Introduction
Lung cancer is the most frequent type of primary
cancer for men in China, and it has the highest mortality rate for
any type of cancer worldwide (1).
Despite the lower incidence of lung cancer than breast cancer for
women, its mortality rate is also the highest for women (2). There are three major types of lung
cancer, including non-small cell lung cancer (NSCLC), small cell
lung cancer and carcinoid lung cancer. NSCLC is the most common
type of lung cancer, accounting for 85% of all lung cancer cases.
NSCLC can be further divided into three major histological
subtypes: Adenocarcinoma (ADC), squamous cell carcinoma (SCC) and
large cell carcinoma (LCC) (3,4). Patients
with lung cancer do not always present distinct symptoms, and are
commonly diagnosed at an advanced stage or after the primary cancer
has metastasized. This causes a poor prognosis and high mortality
rate for patients with lung cancer (4). The prevention and treatment of lung
cancer urgently requires improvement through further understanding
the molecular origins and development of the disease.
An increased exposure to smoking is associated with
an increased risk of developing NSCLC (5,6). However,
only 10–24% of smokers develop NSCLC, indicating that other
environmental and genetic factors also contribute to NSCLC
development (7,8). Mutations in the epidermal growth factor
receptor (EGFR) gene are common in NSCLC patients, with mutation
rates differing in males, females, smokers and non-smokers
(9,10). EGFR mutations affect the
EGF-EGFR-RAS-RAF signaling pathway, and are usually driver
mutations for NSCLC development (11). EGFR is, therefore, one of the most
important targets in NSCLC treatment. Small molecule tyrosine
kinase inhibitors (TKIs) that target EGFR, including gefitinib and
erlotinib, have significantly improved the overall survival rate of
patients with EGFR-activating mutations. The efficacy of TKI drugs
differs depending on the region of the EGFR kinase domain in which
the mutation is located (12,13). Among the NSCLC patients with EGFR
mutations, the overall response rate for treatment with gefitinib
is ~75%, with a progression-free survival time of 9–13 months
(14). Despite their low prevalence,
new targetable EGFR mutations may improve the treatment and
elongate the overall survival rate of patients with NSCLC.
In this study, EGFR mutations were detected in 354
patients with NSCLC of Chinese ethnicity by sequencing exons 18–21
from tumor samples. Further analysis was performed to determine the
association between EGFR mutations and other variables, including
age, gender, smoking status, and histology groups. A novel EGFR
mutation, M793K, was detected in 7 patients with potential
resistance to gefitinib.
Patients and methods
Patients
A total of 354 patients with NSCLC at the 307th
Hospital of the Chinese People's Liberation Army (Beijing, China)
and the General Hospital of the Chinese People's Liberation Army
(Beijing, China) were enrolled in this study between November 2012
and April 2016. Informed consent was obtained from all individual
participants included in the study, which was approved by the
Ethics Committee of the Affiliated Hospital of Academy of Military
Medical Sciences and the Ethics Committee of the General Hospital
of Chinese People's Liberation Army. Formalin-fixed and
paraffin-embedded (FFPE) tumor samples were prepared from primary
surgical or biopsy specimens from patient lung tissue. All FFPE
tissue specimens were identified by pathologists as primary
NSCLC.
Detection of EGFR mutations
Tumor genomic DNA from each FFPE sample was
extracted using the ALLPrep DNA/RNA FFPE kit (Qiagen, Inc.,
Valencia, CA, USA) according to the manufacturer's instructions.
The DNA samples were examined for purity and concentration, and
were diluted to a working concentration of 10 ng/µl. The detection
of EGFR mutations was performed using Sanger sequencing with the
ABI 3130 genetic analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). The primers used are listed in
Table I and were generated according
to the manufacturer's protocol (Tianyi Huiyuan LLC, Beijing,
China).
 | Table I.Primers for the detection of EGFR
mutations in patients with non-small cell lung cancer. |
Table I.
Primers for the detection of EGFR
mutations in patients with non-small cell lung cancer.
| Primer name | Primer sequence,
5′-3′ |
|---|
| EGFR(E18)-F |
GAAGCTCCCAACCAAGCTCT |
| EGFR(E18)-R |
CTCCCCACCAGACCATGAGA |
| EGFR(E19)-F |
TGCCAGTTAACGTCTTCCTTC |
| EGFR(E19)-R |
CCCACACAGCAAAGCAGAAA |
| EGFR(E20)-F |
CCAGGAAGCCTACGTGATGG |
| EGFR(E20)-R |
GACATAGTCCAGGAGGCAGC |
| EGFR(E21)-F |
GTGAAAACACCGCAGCATGT |
| EGFR(E21)-R |
GCCACCTCCTTACTTTGCCT |
Statistical analysis
χ2 tests for univariate analysis were
performed to investigate the association between EGFR mutation
frequency and clinicopathological features. Associations between
EGFR mutation status and sex, age, smoking history and
clinicopathological characteristics were further evaluated by
multivariate logistic regression analysis. The adjusted odds ratio
(OR) and 95% confidence interval (CI) were then identified.
