Introduction
Bladder cancer (BC), one of the most prevalent
carcinomas worldwide, has been identified as the fourth and tenth
leading cause of cancer-related deaths in males and females,
respectively (1). Despite many
efforts have been made, the prognosis still remains unsatisfied.
The initiation and progression of BC involves changes about a
variety of oncogenes and tumor suppressors. Therefore,
investigating the molecular mechanisms underlying the tumorigenesis
of bladder cancer cells is essential for exploring novel treatment
targets.
Currently, long non-coding RNAs (lncRNAs), newly
identified members of the noncoding RNA family with length >200
nucleotides (nt), have been proposed (2–4).
Accumulating documents have revealed that lncRNAs play a critical
role in tumorigenesis and can be used as biomarkers for diagnosis
or prediction of survival and recurrence in multiple cancers
(5–9).
For instance, in 2017, Idogawa et al reported that long
non-coding RNA NEAT1 was a transcriptional target of
p53 and modulated p53-induced transactivation and
tumor-suppressor function (10). Zhou
et al demonstrated that downregulation of lncRNA MEG3
mediated by DNMT3b contributed to nickel malignant
transformation of human bronchial epithelial cells via modulating
PHLPP1 transcription and HIF-1α translation (11). Wang et al uncovered that
2-O-Methylmagnolol upregulated the long non-coding RNA,
GAS5, and enhanced apoptosis in skin cancer cells (12). Several other studies also demonstrated
the function of lncRNAs in BC (13–15).
Despite so many lncRNAs have been reported to be associated with
BC, still many lncRNAs need to be investigated.
Small nucleolar RNA host gene 5 (SNHG5), a
SnoRNA-U50-associated lncRNA, has been demonstrated downregulated
in bladder cancer (BC) and colorectal carcinoma (CRC) (16,17).
However, its biological function in BC has not been investigated.
The aim of our present study is to investigate whether SNGH5
is associated with BC progression and to identify the role of
SNHG5 in the prognosis of BC. Herein, we uncovered that
SNHG5 was significantly overexpressed in BC tissues which
was associated with larger tumor range, metastasis, lymph nodes,
pathological stage and poor prognosis. In addition, we demonstrated
that silenced SNHG5 suppressed cell proliferation through
influencing cell cycle and apoptosis rate. Therefore, the results
indicated that SNHG5 acted as an oncogene in BC.
Materials and methods
Patients and clinical samples
collection
BC tissues (n=67) and pair-matched noncancerous
tissues were obtained through tissue biopsy from patients diagnosed
with BC at the Department of Urology, Yidu Central Hospital of
Weifang between March 2013 and September 2016. Informed consent was
obtained from patients. All procedures involving human participants
were in accordance with the ethical standards of the Human Research
Ethics Committee at the Department of Urology, Yidu Central
Hospital of Weifang.
Cell lines
Bladder cancer SW780, UMUC3, 5637, T-24 and one
normal urothelial cell line SVHUC-1 utilized in present study were
purchased from the Tumor Cell Bank of the Chinese Academy of
Medical Science (Shanghai, China). The UMUC3, T24 and SV-HUC-1
cells were cultured in Dulbecco's modified Eagle's medium (DMEM;
Invitrogen Life Technologies, Carlsbad, CA, USA) plus 10% fetal
bovine serum and ampicillin and streptomycin at 37°C in a
humidified atmospherewith 95% air and 5% CO2. The 5637
and SW780 cells were cultured in RPMI-1640 medium (Invitrogen Life
Technologies) plus 10% fetal bovine serum and ampicillin and
streptomycin at 37°C in a humidified atmosphere with 95% air and 5%
CO2.
Reverse-transcription quantitative
polymerase chain reaction (RT-qPCR)
Total RNAs from tissues and cells were isolated with
TRIzol reagent (Invitrogen Life Technologies) under the
manufacturer's instructions. Reverse transcription was performed
with PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan)
according to the manufacturer's instructions. RT-qPCR was performed
with SYBR Prime Script RT-PCR kits (Takara Bio, Inc.) based on the
manufacturer's instructions. The SNHG5 level was calculated
with the 2−ΔΔCt method, which was normalized to
GAPDH mRNA. The primers for SNHG5 were as the
following: forward, 5′-CGCTTGGTTAAAACCTGACACT-3′ and reverse,
5′-CCAAGACAATCTGGCCTCTATC-3′; the primers for GAPDH were as
the listed: Forward, 5′-ACGGGAAGCTCACTGGCATGG-3′ and reverse,
5′-GGTCCACCACCCTGTTGCTGTA-3′. All assays were performed in
triplicate. The expression levels were relative to the fold change
of the corresponding controls, which were defined as 1.0.
siRNA transfections
Cells were transfected with siRNAs for SNHG5
by using Lipofectamine 2000 (Invitrogen Life Technologies)
according to the manufacturer's instructions. After transfection
(48 h), cells were harvested for the following experiments
including RT-qPCR, western blot analysis, proliferation assays and
flow cytometry. RNA oligonucleotides were purchased from GenePharma
(Shanghai, China). The siRNA sequence for SNHG5 was
si-SNHG5, 5′-CCTCTGGTCTCATCTGCATATTGACTTA-3′.
