plate, cells were cultured in
an incubator for 24 h at 37°C in an atmosphere containing 5%
CO2. An appropriate amount (10 µl) of CCK-8 solution was
then added to each well, and subsequently, the cells were kept in
the incubator for 1–4 h. Eventually, cell proliferation was
analyzed by measuring the absorbance at 450 nm with a microplate
reader (Thermo Fisher Scientific, Inc.).
Statistical analysis
All experiments were performed at least three times
independently. All results shown in our study were analyzed using
SPSS statistical software (version 22.0; IBM Corp., Armonk, NY,
USA). The results are expressed as the mean ± standard deviation.
Statistical analyses were performed using Student's t and
χ2 tests. P<0.05 was considered to indicate a
statistically significant difference.
Results
PDGF-D is highly expressed in CRC
tissues
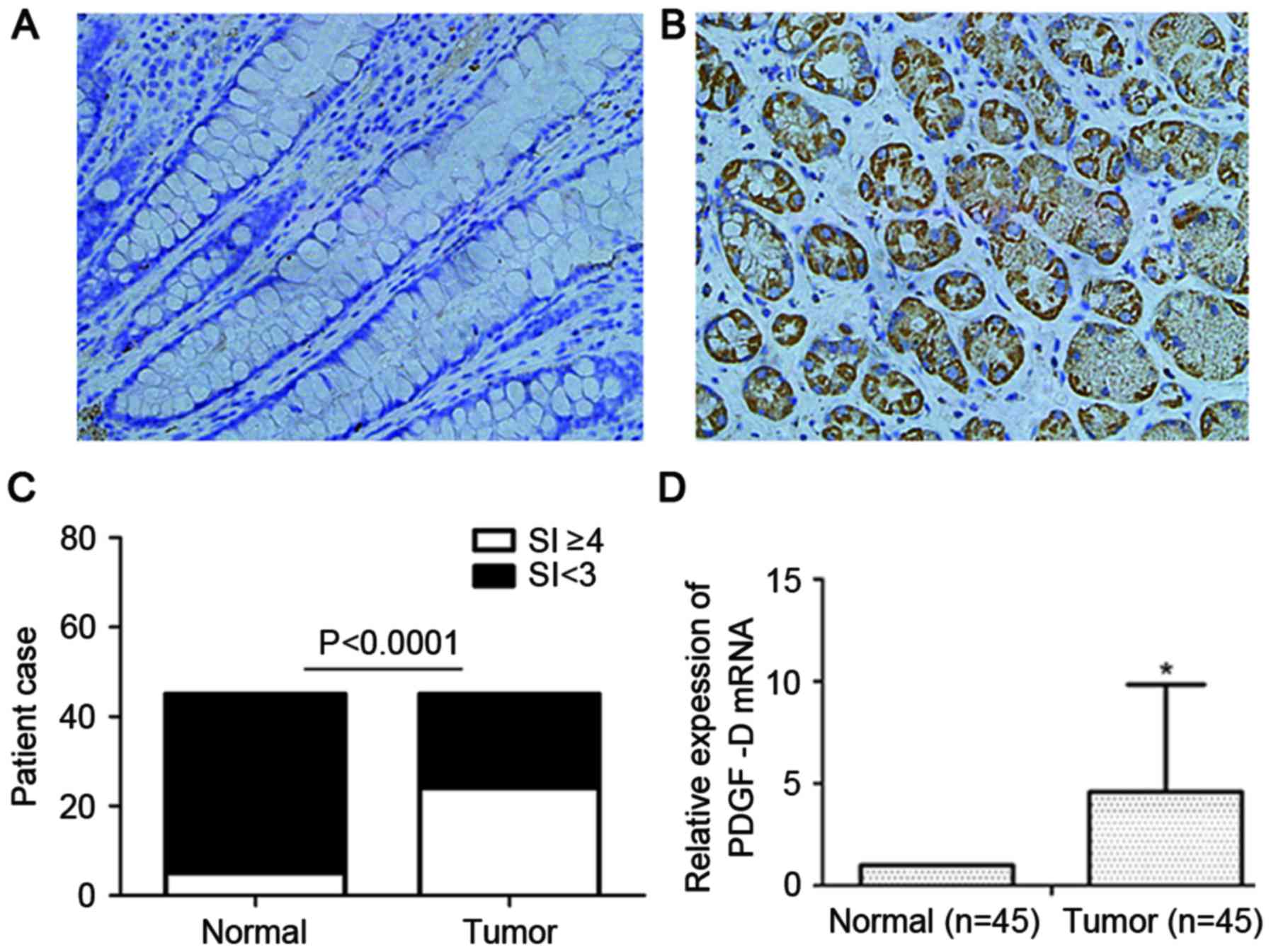
PDGF-D protein expression was investigated through
IHC detection, which revealed that PDGF-D was highly expressed in
CRC tissues (Fig. 1A and B). In
total, 53.3% (24/45) of CRC tissues were positive for PDGF-D
protein expression in the cytoplasm, while 11.1% (5/45) of the
paired normal colorectal tissues were positive for PDGF-D protein
expression (P<0.0001; Fig. 1C).