Associations between sex, age, smoking history and pathology with
specific mutations were analyzed using an exact binomial test. A
two-sided P<0.05 was considered to indicate a statistically
significant difference. All statistical analysis was performed
using R (version 3.2.3; http://www.r-project.org/).
3D model protein building and
EGFR-gefitinib affinity estimation
The EGFR kinase domain with M793K mutation was
constructed using structure 2JIT from the Protein Data Bank
(originally the EGFR kinase domain including a T790M mutation; 3.1
Å, complete from 696–986) as a template using the SWISS-model
server (https://swissmodel.expasy.org/). The structure for
wild type EGFR domain was from 1M14 from the Protein Data Bank (2.6
Å, complete from 672–960). SWISS-DOCK (http://www.swissdock.ch/) was used to test the binding
energy of gefitinib (from ZINC; 19632614) to the 3 structures using
the CHARMM energy field method.
Results
Demographic profile of NSCLC
patients
The EGFR mutation status was analyzed in 354 NSCLC
patients. Of these patients, 43.22% were female and 56.78% were
male. The patient age ranged from 32–92 years, with a median age of
62. A total of 59.04% of the patients were ≥60 years old and 40.96%
<60 years old. Of the 354 patients, 50.85% had never smoked.
Pathological analysis revealed that 81.92% of the samples were from
ADC, 16.95% from SCC, and 1.13% from LCC (Table II).
 | Table II.Characteristics of 354 patients with
non-small cell lung cancer. |
Table II.
Characteristics of 354 patients with
non-small cell lung cancer.
| Characteristic | Patients, n (%) |
|---|
| Sex, n (%) |
|
| Male | 201 (56.78) |
|
Female | 153 (43.22) |
| Age, years |
|
| ≥60, n
(%) | 209 (59.04) |
| <60, n
(%) | 145 (40.96) |
| Median
(range) | 62 (32–92) |
| Smoking status, n
(%) |
|
|
Smokers | 174 (49.15) |
|
Non-smokers | 180 (50.85) |
| Histology type, n
(%) |
|
|
Adenocarcinoma | 290 (81.92) |
|
Squamous cell carcinoma | 60 (16.95) |
| Large
cell carcinoma | 4 (1.13) |
EGFR mutation distributions
EGFR mutations were identified in 48.02% (170)
patients, with a single mutation identified in the majority of
these patients (121 out of 170, 71.18%). Patients with more than
one EGFR mutation were relatively uncommon: 13.53% (23/170)
exhibited double mutations, 6.74% (11/170) triple mutations, 2.94%
(5/170) quadruple mutations, 1.76% (2/170) quintuple mutations,
1.76% (3/170) sextuple mutations, 1.18% (2/170) septuple mutations
and 1.18% (3/170) nonuple mutations. Of the 170 patients with EGFR
mutations, 97 patients had point mutations in exon 19 and 92 in
exon 21. The remaining mutations were located in exon 20 (58
patients) and exon 18 (38 patients). The most common mutation in
exon 19 was E746-A750del (30 patients), and the most common in exon
21 was L858R (36 patients). In exon 20, Q787Q and M793K were
detected in 11 and 7 patients, respectively (Table III).
 | Table III.Summary of epidermal growth factor
receptor exon 18–21 mutations in 354 non-small cell lung cancer
tissue samples. |
Table III.
Summary of epidermal growth factor
receptor exon 18–21 mutations in 354 non-small cell lung cancer
tissue samples.