Cell viability
Cell viability was assessed via
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-trtrazolium bromide (MTT)
assay. 5×103 cells/well transfected with indicated
vector were seeded in a 96-well flat-bottomed plate for 24 h and
cultured in a normal medium. At 0, 24, 48, 72 and 96 h after
transfection, the MTT solution (5 mg/ml, 20 µl) was added to each
well. Following incubation for 4 h, the media was removed and 100
µl of DMSO was added to each well. The relative number of surviving
cells was assessed by measuring the optical density (OD) of cell
lysates at 560 nm. All assays were performed in triplicate.
Colony formation assay
Cells (500 cells/well) transfected with indicated
vector were plated in 6-well plates and incubated in RPMI-1640 with
10% FBS at 37°C. Two weeks later, the cells were fixed and stained
with 0.1% crystal violet. The number of visible colonies was
counted manually.
Flow cytometric analysis of apoptosis
and cell cycle distribution
Apoptosis was performed by using flow cytometric
analysis with Annexin V: FITC Apoptosis Detection kits (BD
Biosciences, San Diego, CA, USA), according to the manufacturer's
instructions. For cell cycle distribution, cells were collected
directly or 48 h after transfection and washed with ice-cold
phosphate-buffered saline (PBS), and fixed with 70% ethanol
overnight at −20°C. Fixed cells were rehydrated in PBS for 10 min
and incubated in RNase A (1 mg/ml) for 30 min at 37°C, then the
cells were subjected to PI/RNase staining followed by flow
cytometric analysis with a FACScan instrument and CellQuest
software (both from Becton-Dickinson, Mountain View, CA, USA) as
described (20).
Western bolt analysis and
antibodies
Total protein lysates were separated in 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and
were electrophoretically transferred to polyvinylidene difluoride
membranes (Roche Diagnostics, Indianapolis, IN, USA). Protein
loading was estimated by using mouse anti-GAPDH monoclonal
antibody. The membranes were blotted with 10% non-fat milk in TBST
for 2 h at room temperature, washed and then probed with the rabbit
anti-human p27 (1:2,000 dilution), CDK2 (1:2,000
dilution), activated caspase-3 (1:2,000 dilution), activated
caspase-9 (1:2,000 dilution), and GAPDH (1:3,000 dilution),
overnight at 4°C, followed by treatment with secondary antibody
conjugated to horseradish peroxidase for 2 h at room temperature.
The proteins were detected by using an enhanced chemiluminescence
system and then exposed to x-ray film. All antibodies were
purchased from Abcam (Cambridge, MA, USA).
Statistical analysis