Subsequently, the mRNA level of PDGF-D was detected in CRC tissues
and paired normal colorectal tissues by qPCR analysis. The results
demonstrated that PDGF-D mRNA expression was averagely 4.6-fold
higher in CRC tissues than in normal tissues (P<0.05; Fig. 1D).
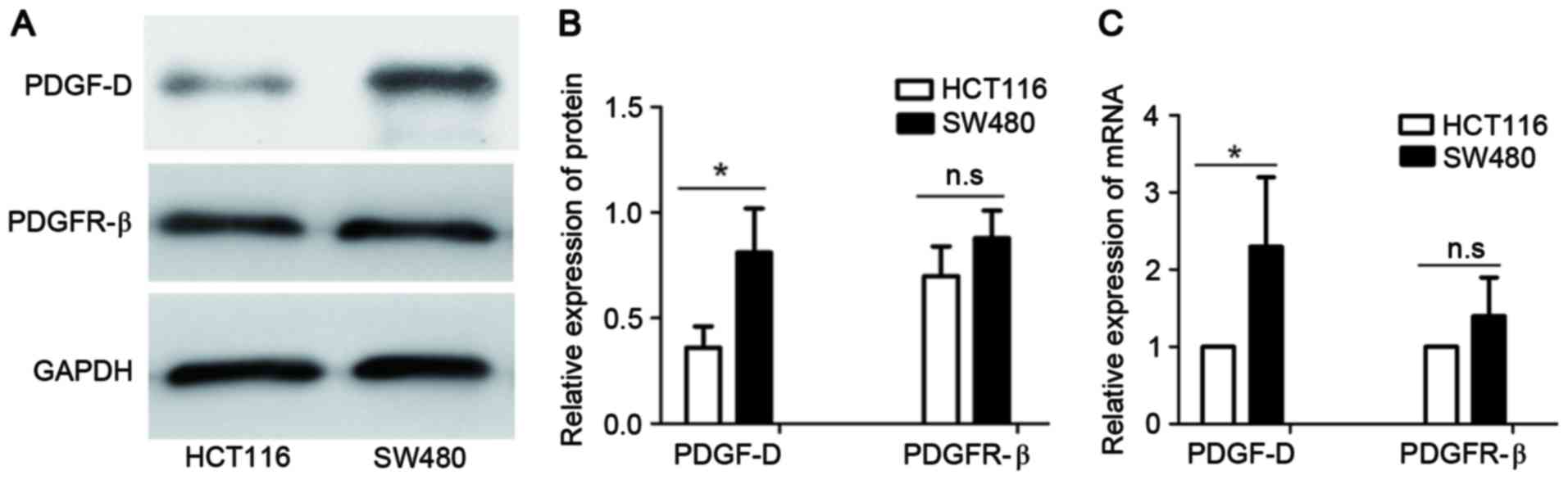
PDGF-D and PDGFR are expressed at a
high level in the SW480 cell line and a low level in HCT116
cells
Several colon cell lines were screened for the
expression of PDGF-D, including SW480, SW620, HCT116, DLD-1 and
LoVo cells (data of SW620, DLD-1 and LoVo cells are not shown).