| Exon | Mutation | Frequency (%) |
|---|
| 18 | V689M | 0.28 |
| 18 | P691S | 0.28 |
| 18 | P694L | 0.28 |
| 18 | Q701L | 0.56 |
| 18 | Q701R | 0.28 |
| 18 | Q701X | 0.28 |
| 18 | L703P | 0.28 |
| 18 | R705G | 0.56 |
| 18 | L707S | 0.56 |
| 18 | K708E | 0.56 |
| 18 | K708R | 0.28 |
| 18 | E709_710T>D | 0.28 |
| 18 | E709K | 0.28 |
| 18 | E711A | 0.28 |
| 18 | E711V | 0.28 |
| 18 | F712L | 0.28 |
| 18 | F712Q | 0.28 |
| 18 | K713H | 0.56 |
| 18 | I715V | 0.56 |
| 18 | V717A | 0.28 |
| 18 | G719A | 0.28 |
| 18 | G719D | 0.28 |
| 18 | G719V | 0.28 |
| 18 | S720P | 0.56 |
| 18 | S720T | 0.28 |
| 18 | G721S | 0.28 |
| 18 | F723C | 0.28 |
| 18 | F723L | 0.56 |
| 18 | K728M | 0.28 |
| 18 | K728T | 0.28 |
| 19 | L730I | 1.13 |
| 19 | I732V | 0.28 |
| 19 | E734A | 0.28 |
| 19 | E734R | 0.85 |
| 19 | G735D | 0.28 |
| 19 | E736V | 0.28 |
| 19 | K737R | 0.28 |
| 19 | V738L | 0.28 |
| 19 | I740F | 0.28 |
| 19 | P741R | 0.28 |
| 19 | P741S | 0.28 |
| 19 | V742A | 0.56 |
| 19 | I744V | 0.28 |
| 19 | K745R | 0.28 |
| 19 | E746_A750del | 8.47 |
| 19 | E746_E749del | 0.28 |
| 19 | E746_S752>A | 0.28 |
| 19 | E746_S752>D | 0.56 |
| 19 | E746_T751>A | 1.69 |
| 19 | E746_T751del | 0.28 |
| 19 | E746K | 0.56 |
| 19 | L747_750A>S | 0.28 |
| 19 | L747_A750del | 0.85 |
| 19 | L747_E749del | 0.56 |
| 19 | L747_P753>S | 1.98 |
| 19 | L747_T751del | 1.13 |
| 19 | R748G | 0.28 |
| 19 | E749G | 0.28 |
| 19 | S752_I759del | 0.85 |
| 19 | P753T | 0.28 |
| 19 | K754R | 0.28 |
| 19 | A755V | 0.28 |
| 19 | K757E | 0.28 |
| 19 | K757T | 0.28 |
| 19 | E758K | 0.28 |
| 19 | L760P | 0.85 |
| 19 | D761H | 0.28 |
| 19 | D761Q | 0.56 |
| 20 | A763T | 0.28 |
| 20 | V765A | 0.28 |
| 20 | A767S | 0.28 |
| 20 | A767V | 0.28 |
| 20 |
S768_V769insGGQ | 0.28 |
| 20 | S768I | 0.28 |
| 20 | S768R | 0.28 |
| 20 | D770_N771insG | 0.28 |
| 20 | D770_N771insTP | 0.28 |
| 20 | N771>KT | 0.28 |
| 20 | C775Y | 0.28 |
| 20 | R776A | 0.28 |
| 20 | R776H | 0.28 |
| 20 | L777C | 0.28 |
| 20 | L777P | 0.28 |
| 20 | L778Q | 0.28 |
| 20 | G779D | 0.28 |
| 20 | I780S | 0.28 |
| 20 | T783A | 0.28 |
| 20 | S784P | 0.28 |
| 20 | V786A | 0.28 |
| 20 | Q787Q | 3.11 |
| 20 | L788Q | 0.28 |
| 20 | I789F | 0.28 |