Data were shown as the means ± standard error of at
least three independent experiments. The SPSS 17.0 software (SPSS
Inc., Chicago, IL, USA) was used for statistical analysis. Two
group comparisons were performed with a Student's t-test. Multiple
group comparisons were analyzed with one-way ANOVA. The Pearson
χ2 test was used to evaluate the relationship between
SNHG5 expression and clinical features. Kaplan-Meier method
was used to compare the overall survival curves between
high-SNHG5 and low-SNHG5 expression groups via the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference.
Results
SNHG5 was upregulated in BC tissues
and cell lines
To explore the biological function of SNHG5
in BC, we first measured the level of SNHG5 in BC tissues
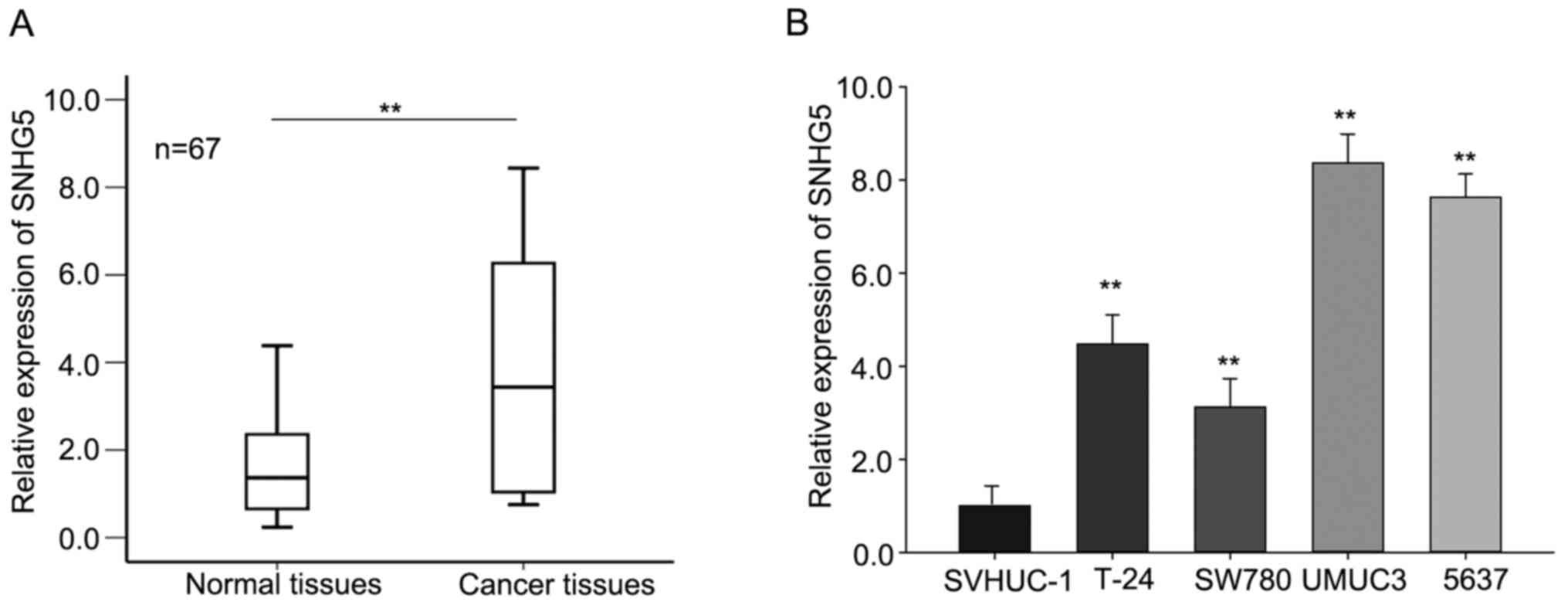
and corresponding normal tissues (n=67) by RT-qPCR. As shown in
Fig. 1A, SNHG5 was aberrantly
increased (P<0.01) in tumor tissues compared with that in
corresponding normal tissues. Furthermore, the levels of
SNHG5 in four bladder cancer cells SW780, UMUC3, 5637, T-24
and one normal urothelial cell line SVHUC-1 were assessed. As
presented in Fig. 1B, the level of
SNHG5 was significantly increased in four BC cell lines in
comparison to that in the normal urothelial cell line. And among
these cell lines, the expression level of SNHG5 in UMUC3 and
5637 cell was relative higher than that in SW780 and T-24 cells;
therefore, we chose UMUC3 and 5637 as the study object in the
following assays. These data revealed that SNHG5 may play a
pivotal role in BC progression.
Correlation of SNHG5 expression with
clinicopathological features and prognosis
Then we investigated the relationship between
SNHG5 expression and clinicopathological features in BC, the
mean expression level of SNHG5 in all BC tissues was used as
a cutoff value, and all samples were divided into two groups (high
expression group, n=36 vs. low expression group, n =31). As
illustrated in Table I, high
expression level of SNHG5 was significantly correlated with
larger tumor range (P=0.001), metastasis (P=0.013), lymph nodes
(P=0.001) and pathological stage (P=0.003), but it had no
significant correlation with age, sex and smoking (P>0.05).
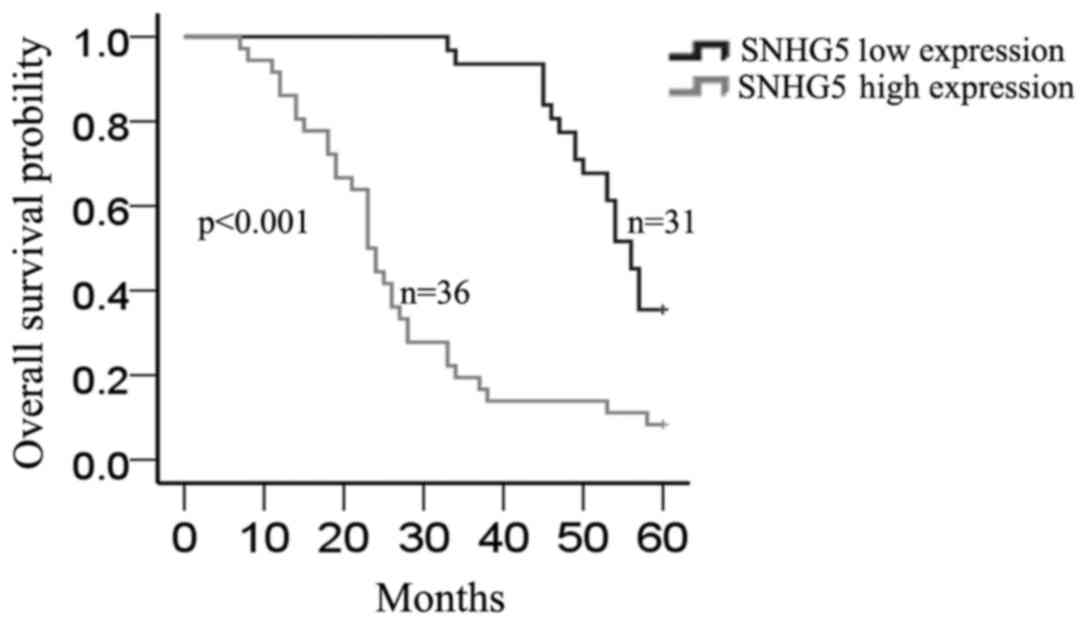
Furthermore, Kaplan-Meier method analysis (log-rank test) was
performed to determine the association between SNHG5
expression and overall survival of patients. As shown in Fig. 2, patients with high expression level
of SNHG5 had a significantly shorter overall survival than
those with low level of SNHG5 (P=0.000).
 | Table I.Correlation between SNHG5
expression and clinical features (n=67). |
Table I.