Finally, SW480 was selected for its stable high expression of
PDGF-D protein and HCT116 was selected for its relatively low
expression of PDGF-D (Fig. 2A and B).
The corresponding mRNA levels were detected by qPCR and the results
were similar to the expression of PDGF-D protein (Fig. 2C). Furthermore, the expression of
PDGFR-β in SW480 and HCT116 cell lines was detected by qPCR and
western blot analysis. The results demonstrated that PDGFR-β was
highly expressed in SW480 cells and expressed at a lower level in
HCT116 cells (Fig. 2).
Knockdown of PDGF-D in SW480 cells
decreases the migration, invasion and proliferation capacity of
cancer cells by downregulation of Notch1 and MMP-9
Knockdown of PDGF-D in the SW480 cell line was
successfully established by lentiviral transfection (Fig. 3A). The capacity of migration, invasion
and proliferation of SW480 cells was significantly decreased
compared with that of the control (Fig.
3B-D). In order to study the potential mechanism, the
associated signaling pathways reported previously, including mTOR,
Notch, NF-κB, Bcl-2 and CXCR4, were screened by western blot
analysis (10). The expression of
Notch1 and MMP-9 was observed to decline following knockdown of
PDGF-D in SW480 cells (Fig. 3A).
Furthermore, the apoptotic rate of transfected SW480 cells was
determined. No clear difference was observed between the
shPDGF-D-lentivirus-transfected group and the negative control
group (Fig. 3E and F; negative
control vs. shPDGF-D-lentivirus=6.5 vs. 6.8%).
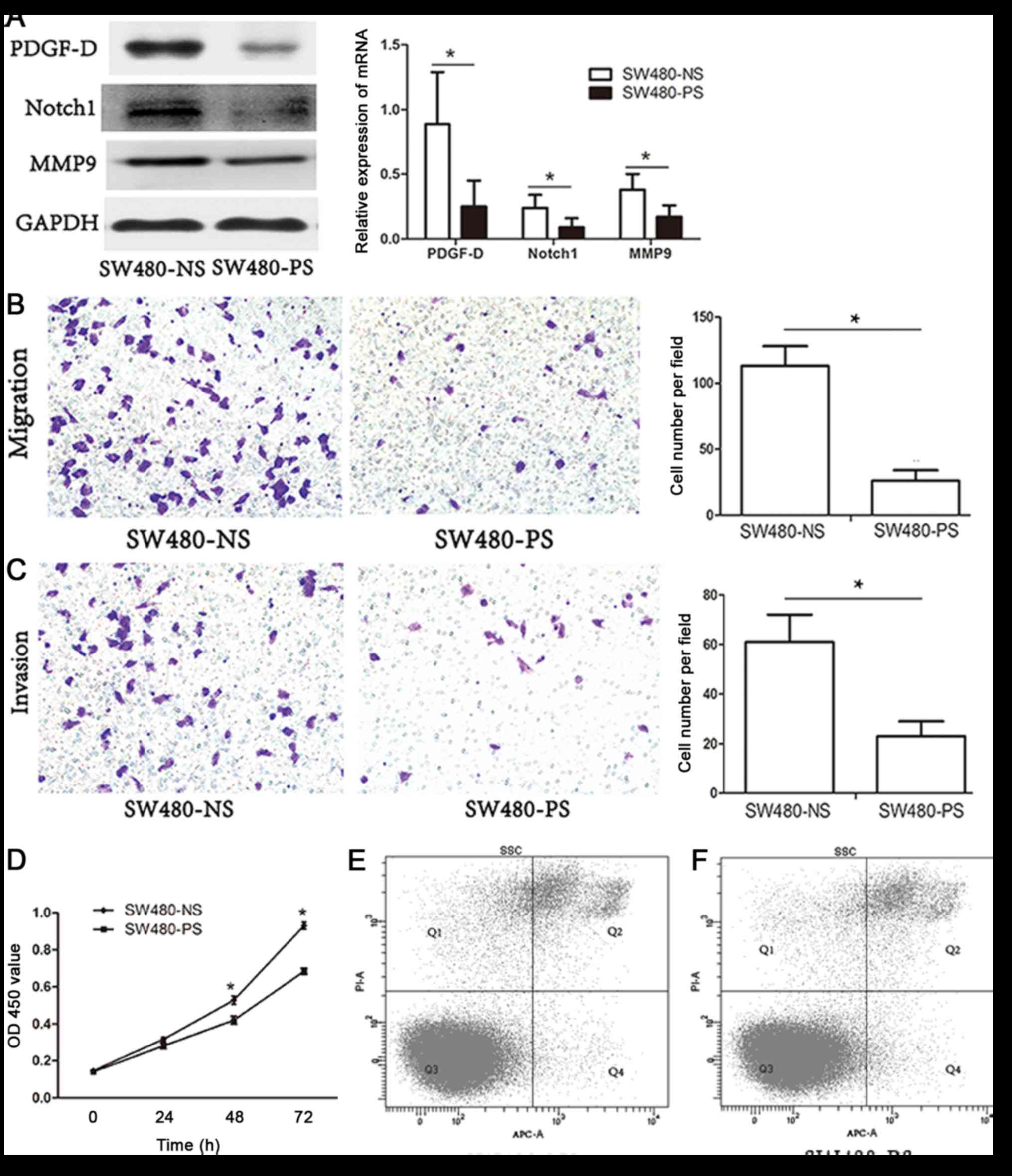 | Figure 3.Knockdown of PDGF-D in SW480 cells
declines the migration, invasion and proliferation capacity of
cancer cells by downregulation of Notch1 and MMP-9. (A) Expression
of PDGF-D, Notch1 and MMP-9 in SW480 cells infected with
shPDGF-D-lentivirus and negative control by western blot analysis.
(B) SW480 cells were transfected with shPDGF-D-lentivirus or