| 20 | T790M | 1.13 |
| 20 | Q791L | 0.28 |
| 20 | M793K | 1.98 |
| 20 | M793L | 0.28 |
| 20 | P794L | 0.28 |
| 20 | F795L | 0.56 |
| 20 | C797S | 0.28 |
| 20 | L799Q | 1.41 |
| 20 | V802A | 0.28 |
| 20 | L814M | 0.28 |
| 21 | M825T | 0.28 |
| 21 | N826H | 0.28 |
| 21 | N826S | 0.28 |
| 21 | E829D | 0.28 |
| 21 | V834A | 0.56 |
| 21 | V834M | 0.28 |
| 21 | R836H | 0.28 |
| 21 | A840T | 0.28 |
| 21 | V845A | 0.28 |
| 21 | K846I | 1.13 |
| 21 | Q849L | 0.28 |
| 21 | Q849R | 0.28 |
| 21 | H850Y | 0.28 |
| 21 | I853T | 0.28 |
| 21 | T854A | 0.28 |
| 21 | D855A | 0.28 |
| 21 | F856L | 1.13 |
| 21 | G857E | 0.28 |
| 21 | G857W | 0.56 |
| 21 | L858H | 0.28 |
| 21 | L858P | 0.28 |
| 21 | L858R | 10.17 |
| 21 | K860R | 0.28 |
| 21 | K860Tfs | 0.28 |
| 21 | L861Q | 0.56 |
| 21 | L862Q | 0.28 |
| 21 | G863C | 0.28 |
| 21 | G863S | 0.28 |
| 21 | G863V | 0.28 |
| 21 | E865G | 0.28 |
| 21 | E865K | 0.56 |
| 21 | E866G | 0.28 |
| 21 | E866R | 0.56 |
| 21 | E866X | 0.28 |
| 21 | K867D | 0.28 |
| 21 | K867I | 0.28 |
| 21 | K867N | 0.85 |
| 21 | E868D | 0.28 |
| 21 | E868V | 0.28 |
| 21 | H870Q | 0.28 |
| 21 | H870R | 0.28 |
| 21 | A871T | 0.28 |
| 21 | A871V | 0.28 |
| 21 | G873E | 0.28 |
Associations between EGFR mutation
occurrence and clinicopathological features
Patients were divided into two groups (with and
without EGFR mutations) for clinicopathological feature association
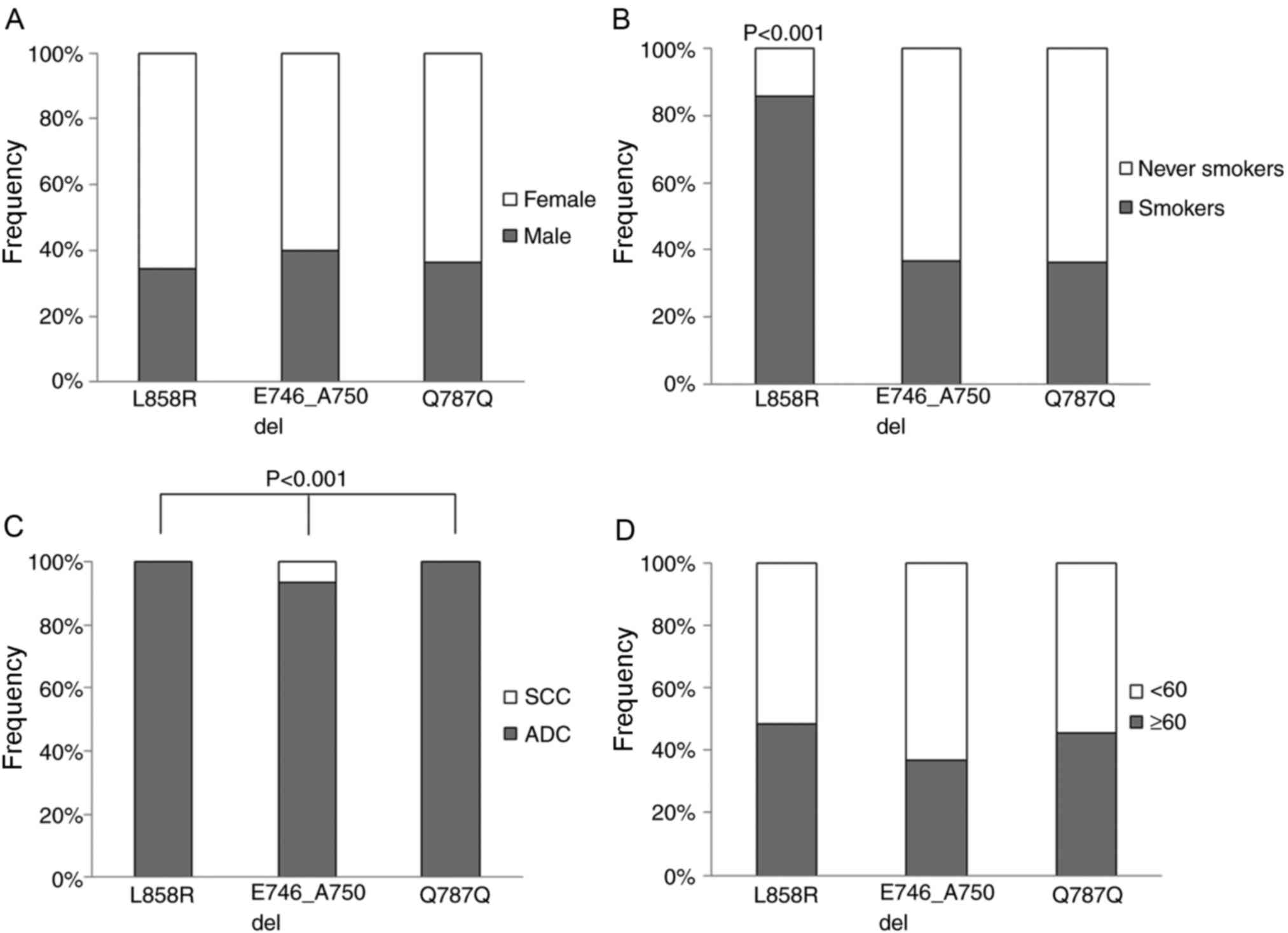
analysis (Fig. 1). Multivariate
logistic regression analysis revealed that EGFR mutations were more
frequently detected in females than in males (female vs. male;
60.13 vs. 38.81%; adjusted OR, 1.93; 95% CI, 1.07–3.51; P=0.029).
Patients ≥60 were more likely to have EGFR mutations than patients
<60 years old (<60 vs. ≥60; 58.62 vs. 40.67%; adjusted OR,
1.87; 95% CI, 1.20–2.92; P=0.006). ADC patients had a higher chance
of exhibiting EGFR mutations than non-ADC patients (ADC vs.
non-ADC; 52.76 vs. 26.56%; adjusted OR, 2.35; 95% CI, 1.28–4.50;
P=0.007). There was no significant difference between smokers and
non-smokers in the EGFR mutation rate (smokers vs. non-smokers;
55.56 vs. 40.23%; adjusted OR, 1.02; 95% CI, 0.56–1.82; P=0.952).