Correlation between SNHG5
expression and clinical features (n=67).
|
| SNHG5
expression |
|
|---|
|
|
|
|
|---|
| Variable | Low | High | P-value |
|---|
| Age (years) |
|
| 0.142 |
|
<60 | 9 | 17 |
|
|
≥60 | 22 | 19 |
|
| Sex |
|
| 0.088 |
|
Male | 19 | 14 |
|
|
Female | 12 | 22 |
|
| Smoking |
|
| 0.820 |
| No
smoking | 8 | 17 |
|
|
Smoking | 23 | 19 |
|
| Tumor range |
|
| 0.001 |
|
T1-T3 | 22 | 10 |
|
|
≥T4 | 9 | 26 |
|
| Metastasis |
|
| 0.013 |
|
Negative | 23 | 15 |
|
|
Positive | 8 | 21 |
|
| Lymph nodes |
|
| 0.001 |
|
Negative | 20 | 9 |
|
|
Positive | 11 | 27 |
|
| Pathological
stage |
|
| 0.003 |
|
<IV | 21 | 11 |
|
|
≥IV | 10 | 25 |
|
Silenced SNHG5 suppresses the
proliferation of BC cells
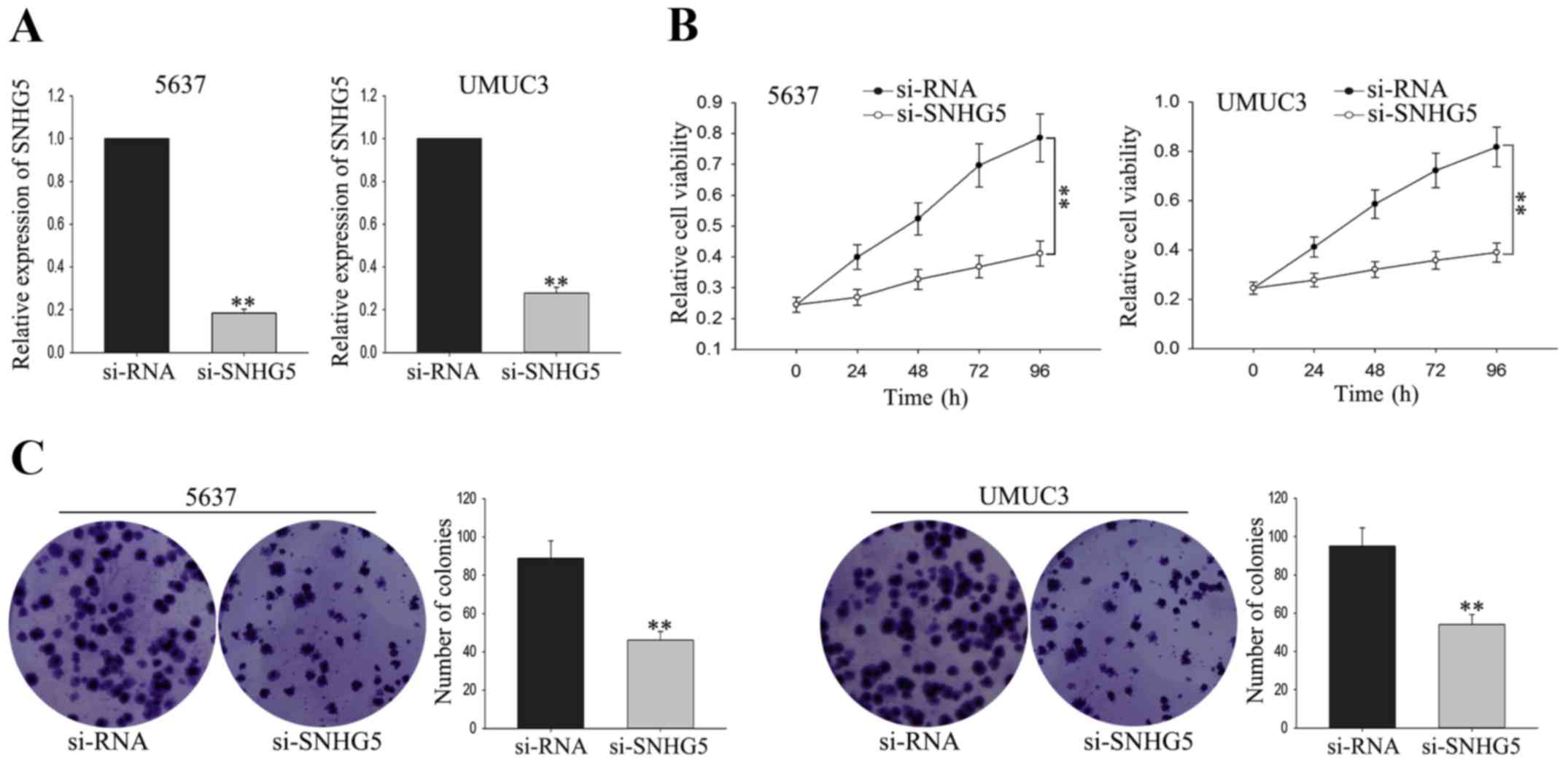
To investigate the biological function of
SNHG5 on the proliferation of BC cells, UMUC3 cell and 5637
cell were transfected with si-SNHG5 andsiRNA was used as as
negative control (NC). The satisfactory transfection efficiency was
obtained at 48 h post-transfection (Fig.