negative control for 48 h, and a Transwell migration assay was then
performed. (C) SW480 cells were transfected with
shPDGF-D-lentivirus or negative control for 48 h, and then used for
Transwell invasion assay. (D) SW480 cells were transfected with
shPDGF-D-lentivirus or negative control for 48 h, and a Cell
Counting kit-8 assay was then performed. (E) The apoptotic rate of
the negative control group was 6.5% by Annexin V/PI staining. (F)
The apoptotic rate of the shPDGF-D-lentivirus group was 6.8% by
Annexin V/PI staining. *P<0.05. NS, non-specific shRNA; PS,
PDGF-D shRNA; PDGF-D, platelet-derived growth factor-D; MMP-9,
matrix metalloproteinase-9; sh, short hairpin; PI, propidium
iodide; OD, optical density; mRNA, messenger RNA; Q, quadrant; APC,
allophycocyanin; SSC, side scatter. |
Upregulation of PDGF-D expression in
HCT116 cells enhances the capacity of migration, invasion and
proliferation of cancer cells by increasing Notch1 and MMP-9
levels
Upregulation of PDGF-D expression in the HCT116 cell
line was successfully established by lentiviral transfection
(Fig. 4A). Compared with that of the
control HCT116 cells, the capacity of migration, invasion and
proliferation of PDGF-D upregulated HCT116 cells were identified to
be markedly increased (Fig. 4B-D).
Our previous study results revealed that Notch1 and MMP-9 are
associated with the migration, invasion and proliferation ability
of colon cancer cells; thus, the expression of Notch1 and MMP-9 was
detected in the PDGF-D-upregulated HTC116 cells. Notably, a
significantly increased level of Notch1 and MMP-9 expression was
observed in the PDGF-D-upregulated cells (Fig. 4A). Furthermore, the apoptotic rate of
transfected HCT116 cells was detected. The results revealed that
there was no clear difference between the
cDNA-PDGF-D-lentivirus-transfected group and the negative control
group (Fig. 4E and F; negative
control vs. cDNA-PDGF-D-lentivirus=9.1 vs. 8.6%).