However, if only patients <60 are considered, EGFR mutation rate
in non-smokers was significantly higher than in smokers
(non-smokers vs. smokers; 66.27 vs. 48.39%; adjusted OR, 2.10; 95%
CI, 1.07–4.11; P=0.046). Similar results were identified between
non-smokers and smokers with ADC (non-smokers with ADC vs. smokers
with ADC; 58.64 vs. 45.31%; adjusted OR, 1.71; 95% CI, 1.07–2.73;
P=0.032; Table IV).
 | Table IV.Association of EGFR mutations with
the clinicopathological features of patients with non-small cell
lung cancer. |
Table IV.
Association of EGFR mutations with
the clinicopathological features of patients with non-small cell
lung cancer.
|
| EGFR status, n | Univariate | Multivariate |
|---|
|
|
|
|
|
|---|
| Variable | Mutant | Wild type | Crude OR (95%
CI) | P-value | Adjusted OR (95%
CI) | P-value |
|---|
| Sex |
|
|
|
|
|
|
|
Male | 78 | 123 | Ref. |
| Ref. |
|
|
Female | 92 | 61 | 2.378
(1.547–3.657) | <0.001 | 1.929
(1.072–3.506) | 0.029 |
| Age, years |
|
|
|
|
|
|
|
≥60 | 85 | 124 | Ref. |
| Ref. |
|
|
<60 | 85 | 60 | 2.067
(1.344–3.179) | 0.001 | 1.869
(1.200–2.923) | 0.006 |
| Smoking status |
|
|
|
|
|
|
|
Smoker | 70 | 104 | Ref. |
| Ref. |
|
|
Non-smoker | 100 | 80 | 1.857
(1.218–2.833) | 0.005 | 1.018
(0.564–1.815) | 0.952 |
| Pathology |
|
|
|
|
|
|
|
Non-ADC | 17 | 47 | Ref. |
| Ref. |
|
|
ADC | 153 | 137 | 3.088
(1.693–5.630) | <0.001 | 2.352
(1.275–4.497) | 0.007 |
| Smokers |
|
|
|
|
|
|
|
Male | 63 | 94 | Ref. |
|
|
|
|
Female | 7 | 10 | 1.044
(0.378–2.888) | 1 |
|
|
| Non-smokers |
|
|
|
|
|
|
|
Male | 15 | 29 | Ref. |
|
|
|
|
Female | 85 | 51 | 3.222
(1.579–6.577) | 0.002 |
|
|
| ADC |
|
|
|
|
|
|
|
Male | 65 | 82 | Ref. |
|
|
|
|
Female | 88 | 55 | 2.019
(1.264–3.225) | 0.005 |
|
|
| SCC |
|
|
|
|
|
|
|
Male | 12 | 38 | Ref. |
|
|
|
|
Female | 4 | 6 | 2.111
(0.509–8.751) | 0.514 |
|
|
| Non-smokers with
ADC |
|
|
|
|
|
|
|
Male | 14 | 20 | Ref. |
|
|
|
|
Female | 81 | 47 | 2.462
(1.138–5.327) | 0.033 |
|
|
| Male |
|
|
|
|
|
|
|
Smokers | 63 | 94 | Ref. |
|
|
|
|
Non-smokers | 15 | 29 | 0.772
(0.383–1.555) | 0.582 |
|
|
| Female |
|
|
|
|
|
|
|
Smokers | 7 | 10 | Ref. |
|
|
|
|
Non-smokers | 85 | 51 | 2.381
(0.853–6.645) | 0.153 |
|
|
| ≥60 |
|
|
|
|
|
|
|
Smokers | 40 | 72 | Ref. |
|
|
|
|
Non-smokers | 45 | 52 | 1.558
(0.894–2.715) | 0.154 |
|
|
| <60 |
|
|
|
|
|
|
|
Smokers | 30 | 32 | Ref. |
|
|
|
|
Non-smokers | 55 | 28 | 2.096
(1.067–4.114) | 0.046 |
|
|
| ADC |
|
|
|
|
|
|
|
Smokers | 58 | 70 | Ref. |
|
|
|
|
Non-smokers | 95 | 67 | 1.711
(1.072–2.732) | 0.032 |
|
|
| SCC |
|
|
|
|
|
|
|
Smokers | 11 | 31 | Ref. |
