Introduction
Esophageal cancer, one of the most common upper
digestive tract tumors, may be divided into esophageal squamous
carcinoma and adenocarcinoma (1).
According to statistical data, esophageal squamous carcinoma is the
major subtype in developing countries, and adenocarcinoma of the
esophagus is more common in developed countries (2). China is located in the ‘Asian area with
high esophageal cancer occurrence’ and is the country with the
highest mortality rate of esophageal cancer. However, the
pathogenesis of esophageal cancer is not yet clear (3). Previously, an association between human
papillomavirus (HPV) and cervical cancer has been established and
relevant preventive vaccines have been released; scholars from
different countries have gradually turned toward studies on HPV
infection and non-genital cancers (4). It has been indicated in clinical
literature that HPV16 may be detected in specimens of esophageal
cancer, suggesting that HPV16 is involved in the occurrence and
progression of esophageal cancer (5,6). Nested
polymerase chain reaction (PCR) was adopted in a preliminary study
to test HPV infection subtype in Kazak esophageal squamous
carcinoma, indicating high expression of HPV16 in esophageal
squamous carcinoma (7). Also, it was
suggested that the level of HPV16E6 protein expression was a key
factor maintaining the malignant phenotype of cancer, and it may
serve a notable function in the occurrence and course of esophagus
cancer (8,9). As the mechanism of HPV16 in causing and
promoting cancer is relatively clear in studies concerning cervical
cancer (10), detection of HPV in
esophageal cancer cases is worthy of further study, and may have a
notable impact on the diagnosis and treatment of esophageal
cancer.
HPVs are a type of small, circular and
double-stranded DNA virus without a cell membrane (11). They are highly specific to their
host-species. High-risk HPV types are of tensaid to be associated
with malignant squamous cell tumors such as reproductive system
tumors, esophageal cancer and mouth neoplasm (12). Based on function, the genomic
structure of HPV can be divided into three coding regions,
including the early, late and upstream regulation regions (13). E6 and E7 are viral oncoproteins; the
latter majorly involves cell transformation during early cancer
stage and the former involves malignant transformation during late
stage (14). Although high expression
of HPV16 in pathological specimens of esophageal cancer has been
indicated in numerous studies (15),
the effects of HPV16 on proliferation, invasion, migration and
other biological features of esophageal squamous carcinoma remain
unclear.
Materials and methods
Cell culture
Human esophageal cancer cells, Eca109 and Eca9706
(Cell Bank of The Chinese Academy of Sciences, Shanghai, China),
were routinely cultured in Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
culture solution (including 1% penicillin-streptomycin mixture)
with 10% fetal calf serum (Hyclone; GE Healthcare Life Sciences,
Logan, UT, USA). Cells were cultured in an incubator at 37°C and 5%
CO2. When cell fusion reached 80–90%, trypsinization was
used for sub-culture. Cells sub-cultured in logarithmic phase were
used for the experiments.
Plasmid extraction
A total of 5 µl pcDNA3.1+ (Carrying HPV16-E6 gene
fragments; Sangon Biotech Co., Ltd., Shanghai, China) recombinant
plasmid-containing competent cells (Escherichia coli cells; Sangon
Biotech Co., Ltd., Shanghai, China) were selected and cultured in
Luria-Bertani (LB) solid medium (16)
at 37°C for 12 h. Monoclonal cells were selected overnight and
inoculated in 40 ml LB fluid medium (17). Ampicillin (30 µg/ml) was added to the
medium at a ratio of 1:100. The conical flask was placed in a
shaker (37°C; rotation speed of 2.96 × g) for overnight vibration.
The next day, a Column Plasmid Mini Preparation kit (Tiangen
Biotech Co., Ltd., Beijing, China) was used to extract plasmids
according to the manufacturer's protocol. The purity and
concentration of the extracted plasmids was determined with a
micro-volume spectrophotometer (NanoDrop 2000; Thermo Fisher
Scientific, Inc.).
Cell transfection
Human esophageal cancer Eca109 and Eca9706 cells
were evenly plated onto a 6-well plate in Opti-MEM (Gibco; Thermo
Fisher Scientific, Inc.) and cultured at 37°C. Once cell fusion
reached 70–80%, Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) was used as the transfection reagent to transfect
the plasmids (10 µg; containing HPV16E6 genes) into esophageal
cancer cells, which were named Eca109-1 and Eca9706-1. Esophageal
cancer cells transfected with nonsense segments were named as
Eca109-0 and Eca9706-0 as negative controls. Untreated esophagus
cancer cells, Ecal109-b and Eca9706-b, were adopted as blank
controls.
Quantitative-PCR (qPCR)
Total DNA of HPV16E6-transfected esophageal cancer
cells was extracted using TRIzol (Qiagen GmbH, Hilden, Germany),
according to the manufacturer's protocol. The fluorophore used was
SYBR Green kit (Qiagen GmbH) and β-actin was used as the reference
gene. PCR conditions were as follows: Pre-denaturation at 94°C for
10 min; 30 cycles of thermo cycling (denaturation at 94°C for 30
sec, annealing at 55°C for 1 min and extension at 72°C for 3 min);
extension at 72°C for 10 min; and completion at 4°C. The presence
of E6 DNA was determined with PCR extension. For E6mRNA
amplification (Sangon Biotech, China), the sequences of the primers
were as follows: Forward, 5′-CGGAATTCATGCACCAAAAGAG-AACTGCA-3′ and
reverse, 5′-CCCAAGCTTACAGCTGGGTT-TCTCTACG-3′. GAPDH was used as an
internal control, with the sequences of primers as follows:
Forward, 5′-GAPDH-FCAAGGTCATCCATGACAACTTTG-3′ and Reverse,
5′-GAPDH-RGTCCACCACCCTGTTGCTGTA-3′. The qPCR thermo cycling
conditions were as follows: Initial heat activation at 95°C for 2
min, denaturation at 95°C for 5 sec and combined annealing 60°C for
10 sec for 40 cycles. The experiment was repeated 3 times. Relative
expression of HPV16E6 was calculated using the 2−∆∆Cq
method (18).
Immunofluorescence test
Glass slides were horizontally placed in a 6-well
plate for cell culture. When cell growth reached 50–60%,
transfection in groups was conducted. At 24 h, transfection
efficiency (number of fluorescent cells/total number of cells) was
determined with a fluorescence microscope and the transfection was
terminated by DMEM, supplemented with 10% fetal calf serum after
cell fixation. Briefly, the slides were washed with pre-heated PBS
three times, and then 4% paraformaldehyde (2 ml/well) was added for
10 min at room temperature. The slides were washed three times with
PBS. Then, 0.1% Triton X-100 (2 ml/well) was added and incubated at
room temperature for 5 min for cell permeability. The slides were
washed with PBS three times. Once cell fusion reached 70–80%, PBS
was used to dilute primary antibodies (1:50) and then the slides
were incubated with goat anti-mouse HPV16E6 polyclonal
immunoglobulin G (IgG) (primary antibodies, cat no sc-1583, Santa
Cruz Biotechnology, Inc., Dallas, TX, USA) at 4°C overnight. The
slides were washed with PBS three times. Subsequently, goat
anti-mouse IgG, tetramethylrhodamine-conjugated secondary
antibodies, (cat no 6921–100, BioVision, Inc., Milpitas, CA, USA)
were diluted with PBS (1:50) and were incubated with the slides at
room temperature for 1 h. The slides were washed with PBS three
times. Cell nuclei were stained with DAPI at room temperature for
10 min and the slides were observed under a fluorescence microscope
(×100).