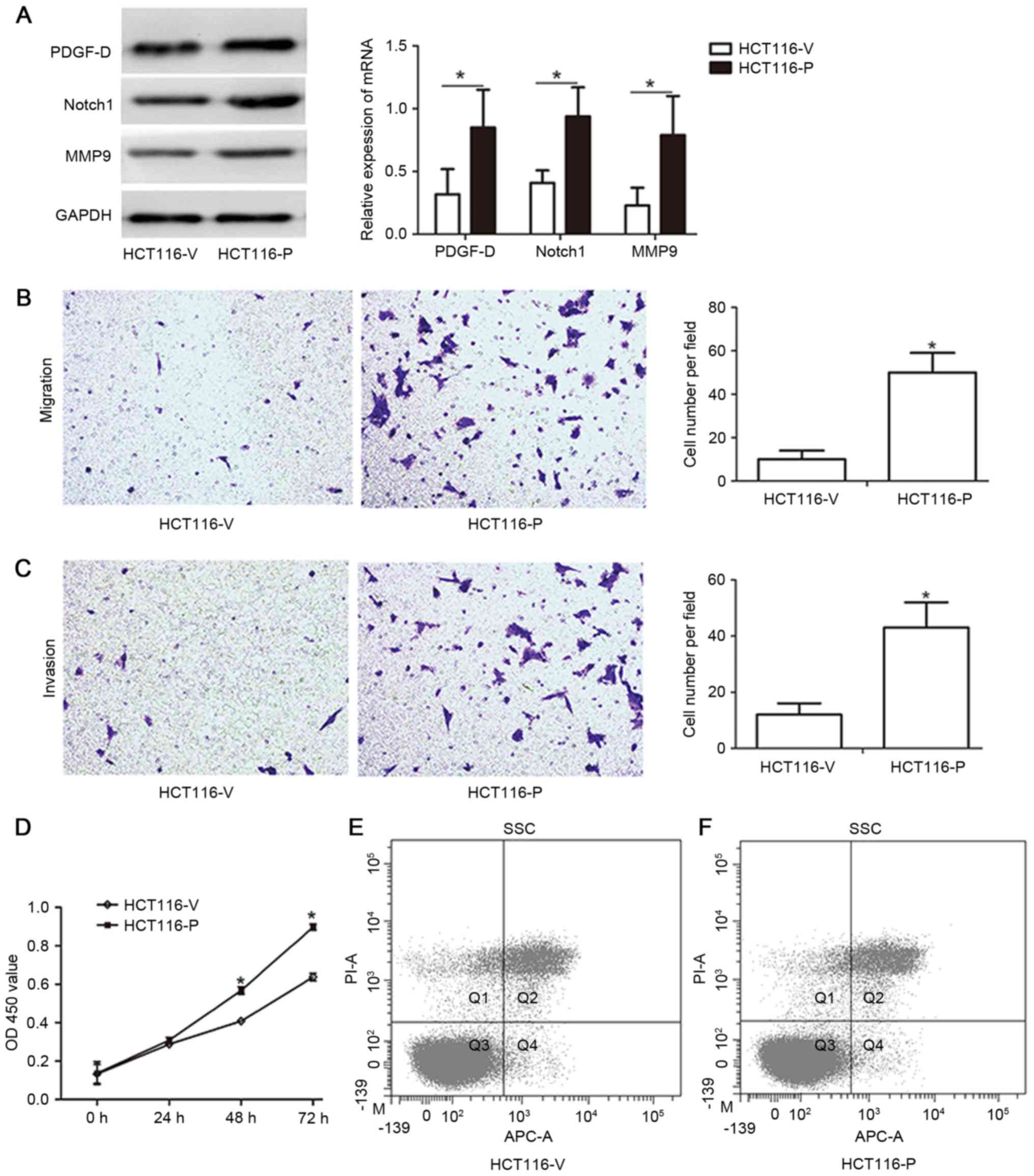 | Figure 4.Upregulation of PDGF-D in the HCT116
cell line enhances the capacity of migration, invasion and
proliferation of cancer cells by increasing Notch1 and MMP-9
expression. (A) Expression of PDGF-D, Notch1 and MMP-9 in HCT116
cells infected with cDNA-PDGF-D-lentivirus and negative control by
western blot analysis. (B) HCT116 cells were transfected with
cDNA-PDGF-D-lentivirus or negative control for 48 h, and then used
for Transwell migration assay. (C) HCT116 cells were transfected
with cDNA-PDGF-D-lentivirus or negative control for 48 h, and then
used for Transwell invasion assay. (D) HCT116 cells were
transfected with cDNA-PDGF-D-lentivirus or negative control for 48
h, and then used for Cell Counting kit-8 assay. (E) The apoptotic
rate of the negative control group was 9.1% by Annexin V/PI
staining. (F) The apoptotic rate of the cDNA-PDGF-D-lentivirus
group was 8.6% by Annexin V/PI staining. *P<0.05. V, vector
plasmid alone; P, plasmid with PDGF-D clone; PDGF-D,
platelet-derived growth factor-D; MMP-9, matrix
metalloproteinase-9; PI, propidium iodide; OD, optical density;
cDNA, complementary DNA; mRNA, messenger RNA; Q, quadrant; APC,
allophycocyanin; SSC, side scatter. |
Discussion
CRC is the one of the most common types of
malignancy around the world, with >1.35 million novel cancer
cases and ~690,000 cancer-associated mortalities in 2012 (19). It has been widely accepted that
tumorigenesis and tumor progression of CRC are associated with
multiple epigenetic changes and molecular alterations (20,21).
Identifying more novel biomarkers and their corresponding molecular
mechanism of carcinogenesis facilitates the development of new
approaches for the prevention and treatment of CRC (22).
Several previous studies indicated that the
expression of PDGF-D is upregulated in various malignancies,
including prostate, breast, pancreatic and gastric cancer (7–9,11). Furthermore, PDGF-D has been shown to