|
|
|
|
Non-smokers | 5 | 13 | 1.084
(0.314–3.745) | 1 |
|
|
| Males with ADC |
|
|
|
|
|
|
|
Smokers | 51 | 62 | Ref. |
|
|
|
|
Non-smokers | 14 | 20 | 0.851
(0.391–1.851) | 0.833 |
|
|
| Females with
ADC |
|
|
|
|
|
|
|
Smokers | 7 | 8 | Ref. |
|
|
|
|
Non-smokers | 81 | 47 | 1.970
(0.671–5.778) | 0.331 |
|
|
| ≥60 male |
|
|
|
|
|
|
|
Smokers | 36 | 63 | Ref. |
|
|
|
|
Non-smokers | 7 | 22 | 0.557
(0.217–1.431) | 0.316 |
|
|
| <60 female |
|
|
|
|
|
|
|
Smokers | 3 | 1 | Ref. |
|
|
|
|
Non-smokers | 47 | 21 | 0.746
(0.073–7.598) | 1 |
|
|
| ≥60 ADC |
|
|
|
|
|
|
|
Smokers | 33 | 45 | Ref. |
|
|
|
|
Non-smokers | 43 | 43 | 1.364
(0.736–2.527) | 0.407 |
|
|
| <60 ADC |
|
|
|
|
|
|
|
Smokers | 25 | 25 | Ref. |
|
|
|
|
Non-smokers | 52 | 24 | 2.167
(1.038–4.522) | 0.059 |
|
|
Association between specific mutations
and clinicopathological features
The association between specific mutations and
clinicopathological features was analyzed using an exact binomial
test. Between patients with ADC and SCC, the frequencies of
E746_A750del, Q787Q and L858R mutations were significantly
different (P<0.001). Furthermore, the L858R mutation was
significantly more frequent in smokers than in non-smokers. No
significant associations were identified between specific mutations
and other clinicopathological features (Fig. 1).
Analysis of the M793K mutation
The EGFR mutation M793K was detected in 7 out of 354
patients with NSCLC, including five smokers. No KRAS mutations or
EGFR drug-resistance mutations, including T790M or C797S, were
detected in these 7 patients. However, the follow-up information
for these patients demonstrated that they responded poorly to
treatment with gefitinib. Similar to the T790M mutation, M793K also
occurs in the inhibitor-binding cleft between the N-lobe and C-lobe
of the EGFR kinase domain, indicating that it is likely to be a
novel drug-resistance mutation.
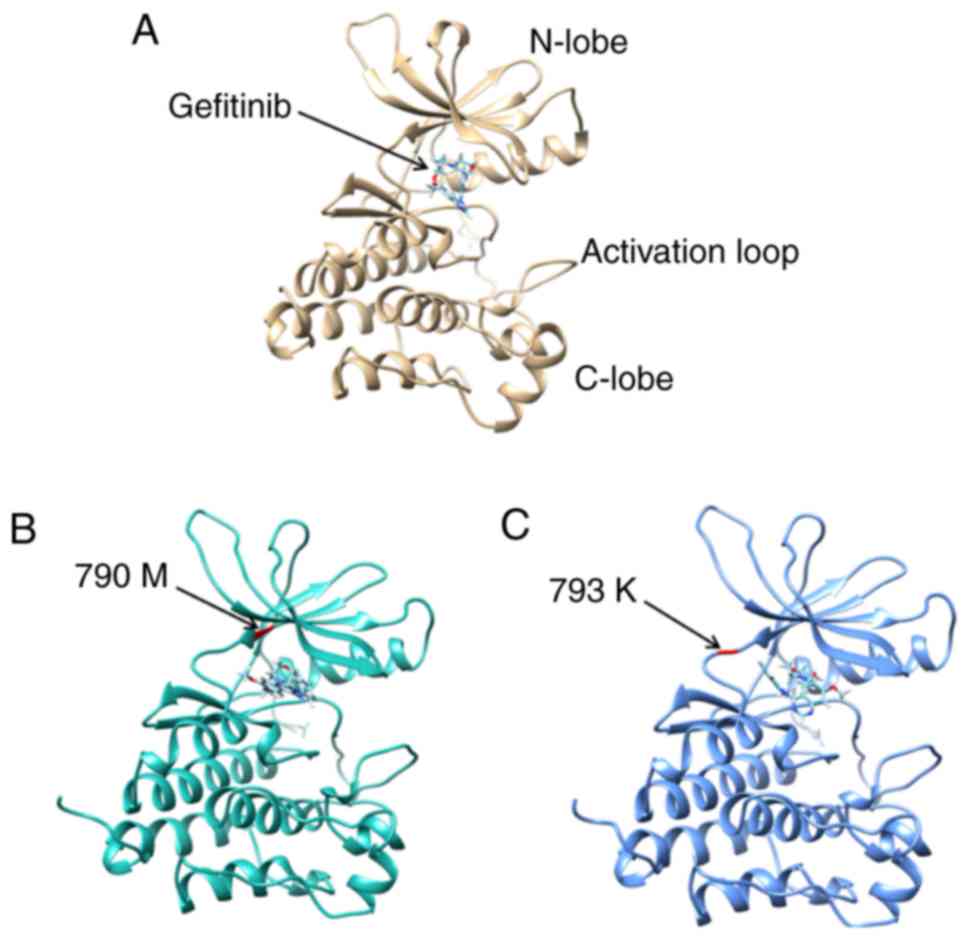
Gefitinib was always identified in the cleft between
the N-lobe and C-lobe in the 3D models of wild type EGFR, EGFR with
T790M and EGFR with M793K (Fig. 2). A
total of 256 binding poses for wild type EGFR, 256 for EGFR with
T790M and 252 for EGFR with M793K were determined. The full fitness
scores between gefitinib and EGFR ranged from −2196.87 to −2160.79
kcal/mol for wild type EGFR, from −1791.71 to −1756.65 kcal/mol for
M793K-mutated EGFR, and from −1805.53 to-1759.77 kcal/mol for
T790M-mutated EGFR (Table V). The
reduced affinity between gefitinib and EGFR with M793K may be
reflective of the gefitinib drug resistance of patients with
M793K.