3A). MTT assay was performed to measure the function of
SNHG5 on cell viability. As shown in Fig. 3B, weakened proliferation ability was
obtained from UMUC3 cell and 5637 cell transfected with
si-SNHG5 in comparison to the NC-transfected cells.
Consistent with the results of MTT, colony formation assay revealed
a growth-inhibition effect mediated by si-SNHG5 (Fig. 3C). The findings indicated that
silenced SNHG5 could suppress the proliferation of BC
cells.
Silenced SNHG5 induces cell cycle
arrest at G1 phase and promotes cell apoptosis
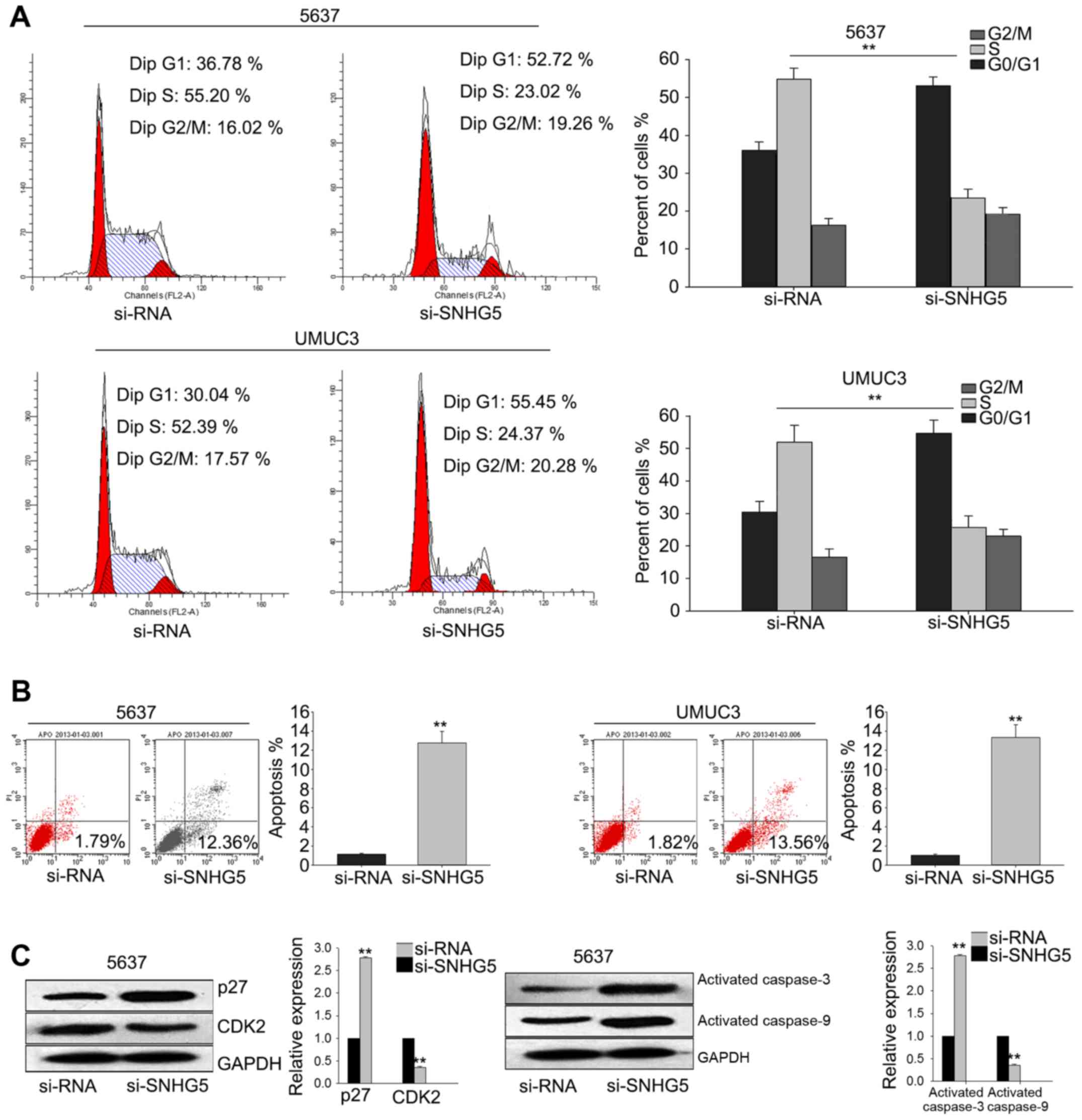
To investigate the underlying mechanism of
si-SNHG5-mediated growth-inhibition, flow cytometric
analysis of cell cycle distribution were performed. As illustrated
in Fig. 4A, silenced SNHG5 in
UMUC3 cell and 5637 cell obviously induced cell cycle arrest at G1.
And flow cytometric analysis of apoptosis revealed that silenced
SNHG5 significantly increased the apoptosis rate of UMUC3
cell and 5637 cell (Fig. 4B).
Furthermore, western blot assay demonstrated that the level of
cyclin-dependent kinase 2 (CDK2) was decreased while the
level of p27 was significantly increased; and
apoptosis-related proteins (activated caspase-3 and activated
caspase-9) were increased when SNHG5 was knockdown (Fig. 4C). These data indicated that
SNHG5 contributed to the proliferation ability of BC cells,
which might be attributed to its influence on cell cycle and
apoptosis.