Western blot analysis
After 48 h of transfection, the total protein was
extracted from cells using radioimmunoprecipitation assay and
phenylmethane sulfonyl fluoride protein lysis buffer for 20 min at
4°C (Applygen Technologies, Inc., Beijing, China). Subsequently,
samples were a centrifuged at 1,200 × g for 20 min at 4°C to obtain
the total protein. Protein concentration was measured on NanoDrop
2000 (Thermo Fisher Scientific, Inc.) Proteins (20 µl, 40 mg/ml)
were separated by 15% SDS-PAGE and transferred to polyvinylidene
difluoride membranes. The membranes were blocked with 5% skim milk
powder for 2 h. Goat anti-mouse HPV16E6 polyclonal IgG (dilution,
1:200; cat no. sc-1583, Santa Cruz Biotechnology, Inc.) were
applied to the membrane and incubated at 4°C overnight. The
membranes were washed with PBS-Tween-20 (PBST). Goat anti-mouse IgG
horseradish peroxidase-conjugated antibodies (dilution, 1:3,300;
cat no. ZB-2306, OriGene Technologies, Inc., Beijing, China) were
added and incubated at room temperature for 2 h, prior to the
membrane being washed with PBST. HPV16E6 protein expression was
evaluated using an enhanced chemiluminescence kit (West Pico PLUS
Chemiluminescent Substrate; Thermo Fisher Scientific, Inc.) and
β-actin was adopted as an internal reference: Mouse anti-β-actin as
primary antibodies (dilution, 1:1,000; cat no. TA-09, OriGene
Technologies, Inc.) and Go at anti-mouse IgG horseradish
peroxidase-conjugated antibodies (dilution, 1:2,000; cat no.
ZB-2305, OriGene Technologies, Inc.). The experiment was repeated
three times.
Cell proliferation analysis
Cells of logarithmic phase were transferred to a
6-well plate for 24 h cell transfection. A cell suspension using
DMEM was prepared by dissociation and re-suspension and was then
transferred onto a 96-well plate. A volume of 100 µl cell
suspension was added to each well (4,000 cells/well). The plate was
placed in an incubator (37°C, 5% CO2) for 3 h for
coherence (regarded as ‘0 h’), and 10 µl CCK-8 solution (Dojindo
Molecular Technologies, Inc., Shanghai, China) was added at 0, 12,
24, 48, 72 and 96 h. Three wells were set for each group. The
culture plate was incubated in the incubator for 2 h. The optical
density (OD) at 460 nm was determined with a microplate reader. The
OD values represented the cell proliferation capacity.
Plate colony formation assay
Cells of logarithmic phase transfected for 24 h were
used, along with DMEM, to prepare a cell suspension. The infinite
dilution method was adopted to incubate cells in a 6-well plate
(1,000 cells/well). The plate was agitated gently to spread the
suspension and incubated in an incubator at 37°C (5%
CO2). The medium was changed every 4–5 days, for a total
of 2 weeks. The culture was stopped when visible colonies appeared
on the plate. The culture solution was removed and the plate was
rinsed with PBS buffer solution twice. The colony was fixed by
adding 4% paraformaldehyde for 15–20 min (37°C). Following natural
drying, moderate 0.1% crystal violet was added for 15–20 min
(37°C). The residual dye was washed off with water and the plate
was air-dried. The number of colonies with >50 cells were
counted under a fluorescence inverted microscope (×71
magnification; Olympus Corporation, Tokyo, Japan).
Wound healing assay
Cells of logarithmic phase were transferred to a
6-well plate (~7×105 cells/well) and placed in an
incubator overnight (37°C, 5% CO2). Cells were
transfected in groups when fusion reached 70–80%. At 24 h after
transfection, a scratch was made in the 6-well plate with a 10 µl
micropipette. Serum-free medium was added after washing off
detached cells with PBS. The scratches were observed following
culture for 0, 12, 24, 36 and 48 h using an inverted fluorescence
microscope (×71 magnification; Olympus Corporation, Tokyo, Japan).
Scratch width ratios were calculated as follows: Scratch width
ratio=(end width/initial width) ×100%.
Cell invasion assay
A total of 5 µl Matrigel (BD Biosciences, Franklin
Lakes, CA, USA) and 40 µl serum-free DMEM (1:8) were mixed in an
Eppendorf tube (both the tip and DMEM underwent pre-cooling
treatment). A volume of 45 µl solution was used to cover the bottom
of the transwell insert (Corning Life Sciences, Corning, NY, USA).
The chambers were subsequently placed in an incubator for 2–3 h.
The residual liquid in the upper chamber was removed following
solidification, 80 µl serum-free DMEM was added and the chamber was
placed in the incubator for 30 min hydration. After 24 h
transfection, cells were dissociated with trypsin, re-suspended in
serum-free DMEM and 200 µl was added to the upper chamber
(~8×104 cells/well) prior to the addition of 600 µl
complete medium containing 20% serumto the lower chamber. Following
incubation for 48 h, 100 and 600 µl preheated PBS was added to the
upper and lower chambers respectively, chambers were washed 1–2
times and PBS in the upper chambers was discarded. A total of 600
µl pre-cooled paraformaldehyde (4%) was added to the upper chamber
for cell fixing at 4°C for 20 min. Transwell inserts were placed in
PBS to be washed. Residual liquids were removed from the chamber
and allowed to dry naturally. Crystal violet (0.1%; 600 µl) was
added to the lower chamber to stain the cells in transwell inserts
for 20 min at room temperature. Transwell inserts were then washed
twice with PBS and any un-migrated cells on the surface of the
microporous membrane were removed. Finally, cell migration was
observed using light microscopy (magnification ×100; ×71
magnification; Olympus Corporation, Tokyo, Japan) for subsequent
statistical analysis.
Statistical analysis
All experiments were conducted independently three
times. In the in vitro wound healing and invasion assays, a
total of five fields of view were randomly selected and examined at
×100 magnification for each experimental group. Experimental data
and results were analyzed with SPSS 17.0 software (SPSS, Inc.,
Chicago, IL, USA). Image J (version 1.48) image processing software
(National Institutes of Health, Bethesda, MD, USA) was used for
cell counting. Image-Pro Plus 6.0 (Media Cybernetics, Inc.,
Rockville, MD, USA) was used to measure the widths of scratches.
Experimental data are presented as the mean ± standard deviation.