 | Table V.Full fitness score between gefitinib
and EGFR wild type and EGFR T790M and M793K mutants, as predicted
by Swiss-Dock. |
Table V.
Full fitness score between gefitinib
and EGFR wild type and EGFR T790M and M793K mutants, as predicted
by Swiss-Dock.
| EGFR | Full fitness score
(kcal/mol) |
|---|
| Wild type | (−2,196.87;
−2,160.79) |
| T790M | (−1,805.53;
−1,759.77) |
| M793K | (−1,791.71;
−1,756.65) |
Discussion
EGFR mutations were detected in 170 out of the 354
patients with NSCLC of Chinese ethnicity by Sanger sequencing of
EGFR exons 18–21. Associations between EGFR mutation occurrence and
patient clinicopathological factors were further analyzed. A new
EGFR mutation, M793K, was detected and predicted to be a TKI
resistance mutation.
Of the 354 NSCLC patients, 43.22% were female and
56.78% were male, 50.85% patients were non-smokers and 49.15% were
smokers. Pathological slides were collected and diagnosed for all
patients, with 81.92% samples identified as ADCs, 16.95% as SCCs
and 1.13% as LCCs. It is generally accepted that the median age of
patients with NSCLC worldwide is 71 (15). The lower median age in this study (62
years), supports the indication that the median age of patients
with NSCLC in Asia has lowered (9,16,17).
Less than 30% patients had multiple EGFR mutations
in the present study. The most common mutations were L858R (36
patients) and E746-A750del (30 patients), which is consistent with
previous reports (9,10). Multivariate logistic regression
analysis revealed that EGFR mutations happened more frequently in
females (P=0.029), older patients (≥60 years old; P=0.006) and ADC
(P=0.007), which consolidates the results of previous reports
(18–21). There was no significant difference in
the EGFR mutation rate between smokers and non-smokers (P=0.952).
However, in patients <60 years old, univariate analysis revealed
that non-smokers are much more likely to have EGFR mutations than
smokers (P=0.046). This was also true for patients with ADC,
whereas non-smokers exhibited a higher EGFR mutation rate than
smokers (P=0.032).
A significant association was observed between
histology type and specific EGFR mutation rates as the frequencies
of E746_A750del, Q787Q and L858R mutations were significantly
different between patients with ADC and patients with SCC
(P<0.001). Smokers were also more likely to have the L858R
mutation than non-smokers.
The EGFR gene encodes a receptor protein that
dimerizes upon ligand binding, activating tyrosine kinase activity
and receptor phosphorylation. The kinase activity of EGFR can be
increased by mutations to EGFR, inducing the hyperactivation of
downstream pro-survival signaling pathways (22). Initial studies on the TKIs gefitinib
(Iressa) and erlotinib (Tarceva) demonstrated biological and
clinical significance in a subset of lung cancers (23). Further investigation demonstrated that
patients with advanced NSCLC and EGFR-activating mutations
(particularly exon 19 deletions, L858R in exon 21, and G719X in
exon 18) demonstrated the highest response rates to these TKIs
(24). However, over 6–12 months of
treatment, the majority of tumor cells gained resistance to
EGFR-TKIs through a secondary mutation. Previous studies reported
that T790M occurs in 50% of EGFR-mutated patients with TKI
resistance (25,26) and it is considered a TKI acquired
resistance mutation (27–29). In this study, a new mutation, M793K,
was detected in 7 out of 354 NSCLC patients. These 7 patients
gained drug resistance following their treatment with gefitinib,
and no other previously identified drug resistance mutations were
detected in these patients.
The docking analysis of gefitinib and EGFR kinase
domain with/without T790M or M793K mutations demonstrated that
gefitinib was always identified in the cleft between the N-lobe and
C-lobe of the EGFR kinase domain, which is also the location of
M793K. TKIs form direct H-bonds with M793 and T790 (30–33).