be a critical factor that regulates the processes of cell
proliferation, apoptosis, migration, invasion, angiogenesis and
metastasis (23). As there were few
studies concerning the function and mechanism of PDGF-D in CRC
tumorigenesis and progression, the present study was performed.
According to the present study, 53.3% (24/45) of CRC
tissues exhibited overexpression of PDGF-D protein by
immunohistochemical detection, and similarly, the majority of the
CRC tissues exhibited overexpression of PDGF-D mRNA by qPCR assay,
with a significant difference (P<0.05). These results revealed
that PDGF-D is one of the biomarkers that promotes the process of
CRC oncogenesis, similar to other types of cancer reported
previously (24,25).
In order to unravel the potential mechanism,
subsequent experiments were conducted in vitro. Human colon
cell lines, including SW480, SW620, HCT116, CT26, DLD-1 and LoVo,
were screened for the expression of PDGF-D (data of SW620, DLD-1
and LoVo cells are not shown). Finally, SW480 was selected for its
overexpression of PDGF-D, and HCT116 was selected for its low
expression of PDGF-D. In the two cell lines, the expression of
PDGFR-β exhibited similar changes, predicting that PDGF-D exerts
its cellular functions through PDGFR-β. PDGF-D expression was then
successfully downregulated in SW480 cells and upregulated in HCT116
cells. For the purpose of excluding the possibility that the
following experiments were affected by different apoptosis rates
subsequent to transfection, detection of apoptosis was conducted by
flow cytometry. The results indicated that there was no clear
difference between the experimental and negative control groups. As
expected, the potential of migration, invasion and proliferation of
cancer cells changed accordingly. These results suggest that PDGF-D
promotes CRC tumorigenesis and progression by regulating the
capacity of migration, invasion and proliferation in CRC.