Data were analyzed by one-way analysis of variance followed by
Bonferroni's multiple comparison test. P<0.05 was considered to
indicate a statistically significant difference.
Results
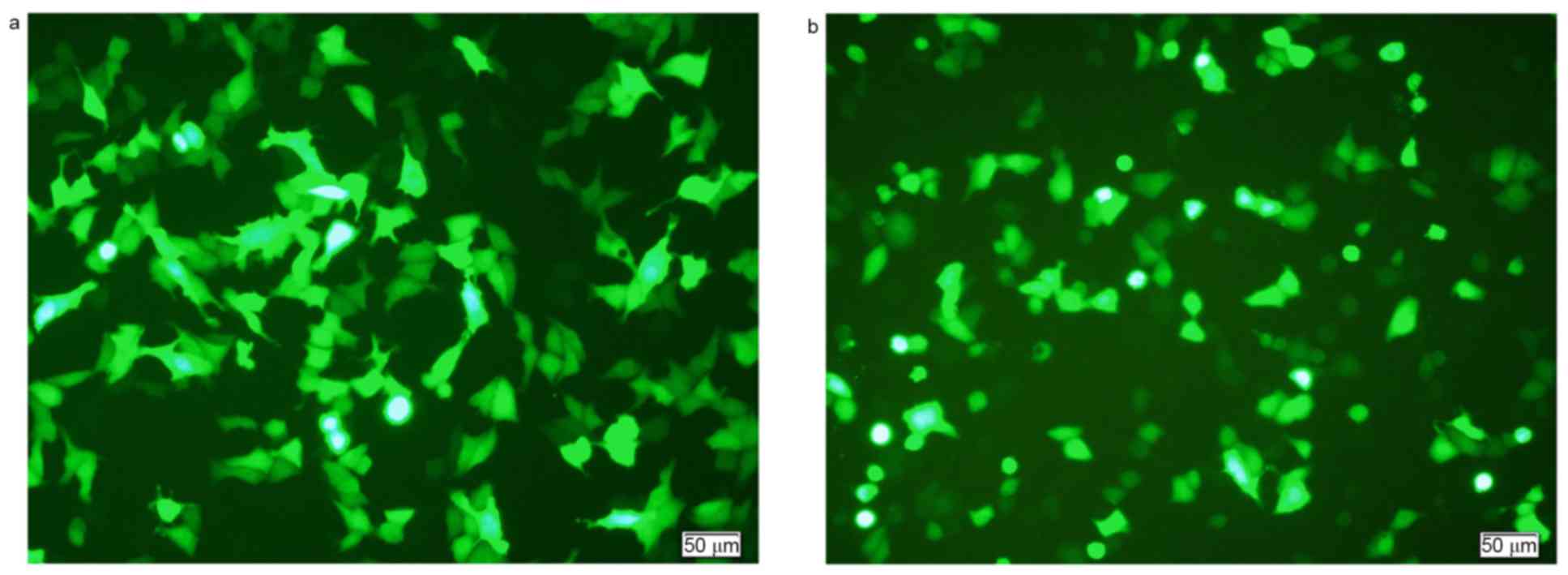
Transfection efficiency
The peak transfection efficiency of Eca109 cells
was~40%. The conditions for highest efficiency transfection were as
follows: 10 µg DNA (plasmid) per well; ratio of Lipofectamine 2000
to DNA of 1:1; 12 h transfection time; and no starvation performed
before transfection. The peak transfection efficiency for Eca9706
cells was ~30%, with the following conditions: 10 µg DNA (plasmid)
per well; ratio of Lipofectamine 2000 to DNA of 1:1; 24 h
transfection time; and 6 h starvation performed before
transfection. Representative images of peak transfection efficiency
are presented in Fig. 1.
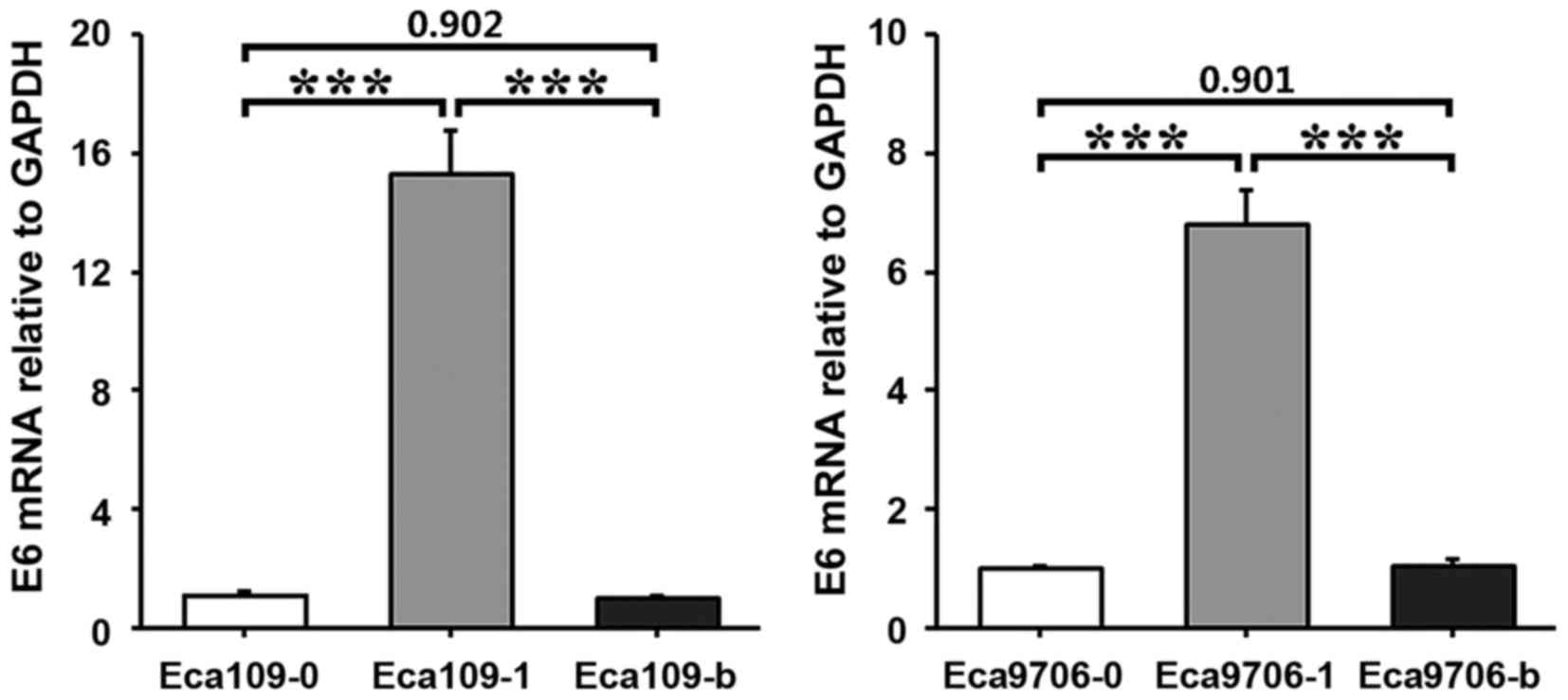
RT-qPCR results
HPV16E6 esophagus cancer cells were treated
separately to establish Eca109-0, Eca109-1, Eca109-b, Eca9706-1,
Eca9706-0 and Eca9706-b cells. Total DNA extracted from transfected
esophageal cancer cells underwent PCR amplification. The amplified
products underwent electrophoresis. This indicated a specific band
at ~500 bp, which was consistent with the target gene HPV16E6 (474
bp). RT-qPCR indicated that E6 mRNA expression levels were
significantly higher in Eca109-1 cells compared with Eca109-0 or
Eca109-b cells (P<0.001; Fig. 2).