The oncogenic function of SNHG5 is in
a p27-dependent manner
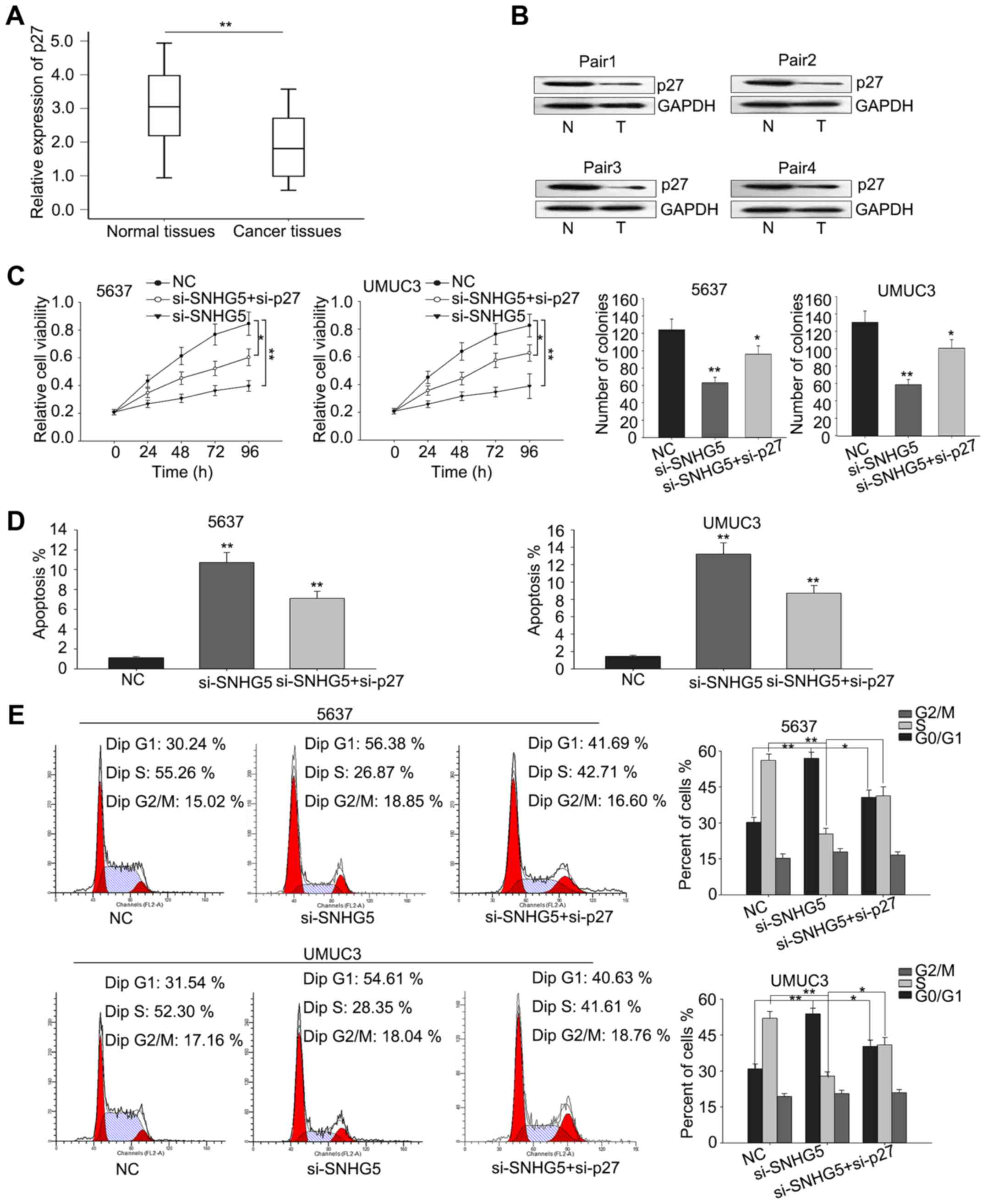
To determine whether p27 was involved in the
si-SNHG5-mediated growth inhibition, we first measured the
level of p27 in BC tissues and corresponding normal tissues
by RT-qPCR and then western blot analysis was performed to
determine the protein level of p27 in four pairs of cancer
tissues and normal tissues. As illustrated in Fig. 5A and B, the mRNA level of p27
was significantly downregulated in BC tissues and the protein level
was also obviously decreased in BC tissues. Furthermore, rescue
assays were performed to verify whether SNHG5 regulated BC
cell proliferation via silencing p27 expression. UMUC3 and
5637 cells were co-transfected with si-SNHG5 and
si-p27, and results from MTT and colony-formation assays
revealed that co-transfection with si-p27 could partially
abolish the si-SNHG5-mediated growth-inhibition (Fig. 5C). Additionally, flow cytometric
analyses of apoptosis and cell cycle distribution showed that
co-transfection with si-p27 could partially rescue the
si-SNHG5-mediated cell cycle arrest and increased apoptosis
rate (Fig. 5D and E). These results
indicated that the effect of SNHG5 on BC was partially
involved with targeting p27.