Accumulating evidence suggests that PDGF-D promotes
tumorigenesis and cancer progression by regulating several
downstream signaling pathways (26–28). In
prostate cancer, upregulated PDGF-D promotes cell proliferation and
tumor growth through the mTOR signaling pathway (by activating the
downstream targets S6 kinase and 4E binding protein) and
upregulation of Bcl-2 (26). In
breast cancer, overexpression of PDGF-D improves the aggression of
cancer cells by upregulating the Notch, NF-κB and CXCR4 signaling
pathways (17,27). In pancreatic cancer, overexpressed
PDGF-D increases the capacity of proliferation and invasion by
activating the Notch and NF-κB signaling pathways (28). In the present study, it was shown that
the expression of Notch1 and MMP-9 markedly decreased when PDGF-D
was successfully downregulated in SW480 cells, and by contrast, the
expression of Notch1 and MMP-9 increased significantly in HCT116
cells when PDGF-D was effectively upregulated. Since MMP-9 is the
known downstream target of the Notch and NF-κB signaling pathways
(29,30), the results may predict that PDGF-D
promotes CRC migration, invasion and proliferation by regulating
the Notch and/or NF-κB signaling pathways. Therefore, future
studies will provide more evidence about the association between
PDGF-D and the Notch and NF-κB signaling pathways, and subsequently
elucidate the complete mechanism.
In summary, the overexpression of PDGF-D, existing
in the majority of human CRC tissues, promotes CRC migration,
invasion and proliferation. In vitro experiments
demonstrated that Notch1 and MMP-9 were upregulated by PDGF-D, and
the invasiveness of cancer cells was distinctly enhanced as PDGF-D
was overexpressed. Thus, the PDGF-D gene may be developed into a
novel therapeutic target of human CRC.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81271199 and
81470789).
Glossary
Abbreviations
Abbreviations:
|
CRC
|
colorectal cancer
|
|
PDGF-D
|
platelet-derived growth factor-D
|
|
PDGFR-β
|
platelet-derived growth factor
receptor-β
|
References
|
1
|
Siegel R, Desantis C and Jemal A:
Colorectal cancer statistics, 2014. CA Cancer J Clin. 64:104–117.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Center MM, Jemal A, Smith RA and Ward E:
Worldwide variations in colorectal cancer. CA Cancer J Clin.
59:366–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Center MM, Jemal A and Ward E:
International trends in colorectal cancer incidence rates. Cancer
Epidemiol Biomarkers Prev. 18:1688–1694. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Morán A, Ortega P, de Juan C,
Fernández-Marcelo T, Frías C, Sánchez-Pernaute A, Torres AJ,
Díaz-Rubio E, Iniesta P and Benito M: Differential colorectal
carcinogenesis: Molecular basis and clinical relevance. World J
Gastrointest Oncol. 2:151–158. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Andrae J, Gallini R and Betsholtz C: Role
of platelet-derived growth factors in physiology and medicine.
Genes Dev. 22:1276–1312. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Li X and Eriksson U: Novel PDGF family
members: PDGF-C and PDGF-D. Cytokine Growth Factor Rev. 14:91–98.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kong D, Banerjee S, Ahmad A, Li Y, Wang Z,
Sethi S and Sarkar FH: Epithelial to mesenchymal transition is
mechanistically linked with stem cell signatures in prostate cancer
cells. PLoS One. 5:e124452010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sethi S, Sarkar FH, Ahmed Q, Bandyopadhyay
S, Nahleh ZA, Semaan A, Sakr W, Munkarah A and Ali-Fehmi R:
Molecular markers of epithelial-to-mesenchymal transition are
associated with tumor aggressiveness in breast carcinoma. Transl
Oncol. 4:222–226. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Z, Ali S, Banerjee S, Bao B, Li Y,
Azmi AS, Korc M and Sarkar FH: Activated K-Ras and INK4a/Arf
deficiency promote aggressiveness of pancreatic cancer by induction
of EMT consistent with cancer stem cell phenotype. J Cell Physiol.