E6 mRNA expression levels were also significantly higher in
Eca9706-1 cells compared with Eca9706-0 or Eca9706-b cells
(P<0.001; Fig. 2).
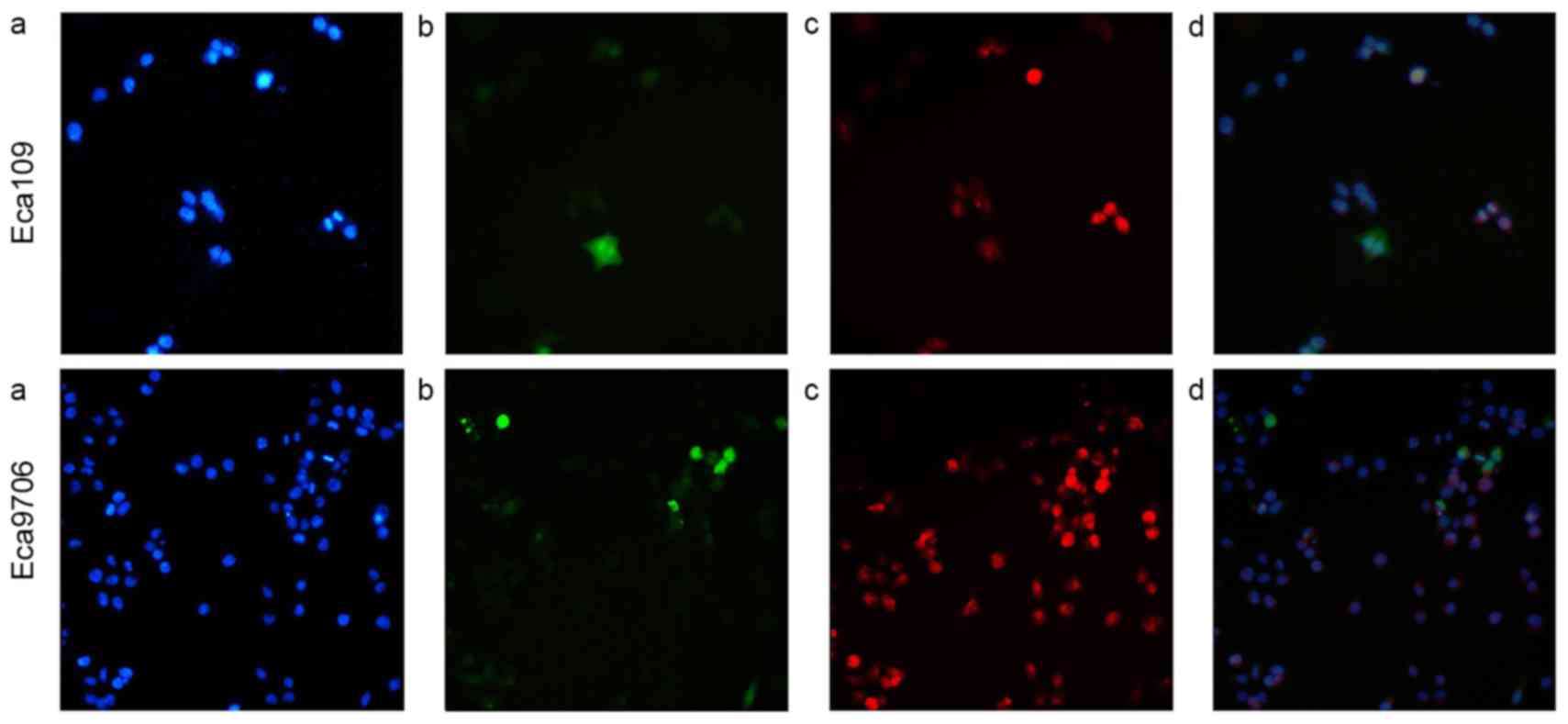
Immunofluorescence assay results
An immunofluorescence assay was used to determine
expression of HPV16E6 in esophageal cancer cells Eca109 and
Eca9706. The results indicated that HPV16E6 was widely distributed
in the cell nuclei and cytoplasm (Fig.
3).
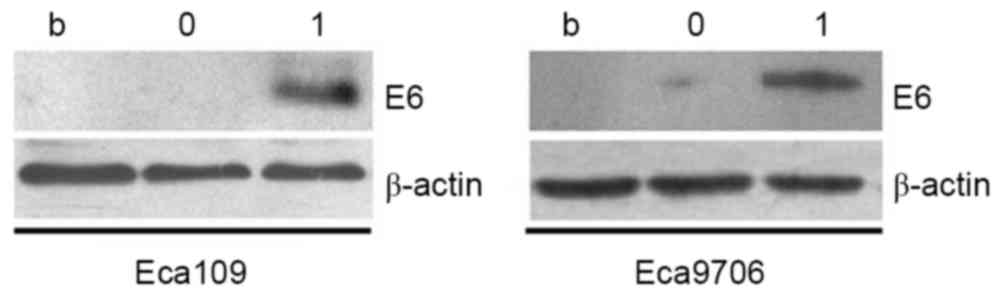
Western blot analysis
After cells had been transfected with plasmid for 48
h, western blot analyses were conducted, with β-actin as the
internal reference (Fig. 4). An
obvious band was observed in the 17 kb position in the transfection
groups, while no obvious bands were indicated in the nonsense
transfected group or the blank control group. These results
indicated that transfection with recombinant plasmid
(HPV16E6pcDNA-3.1) had resulted in upregulated HPV16E6 protein
expression.
Cell proliferation assay result
OD values during different stages and in different
groups measured with microplate reader are presented in Table I. The results at 48 h indicated that
the OD values of Eca109-1 were significantly higher compared with
Eca109-0 and Eca109-b (P<0.05), while no statistically
significant difference in OD was observed between Eca109-b and
Eca109-0 (P=0.071). For Eca9706 cells, statistically significant
differences were observed between different experimental groups
(P<0.05), which were ranked as
Eca9706-1>Eca9706-b>Eca9706-0, with respect to OD value.
 | Table I.Cell proliferation assay. |
Table I.