Discussion
Accumulating documents have demonstrated that the
dysregulation of lncRNAs are associated with tumorigenesis and
progression of malignant tumors (18–23). For
instance, Zhou et al demonstrated that downregulation of
lncRNA MEG3 mediated by DNMT3b contributed to nickel
malignant transformation of human bronchial epithelial cells via
modulating PHLPP1 transcription and HIF-1alpha
translation (11). Zhang et al
revealed that long non-coding RNA FTH1P3 facilitated oral
squamous cell carcinoma progression by acting as a molecular sponge
of miR-224-5p to modulate fizzled 5 expression (24). Cui et al uncovered that
upregulated lncRNA SNHG1 contributed to progression of non-small
cell lung cancer through inhibition of miR-101-3p and
activation of Wnt/β-catenin signaling pathway
(25). SNHG5 is anomalously
expressed in human gastric cancer and colorectal cancer (16,17).
However, its biological function in BC has not been
investigated.
In our present study, we demonstrated that the level
of SNHG5 was significantly increased in BC specimens and BC
cell lines. And analysis of the clinicopathological characteristics
of patients with BC revealed that high level of SNHG5 was
associated with tumor range, metastasis, lymph nodes, pathological
stage and poor prognosis. These results suggested that SNHG5
expression might be associated with the level of malignancy of BC,
and might be involved in the tumorigenesis and progression of BC.
It has been demonstrated that the effect of SNHG5 is related
with its biological function on cell proliferation (16,17).
Therefore, we explored the biological function of SNHG5 in
BC cells. We employed MTT and colony formation assays to measure
the function of SNHG5 on cell proliferation ability and
found that silenced SNHG5 significantly reduced cell growth
in BC cells. Then, flow cytometric analysis revealed that the
si-SNHG5-mediated growth-inhibition was attributed to its
influence on cell cycle and apoptosis rate.
It has been reported that SNHG5 acts as a
tumor suppressor in gastric cancer. Zhao et al demonstrated
that overexpressed SNHG5 significantly represses the
progression of gastric cancer (17,26).
While, Damas et al uncovered that high level of SNHG5
obviously promotes cell survival in colorectal cancer (16). Consistently, our study also presented
tumor-promoting function of SNHG5. As we known, the function
of lncRNAs has the cancer-specificity, means that the role of same
lncRNA was different in different cancer type. The different role
of SNHG5 in different cancer-type profoundly explants
tumor-type specificity of lncRNAs.
Collectively, our study presented that SNHG5
was significantly upregulated in BC tissues and cells lines. And
molecular experiments revealed that SNHG5 exerted an
oncogene functions in the genesis and progression of BC, which
provided a potential attractive therapeutic target for this
malignancy.
References
|
1
|
Li N, Yang L, Wang H, Yi T, Jia X, Chen C
and Xu P: miR-130a and miR-374a function as novel regulators of
cisplatin resistance in human ovarian cancer A2780 cells. PLoS One.
10:e01288862015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yu H, Xue Y, Wang P, Liu X, Ma J, Zheng J,
Li Z, Li Z, Cai H and Liu Y: Knockdown of long non-coding RNA XIST
increases blood-tumor barrier permeability and inhibits glioma
angiogenesis by targeting miR-137. Oncogenesis. 6:e3032017.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Su J, Zhang E, Han L, Yin D, Liu Z, He X,
Zhang Y, Lin F, Lin Q, Mao P, et al: Long noncoding RNA BLACAT1
indicates a poor prognosis of colorectal cancer and affects cell
proliferation by epigenetically silencing of p15. Cell Death Dis.
8:e26652017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu Y, Wang B, Zhang F, Wang A, Du X, Hu P,
Zhu Y and Fang Z: Long non-coding RNA CCAT2 is associated with poor
prognosis in hepatocellular carcinoma and promotes tumor metastasis
by regulating Snail2-mediated epithelial-mesenchymal transition.
Onco Targets Ther. 10:1191–1198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li W, Li H, Zhang L, Hu M, Li F, Deng J,
An M, Wu S, Ma R, Lu J and Zhou Y: Long non-coding RNA LINC00672
contributes p53-mediated gene suppression and promotes endometrial
cancer chemosensitivity. J Biol Chem. 292:5801–5813. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang JZ, Xu CL, Wu H and Shen SJ: lncRNA
SNHG12 promotes cell growth and inhibits cell apoptosis in
colorectal cancer cells. Braz J Med Biol Res. 50:e60792017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mang Y, Li L, Ran J, Zhang S, Liu J, Li L,
Chen Y, Liu J, Gao Y and Ren G: Long noncoding RNA NEAT1 promotes
cell proliferation and invasion by regulating hnRNP A2 expression
in hepatocellular carcinoma cells. Onco Targets Ther. 10:1003–1016.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li H, Jiang X and Niu X: Long non-coding
RNA reprogramming (ROR) promotes cell proliferation in colorectal
cancer via affecting P53. Med Sci Monit. 23:919–928. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Yang J, Sun L, Zhang L, Jiang Z,
Puri P, Gurley EC, Lai G, Tang Y, Huang Z, et al: The role of long
noncoding RNA H19 in gender disparity of cholestatic liver injury
in multidrug resistance 2 gene knockout mice. Hepatology.