228:556–562. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wu Q, Hou X, Xia J, Qian X, Miele L,
Sarkar FH and Wang Z: Emerging roles of PDGF-D in EMT progression
during tumorigenesis. Cancer Treat Rev. 39:640–646. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao L, Zhang C, Liao G and Long J:
RNAi-mediated inhibition of PDGF-D leads to decreased cell growth,
invasion and angiogenesis in the SGC-7901 gastric cancer xenograft
model. Cancer Biol Ther. 9:42–48. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Schläfli AM, Berezowska S, Adams O, Langer
R and Tschan MP: Reliable LC3 and p62 autophagy marker detection in
formalin fixed paraffin embedded human tissue by
immunohistochemistry. Eur J Histochem. 59:24812015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jackson MF, Hoversten KE, Powers JM,
Trobridge GD and Rodgers BD: Genetic manipulation of myoblasts and
a novel primary myosatellite cell culture system: Comparing and
optimizing approaches. FEBS J. 280:827–839. 2013.PubMed/NCBI
|
|
14
|
Yan LH, Wei WY, Cao WL, Zhang XS, Xie YB
and Xiao Q: Overexpression of E2F1 in human gastric carcinoma is
involved in anti-cancer drug resistance. BMC Cancer. 14:9042014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408, 25. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ahmad A, Wang Z, Kong D, Ali R, Ali S,
Banerjee S and Sarkar FH: Platelet-derived growth factor-D
contributes to aggressiveness of breast cancer cells by
up-regulating Notch and NF-κB signaling pathways. Breast Cancer Res
Treat. 126:15–25. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li H, Tang J, Lei H, Cai P, Zhu H, Li B,
Xu X, Xia Y and Tang W: Decreased MiR-200a/141 suppress cell
migration and proliferation by targeting PTEN in hirschsprung's
disease. Cell Physiol Biochem. 34:543–553. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stoffel EM: Screening in GI cancers: The
role of genetics. J Clin Oncol. 33:1721–1728. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Grady WM and Markowitz SD: The molecular
pathogenesis of colorectal cancer and its potential application to
colorectal cancer screening. Dig Dis Sci. 60:762–772. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsujii M: Search for novel target
molecules for the effective treatment or prevention of colorectal
cancer. Digestion. 85:99–102. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang Z, Ahmad A, Li Y, Kong D, Azmi AS,
Banerjee S and Sarkar FH: Emerging roles of PDGF-D signaling
pathway in tumor development and progression. Biochim Biophys Acta.
1806:122–130. 2010.PubMed/NCBI
|
|
24
|
Kong D, Banerjee S, Huang W, Li Y, Wang Z,
Kim HR and Sarkar FH: Mammalian target of rapamycin repression by
3,3′-diindolylmethane inhibits invasion and angiogenesis in
platelet-derived growth factor-D-overexpressing PC3 cells. Cancer
Res. 68:1927–1934. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Z, Kong D, Li Y and Sarkar FH: PDGF-D
signaling: A novel target in cancer therapy. Curr Drug Targets.
10:38–41. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kong D, Wang Z, Sarkar SH, Li Y, Banerjee
S, Saliganan A, Kim HR, Cher ML and Sarkar FH: Platelet-derived
growth factor-D overexpression contributes to
epithelial-mesenchymal transition of PC3 prostate cancer cells.
Stem Cells. 26:1425–1435. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu J, Liao S, Huang Y, Samuel R, Shi T,
Naxerova K, Huang P, Kamoun W, Jain RK, Fukumura D and Xu L: PDGF-D
improves drug delivery and efficacy via vascular normalization, but
promotes lymphatic metastasis by activating CXCR4 in breast cancer.
Clin Cancer Res. 17:3638–3648. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Z, Kong D, Banerjee S, Li Y, Adsay
NV, Abbruzzese J and Sarkar FH: Down-regulation of platelet-derived
growth factor-D inhibits cell growth and angiogenesis through
inactivation of Notch-1 and nuclear factor-kappaB signaling. Cancer
Res. 67:11377–11385. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ranganathan P, Weaver KL and Capobianco
AJ: Notch signalling in solid tumours: A little bit of everything
but not all the time. Nat Rev Cancer. 11:338–351. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Fraser CC: Exploring the positive and
negative consequences of NF-kappaB inhibition for the treatment of
human disease. Cell Cycle. 5:1160–1163. 2006. View Article : Google Scholar : PubMed/NCBI
|