Cell proliferation assay.
|
| Optical density
value |
|---|
|
|
|
|---|
| Group | 0 h | 12 h | 24 h | 36 h | 48 h |
|---|
| Eca109-0 | 0.681±0.069 | 0.802±0.049 | 1.126±0.138 | 1.645±0.098 |
2.057±0.160c |
| Eca109-1 | 0.797±0.046 | 0.850±0.068 | 1.063±0.183 | 1.741±0.197 |
2.277±0.091a,b |
| Eca109-b | 0.844±0.077 | 1.070±0.109 | 1.595±0.103 | 2.285±0.063 | 2.710±0.008 |
| Eca9706-0 | 0.166±0.027 | 0.419±0.055 | 0.769±0.123 | 1.784±0.114 |
2.671±0.163f |
| Eca9706-1 | 0.170±0.028 | 0.349±0.043 | 0.491±0.045 | 1.367±0.087 |
2.232±0.124d,e |
| Eca9706-b | 0.183±0.025 | 0.929±0.102 | 1.042±0.067 | 1.901±0.075 | 3.025±0.025 |
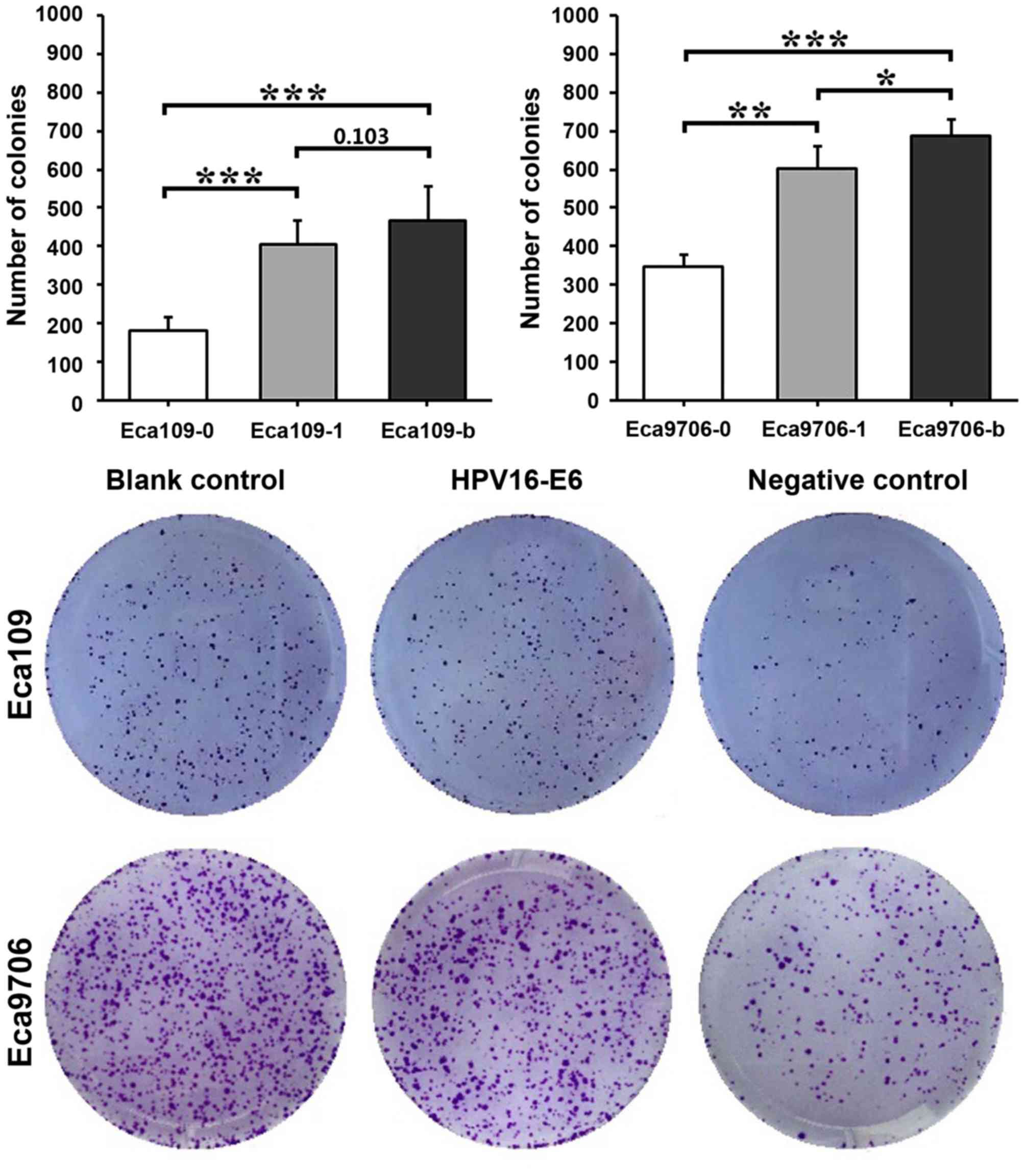
Cell colony formation assay
The results of the colony assay indicated that the
colony formation capacity was strongest in the blank control groups
and weakest in the negative control groups (Fig. 5). In Eca109 cells, Eca109-0 exhibited
significantly fewer colonies compared with Eca109-1 (P<0.001) or
Eca109-b (P<0.001) cells. However, no significant difference was
observed between Eca109-1 and Eca109-b cells (P=0.103). In Eca9706
cells, Eca9706-0 cells exhibited significantly fewer colonies
compared with Eca9706-1 (P<0.01) or Eca9706-b (P<0.001)
cells. Eca9706-1 cells also exhibited significantly fewer colonies
compared with Eca9706-b cells (P<0.05).
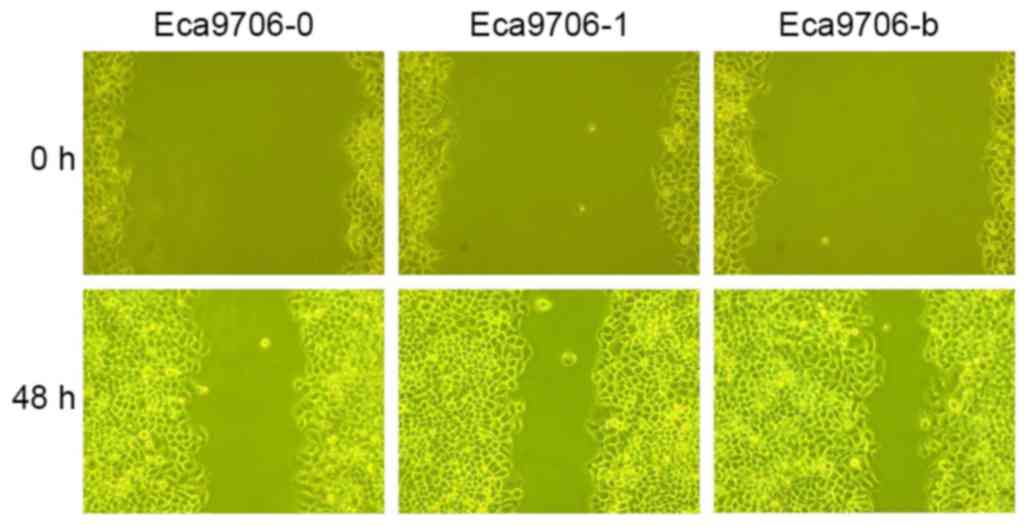
Wound healing assay
Scratch width ratios of experimental groups at
different time points are presented in Table II. The results at 48 h indicated that
lower scratch width ratios were associated with longer growth times
in different cell groups. In Eca109 cells, Eca109-b exhibited
significantly lower scratch width ratio with Eca109-1 (P<0.05)
and Eca109-0 (P<0.05). Meanwhile, the scratch width ratio in
Eca109-1 was significantly lower than that of Eca109-0 (P<0.05).