66:869–884. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Idogawa M, Ohashi T, Sasaki Y, Nakase H
and Tokino T: Long non-coding RNA NEAT1 is a transcriptional target
of p53 and modulates p53-induced transactivation and
tumor-suppressor function. 140:1–2791. 2017.
|
|
11
|
Zhou C, Huang C, Wang J, Huang H, Li J,
Xie Q, Liu Y, Zhu J, Li Y, Zhang D, et al: LncRNA MEG3
downregulation mediated by DNMT3b contributes to nickel malignant
transformation of human bronchial epithelial cells via modulating
PHLPP1 transcription and HIF-1α translation. Oncogene.
36:3878–3889. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang TH, Chan CW, Fang JY, Shih YM, Liu
YW, Wang TV and Chen CY: 2-O-Methylmagnolol upregulates the long
non-coding RNA, GAS5, and enhances apoptosis in skin cancer cells.
Cell Death Dis. 8:e26382017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pei Z, Du X, Song Y, Fan L, Li F, Gao Y,
Wu R, Chen Y, Li W, Zhou H, et al: Down-regulation of lncRNA CASC2
promotes cell proliferation and metastasis of bladder cancer by
activation of the Wnt/β-catenin signaling pathway. Oncotarget.
8:18145–18153. 2017.PubMed/NCBI
|
|
14
|
Liu D, Li Y, Luo G, Xiao X, Tao D, Wu X,
Wang M, Huang C, Wang L, Zeng F and Jiang G: LncRNA SPRY4-IT1
sponges miR-101-3p to promote proliferation and metastasis of
bladder cancer cells through up-regulating EZH2. Cancer Lett.
388:281–291. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pan J, Li X, Wu W, Xue M, Hou H, Zhai W
and Chen W: Long non-coding RNA UCA1 promotes cisplatin/gemcitabine
resistance through CREB modulating miR-196a-5p in bladder cancer
cells. Cancer Lett. 382:64–76. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Damas ND, Marcatti M, Côme C, Christensen
LL, Nielsen MM, Baumgartner R, Gylling HM, Maglieri G, Rundsten CF,
Seemann SE, et al: SNHG5 promotes colorectal cancer cell
survival by counteracting STAU1-mediated mRNA destabilization. Nat
Commun. 7:138752016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao L, Guo H, Zhou B, Feng J, Li Y, Han
T, Liu L, Li L, Zhang S, Liu Y, et al: Long non-coding RNA
SNHG5 suppresses gastric cancer progression by trapping MTA2
in the cytosol. Oncogene. 35:5770–5780. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zou G, Liu T, Guo L, Huang Y, Feng Y,
Huang Q and Duan T: miR-145 modulates lncRNA-ROR and Sox2
expression to maintain human amniotic epithelial stem cell
pluripotency and β islet-like cell differentiation efficiency.
Gene. 591:48–57. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhu H, Lv Z, An C, Shi M, Pan W, Zhou L,
Yang W and Yang M: Onco-lncRNA HOTAIR and its functional genetic
variants in papillary thyroid carcinoma. Sci Rep. 6:319692016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zheng H and Min J: Role of long noncoding
RNA HOTAIR in the growth and apoptosis of osteosarcoma cell MG-63.
Biomed Res Int. 2016:57576412016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang XY, Zhang LX, Tian CJ, Tang XY, Zhao
LM, Guo YL, Cheng DJ, Chen XL, Ma LJ and Chen ZC: LncRNAs BCYRN1
promoted the proliferation and migration of rat airway smooth
muscle cells in asthma via upregulating the expression of transient
receptor potential 1. Am J Transl Res. 8:3409–3418. 2016.PubMed/NCBI
|
|
22
|
Zhang E, Han L, Yin D, He X, Hong L, Si X,
Qiu M, Xu T, De W, Xu L, et al: H3K27 acetylation activated-long
non-coding RNA CCAT1 affects cell proliferation and migration by
regulating SPRY4 and HOXB13 expression in esophageal squamous cell
carcinoma. Nucleic Acids Res. 45:3086–3101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xue M, Pang H, Li X, Li H, Pan J and Chen
W: Long non-coding RNA urothelial cancer-associated 1 promotes
bladder cancer cell migration and invasion by way of the
hsa-miR-145-ZEB1/2-FSCN1 pathway. Cancer Sci. 107:18–27. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang CZ: Long non-coding RNA FTH1P3
facilitates oral squamous cell carcinoma progression by acting as a
molecular sponge of miR-224-5p to modulate fizzled 5 expression.
Gene. 607:47–55. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cui Y, Zhang F, Zhu C, Geng L, Tian T and
Liu H: Upregulated lncRNA SNHG1 contributes to progression of
non-small cell lung cancer through inhibition of miR-101-3p and
activation of Wnt/β-catenin signaling pathway. Oncotarget.
8:17785–17794. 2017.PubMed/NCBI
|
|
26
|
Zhao L, Han T, Li Y, Sun J, Zhang S, Liu
Y, Shan B, Zheng D and Shi J: The lncRNA SNHG5/miR-32 axis
regulates gastric cancer cell proliferation and migration by
targeting KLF4. Faseb J. 31:893–903. 2017. View Article : Google Scholar : PubMed/NCBI
|