In Eca9706 cells, the scratch width ratio in Eca9706-0 was
significantly higher compared with Eca9706-1 (P<0.05) and
Eca9706-b (P<0.05), while no statistically significant
difference was observed between Eca9706-1 and Eca9706-b (P=0.121).
Representative images of the wound healing assay are presented in
Fig. 6.
 | Table II.Wound healing assay. |
Table II.
Wound healing assay.
|
| Scratch width ratio
(%) |
|---|
|
|
|
|---|
| Group | 12 h | 24 h | 36 h | 48 h |
|---|
| Eca109-0 | 87.1±3.7 | 78.8±1.6 | 62.3±2.8 |
40.8±3.9c |
| Eca109-1 | 83.4±1.2 | 72.7±3.8 | 46.2±4.9 |
33.1±1.7a,b |
| Eca109-b | 81.8±4.0 | 63.3±3.1 | 36.8±1.8 | 26.2±2.8 |
| Eca9706-0 | 94.1±3.5 | 85.5±3.8 | 74.0±2.4 |
48.5±2.7f |
| Eca9706-1 | 88.7±1.9 | 77.4±3.6 | 62.2±2.2 |
40.2±2.8d,e |
| Eca9706-b | 91.9±2.9 | 73.5±1.7 | 57.9±2.9 | 31.3±2.6 |
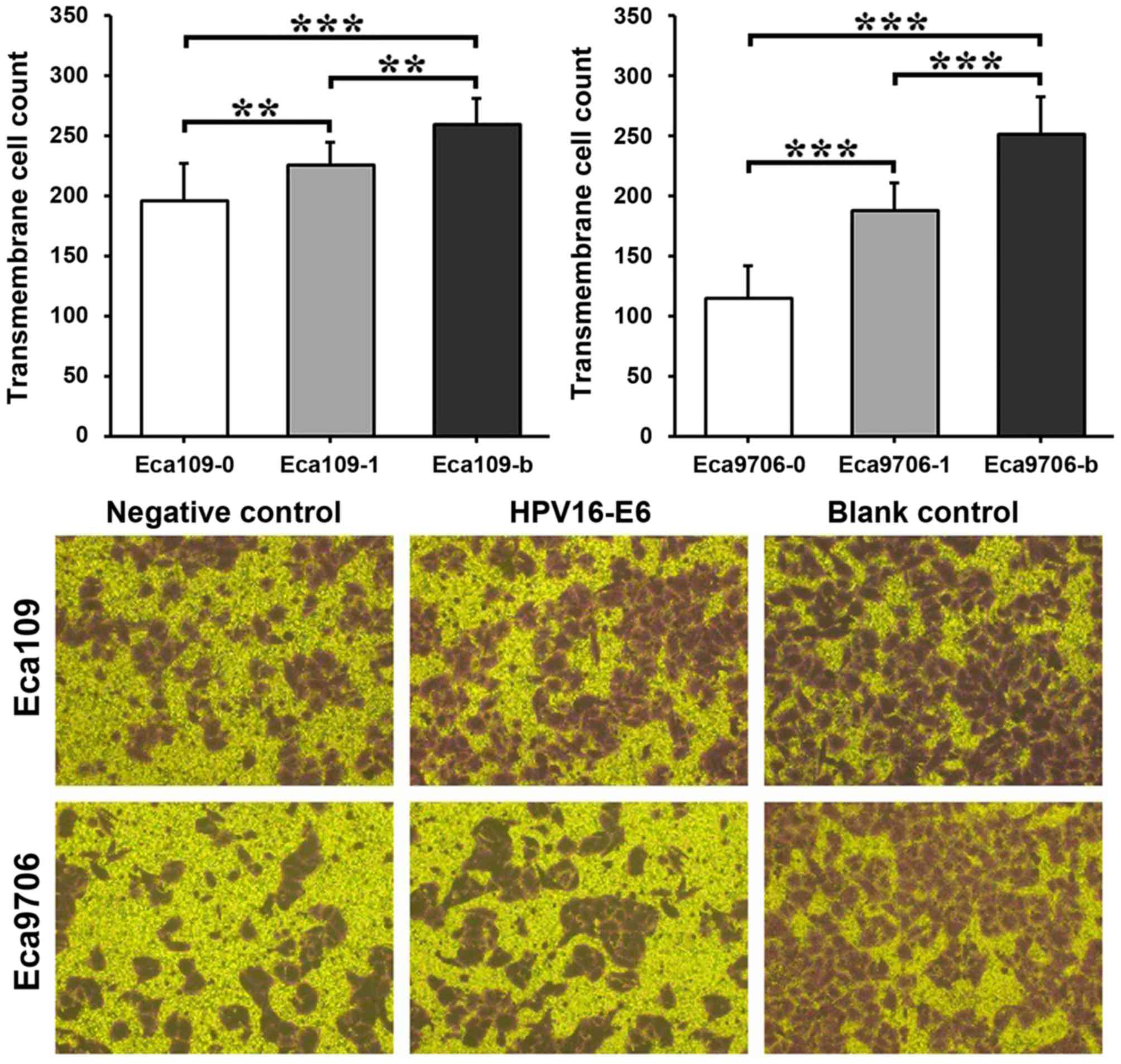
Cell invasion assay results
The invasion capacity was similar in the two
esophageal cancer cell types (Fig.
7). The blank control group exhibited a significantly higher
invasion capacity compared with the transfection group (P<0.01
in Eca109 and P<0.001 in Eca9706) and negative control group
(P<0.001 in the two cell types). In addition, the transfection
group exhibited a significantly higher invasion capacity compared
with the negative control group (P<0.01 in Eca109 and P<0.001
in Eca9706).
Discussion
Syrjanen (19) was the
first to identify that morphological change of esophageal squamous
carcinoma was similar to that of condyloma of the genital system
and speculate that HPV infection maybe a risk factor for esophageal
cancer. Since then, another study demonstrated HPV16E6 is closely
is closely associated with tumorigenesis at the gene, mRNA, protein
and other levels (20). With in-depth
studies on oncogenes, it has been identified that HPV16E6 protein
serves a critical function in carcinogenic development, as it
activates transcription of human telomerase reverse transcriptase,
collaborates with E7 protein to inactivate retinoblastoma protein
and promotes cell immortalization (21,22). It
has been suggested in numerous clinical studies and investigations
that HPV is associated with esophageal cancer and HPV infection may
promote development of esophageal cancer (5–8,23). Tumor development is a complicated
process associated with multiple possible mechanisms. Early
expressed genes of HPV16 may be involved in tumorigenesis and
progression in cell cycles (6,7), but it
has not yet been reported whether E6 regulates and controls
proliferation, invasion and migration of tumor cells in esophageal
cancer. Therefore, in the present study, HPV16E6 was used to
transfect Eca109 and Eca9706 esophageal cancer cells, and the
effects on biological properties of these cell lines were
observed.
In fluorescence microscope images in the present
study, it was demonstrated that positive liposome-mediated HPV16E6
successfully transfected esophageal cancer cell lines Eca109 and
Eca9706, and stable cell lines were achieved by screening. This
laid a foundation for studying the etiology and mechanism of
esophageal cancer. With respect to function, it was demonstrated
with RT-PCR that the transfected esophagus cancer cells contained
the target gene, HPV16E6, and HPV16E6mRNA, which was demonstrated
at the protein level. It was indicated by the immunofluorescence
assay and western blot analysis that HPV16E6 protein was expressed
in the transfection group, but not in the blank or negative control
group. HPV16E6 expression was distributed in the cytoplasm and cell
nucleus. The results also suggested that the transfection
efficiency was higher in poorly-differentiated Eca109 cells
compared with well-differentiated Eca9706 cells. This finding was
consistent with previous research, which identified that HPV16E6
expression is correlated with a decreased degree of tissue
differentiation (24).
In the present study, cell proliferation in in the
target gene-transfected esophageal cancer cell lines was
significantly higher compared with the negative control groups, and
increased over time. Similarly, in the cell colony formation assay,
the number of cell colonies was significantly in the transfected
cell groups compared with negative control groups. These results
suggest that cell proliferation ability is improved after HPV
infection. The results of the wound healing assay indicated that
the migratory capacity was higher in the transfected groups
compared with the negative control groups. Similarly, in the
Transwell Matrigel assay, the number of membrane-crossing cells was
significantly higher in the experimental groups compared with the
negative control groups, suggesting that invasion capacity was
increased in HPV16E6-transfected cells. These results suggested
that expression of HPV16E6 may be associated with proliferation,
invasion and migration of esophageal cancer. Eca109 cells exhibited
higher transfection efficiency but lower proliferation capacity
compared with Eca9706 cells, suggesting that HPV16E6 exerts greater
effects on well-differentiated Eca9706 compared with
poorly-differentiated Eca109. However, Eca109 cells exhibited
stronger invasion and migration abilities compared with Eca9706
cells. The reason for this may be that Eca109 cells are poorly
differentiated esophageal cancer cells with high malignancy, or
that the high transfection efficiency of Eca109 leads to greater
effects of HPV16E6. The underlying mechanisms of these differences
require further investigation. The current results also identified
that non-transfected (blank control) cell lines have slightly
stronger proliferation, invasion and migration capacities compared
with nonsense transfected (negative control) cell lines, suggesting
that transfection causes a certain level of injury to esophageal
cancer cells (25,26).
The effects of HPV16E6 on esophageal cancer cells
may be associated with the following mechanisms. P53 is an
extensively studied cancer suppressor gene and is involved in
almost half of human malignant tumor types (27). It has been indicated in certain
studies that, in HPV16E6-infected esophagus cancer cells, p53
couples with E6 protein closely to inhibit p53 from entering the
cell nucleus. In addition, the ubiquitin-dependent protease system
facilitates p53 protein degradation to disable effective p53
expression. This results in inactivated regulatory functions of
cell cycle-related factors (including P21, proliferating cell
nuclear antigen, cyclins and cyclin-dependent kinases), shortened
G1/S stage, hindered cell apoptosis and cell cycle arrest,
reactivated DNA synthesis mechanisms, and duplicated virus DNA
(28,29). Therefore, the effects of HPV16E6 on
the normal functions of p53 indicate particular significance for
carcinogenicity (30,31). It has also been demonstrated in
previous studies that HPV16E6 can allow cells to escape the
proliferation limit of senescence and immortalize normal cells by
activating telomerase (32–34).
In conclusion, positive liposome-mediated HPV16E6
successfully transfected Eca109 and Eca9706 cell lines and E6
protein was stably expressed in transfected cell lines. Cell lines
with stable expression of HPV16E6, achieved with a screening
process, facilitated studies of the biological functions of E6
proteins at the cellular level. By observing the biological effects
of HPV16E6 transfection in differently differentiated esophageal
cancer cells, it was identified that E6 induces cell proliferation
and promotes malignancy (invasion and migration capacity). These
findings lay a foundation for further study of the association
between HPV and esophageal cancer and provide theoretical guidance
for the prevention and treatment of HPV-associated esophageal
cancer. Further studies are required to identify the underlying
mechanism of HPV16E in tumorigenesis and progression.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81260362).
References
|
1
|
Power DG, O'Sulleabhain C and Murphy TJ:
Preoperative chemoradiotherapy for esophageal cancer. Australasian
Radiol. 43:215–219. 2001.
|
|
2
|
Lin Y, Totsuka Y, He Y, Kikuchi S, Qiao Y,
Ueda J, Wei W, Inoue M and Tanaka H: Epidemiology of esophageal
cancer in Japan and China. J Epidemiol. 23:233–242. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen W, Zheng R, Zhang S, Zeng H, Fan Y,
Qiao Y and Zhou Q: Esophageal cancer incidence and mortality in
China, 2010. Thoracic Cancer. 5:343–348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cao BW, Jing-Lin YU, Huan HE, Shen-ta LI
and Yu-lian Z: Meta-analysis on the Relationship between HPV
infection and esophageal cancer in Chinese Population. J Capital
Med Univ. 97:148–150. 2010.
|
|
5
|
Wang X, Tian X, Liu F, Zhao Y, Sun M, Chen
D, Lu C, Wang Z, Shi X, Zhang Q, et al: Detection of HPV DNA in
esophageal cancer specimens from different regions and ethnic
groups: A descriptive study. BMC Cancer. 10:192010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou Y, Pan Y, Zhang S, Shi X, Ning T and
Ke Y: Increased phosphorylation of p70 S6 kinase is associated with
HPV16 infection in cervical cancer and esophageal cancer. Br J
Cancer. 97:218–222. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen WG, Yang CM, Xu LH, Zhang N, Liu XY,
Ma YG, Huo XL, Han YS, Tian DA and Zheng Y: Gene chip technology
used in the detection of HPV infection in esophageal cancer of
Kazakh Chinese in Xinjiang province. J Huazhong Univ Sci Technolog
Med Sci. 34:343–347. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dong GC: Multiple PCR-mass spectrometry
detection of kazak in xinjiang esophageal cancer HPV infection and
genotype distribution. Xinjiang Shihezi Univ. 26–27. 2014.
|
|
9
|
Chen YW, Wu MF, Wand J, Yeh KT, Goan YG,
Chiou HL, Chen CY and Lee H: Human papillomavirus 16/18 E6
oncoprotein is expressed in lung cancer and related with p53
inactivation. Cancer Res. 67:10686–10693. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Skinner SR, Apter D, De Carvalho N, Harper
DM, Konno R, Paavonen J, Romanowski B, Roteli-Martins C, Burlet N,
Mihalyi A and Struyf F: Human papillomavirus (HPV)-16/18
AS04-adjuvanted vaccine for the prevention of cervical cancer and
HPV-related diseases. Expert Rev Vaccines. 15:367–387.
2016.PubMed/NCBI
|
|
11
|
Moody CA and Laimins LA: Human
papillomavirus oncoproteins: Pathways to transformation. Nat Rev
Cancer. 10:550–560. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang H, Zhang B, Chen W, Zhou SM, Zhang
YX, Gao L, Xu ZG, Qiao YL and Tang PZ: Human papillomavirus
infection and prognostic predictors in patients with oropharyngeal
squamous cell carcinoma. Asian Pac J Cancer Prev. 13:891–896. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Talora C, Sgroi DC, Crum CP and Dotto GP:
Specific down-modulation of Notch1 signaling in cervical cancer
cells is required for sustained HPV-E6/E7 expression and late steps
of malignant transformation. Genes Dev. 16:2252–2263. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rosty C, Sheffer M, Tsafrir D, Stransky N,
Tsafrir I, Peter M, de Crémoux P, de La Rochefordière A, Salmon R,
Dorval T, et al: Identification of a proliferation gene cluster
associated with HPV E6/E7 expression level and viral DNA load in
invasive cervical carcinoma. Oncogene. 24:7094–7104. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu CL, Qian XL, Zhou XS, Zhao QZ and Li
YC: Expression of HPV16-E6 and E7 oncoproteins in squamous cell
carcinoma tissues of esophageal cancer and non-cancer tissues. Ai
Zheng. 23:165–168. 2004.(In Chinese). PubMed/NCBI
|
|
16
|
Rayner MH, Sadler PJ and Scawen MD: NMR
studies of a bacterial cell culture medium (LB broth): Cyclic
nucleotides in yeast extracts. FEMS Microbiol Lett. 56:217–221.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Park CS: Effect of Tryptic Soy Broth (TSB)
and Luria-Bertani (LB) medium on production of subtilisin CP-1 from
Bacillus sp. CP-1 and Characterization of Subtilisin CP-1. Archives
Clin Psychiatry. 22:250–255. 2012.
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Syrjänen KJ: Histological changes
identical to those of condylomatous lesions found in esophageal
squamous cell carcinomas. Arch Geschwustforsch. 52:283–292.
1982.
|
|
20
|
Gu LY, Hou Saliman XL, et al: HPV16
infection and kazak in xinjiang esophageal cancer and clinical
pathology study. Chin J Cancer. 9:68–672. 2010.
|
|
21
|
O'rorke MA, Ellison MV, Murray LJ, Moran
M, James J and Anderson LA: Human papillomavirus related head and
neck cancer survival: A systematic review and meta-analysis. Oral
Oncol. 48:1191–1201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiang LZ, Lv CF, Lu HY, et al: HPV16e6
protein element D_1, cell cycle and telomerase reverse
transcriptase in the tissue of nasopharyngeal carcinoma and
significance. J Sec Military Med Univ. 12:67–1371. 2009.
|
|
23
|
Liu M, Zeng HC, Zhang XL, Zhu J, Huang JF,
Zhang X and Xia M: Study on human papillomavirus infection and loss
of heterozygosity of microsatellite in esophageal cancer. Zhonghua
Liu Xing Bing Xue Za Zhi. 28:1203–1206. 2007.(In Chinese).
PubMed/NCBI
|
|
24
|
Miller D, Puricelli MD and Stack MS:
Virology and molecular pathogenesis of HPV (human
papillomavirus)-associated oropharyngeal squamous cell carcinoma.
Biochem J. 443:339–353. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wu Y, Gao T, Wang X, Hu Y, Hu X, Hu Z,
Pang J, Li Z, Xue J, Feng M, et al: TALE nickase mediates high
efficient targeted transgene integration at the human multi-copy
ribosomal DNA locus. Biochem Biophys Res Commun. 446:261–266. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou YF, Chen XA, Ye M, et al: Orthogonal
design optimization of polyethylene imine mediated liver cancer
cell gene transfection efficiency of study. J Biomed Eng.
1:104–109. 2011.
|
|
27
|
Crook T, Tidy JA and Vousden KH:
Degradation of p53 can be targeted by HPV E6 sequences distinct
from those required for p53 binding and trans-activation. Cell.
67:547–556. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wiest T, Schwarz E, Enders C,
Flechtenmacher C and Bosch FX: Involvement of intact HPV16 E6/E7
gene expression in head and neck cancers with unaltered p53 status
and perturbed pRb cell cycle control. Oncogene. 21:1510–1517. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Havre PA, Yuan J, Hedrick L, Cho KR and
Glazer PM: p53 inactivation by HPV16 E6 results in increased
mutagenesis in human cells. Cancer Res. 55:4420–4424.
1995.PubMed/NCBI
|
|
30
|
Illiano E, Demurtas OC, Massa S, Di Bonito
P, Consalvi V, Chiaraluce R, Zanotto C, De Giuli Morghen C,
Radaelli A, Venuti A and Franconi R: Production of functional,
stable, unmutated recombinant human papillomavirus E6 oncoprotein:
Implications for HPV-tumor diagnosis and therapy. J Transl Med.
14:2242016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang L, Dang YW, Mo XL, et al: The
cervical lesions in h-TERC gene amplification and the relationship
between h-TERT protein expression and the significance. J
Diagnostic Pathol J. 21:561–564. 2014.
|
|
32
|
Zhang H, Jin Y, Chen X, Jin C, Law S, Tsao
SW and Kwong YL: Papillomavirus type 16 E6/E7 and human telomerase
reverse transcriptase in esophageal cell immortalization and early
transformation. Cancer Lett. 245:184–194. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Weijzen S, Zlobin A, Braid M, Miele L and
Kast WM: HPV16 E6 and E7 oncoproteins regulate Notch-1 expression
and cooperate to induce transformation. J Cell Physiol.
194:356–362. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cassetti MC, McElhiney SP, Shahabi V,
Pullen JK, Le Poole IC, Eiben GL, Smith LR and Kast WM: Antitumor
efficacy of Venezuelan equine encephalitis virus replicon particles
encoding mutated HPV16 E6 and E7 genes. Vaccine. 22:520–527. 2004.
View Article : Google Scholar : PubMed/NCBI
|