Computational simulation and prediction methodologies have
predicted that M793 forms a high proportion of these H-bonds with
inhibitors (34) and M793K was
previously predicted to be associated with drug resistance
(resistance score, 0.057) (35). The
full fitness score between gefitinib and the EGFR M793K structure
was higher than that between gefitinib and the wild type EGFR
structure, and between gefitinib and the EGFR T790M structure.
These findings indicate that M793K reduces the binding affinity
between gefitinib and EGFR and may induce the development of the
resistance to TKI treatment.
In coclusion, the present study presents a complete
picture of exon 18–21 EGFR mutations based on 354 Chinese patients
with NSCLC, and investigates the association between EGFR mutations
with sex, age, smoking history, and histology. The EGFR M793K
mutation was identified for the first time in NSCLC patients and
may have been associated with resistance to TKI treatment. This
finding laid the basis for the further investigation of the
association between M793K mutation and TKI treatment clinical
outcomes.
Acknowledgements
The authors wish to thank Mr. Jinglei Bi (Vishuo
MedTech Ltd., Beijing, China) and Dr Dianyun Wu (Vishuo MedTech
Ltd.) for their technical assistance. The present study was
financially supported by Vishuo MedTech Ltd.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Travis WD, Brambilla E, Nicholson AG,
Yatabe Y, Austin JHM, Beasley MB, Chirieac LR, Dacic S, Duhig E,
Flieder DB, et al: The 2015 world health organization
classification of lung tumors: Impact of genetic, clinical and
radiologic advances since the 2004 classification. J Thorac Oncol.
10:1243–1260. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hecht SS: Tobacco carcinogens, their
biomarkers and tobacco-induced cancer. Nat Rev Cancer. 3:733–744.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Thun MJ, Hannan LM, Adams-Campbell LL,
Boffetta P, Buring JE, Feskanich D, Flanders WD, Jee SH, Katanoda
K, Kolonel LN, et al: Lung cancer occurrence in never-smokers: An
analysis of 13 cohorts and 22 cancer registry studies. PLoS Med.
5:e1852008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Thun MJ, Henley SJ and Calle EE: Tobacco
use and cancer: An epidemiologic perspective for geneticists.
Oncogene. 21:7307–7325. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun S, Schiller JH and Gazdar AF: Lung
cancer in never smokers-a different disease. Nat Rev Cancer.
7:778–790. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li S, Li L, Zhu Y, Huang C, Qin Y, Liu H,
Ren-Heidenreich L, Shi B, Ren H, Chu X, et al: Coexistence of EGFR
with KRAS, or BRAF, or PIK3CA somatic mutations in lung cancer: A
comprehensive mutation profiling from 5125 Chinese cohorts. Br J
Cancer. 110:2812–2820. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cai X, Sheng J, Tang C, Nandakumar V, Ye
H, Ji H, Tang H, Qin Y, Guan H, Lou F, et al: Frequent mutations in
EGFR, KRAS and TP53 genes in human lung cancer tumors detected by
ion torrent DNA sequencing. PLoS One. 9:e952282014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang Y, Wang M, Wang Q, Geng Z and Sun M:
Incidence and risk of infections associated with EGFR-TKIs in
advanced non-small-cell lung cancer: A systematic review and
meta-analysis of randomized controlled trials. Oncotarget.
8:29406–29415. 2017.PubMed/NCBI
|
|
12
|
Pao W, Miller V, Zakowski M, Doherty J,
Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al:
EGF receptor gene mutations are common in lung cancers from ‘never
smokers’ and are associated with sensitivity of tumors to gefitinib
and erlotinib. Proc Natl Acad Sci USA. 101:13306–13311. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Toyooka S, Kiura K and Mitsudomi T: EGFR
mutation and response of lung cancer to gefitinib. N Engl J Med.
352:21362005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mitsudomi T, Morita S, Yatabe Y, Negoro S,
Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et
al: Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): An open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Subramanian J, Morgensztern D, Goodgame B,
Baggstrom MQ, Gao F, Piccirillo J and Govindan R: Distinctive
characteristics of non-small cell lung cancer (NSCLC) in the young:
A surveillance, epidemiology, and end results (SEER) analysis. J
Thorac Oncol. 5:23–28. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xu J, He J, Yang H, Luo X, Liang Z, Chen
J, Cai Z, Xu J and Ren-Heidenreich L: Somatic mutation analysis of
EGFR, KRAS, BRAF and PIK3CA in 861 patients with non-small cell
lung cancer. Cancer Biomark. 10:63–69. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang L, Yang H, Zhao Y, Liu W, Wu S, He
J, Luo X, Zhu Z, Xu J, Zhou Q and Ren-Heidenreich L: Detection of
EGFR somatic mutations in non-small cell lung cancer (NSCLC) using
a novel mutant-enriched liquidchip (MEL) technology. Curr Drug
Metab. 13:1007–1011. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim TJ, Park CK, Yeo CD, Park K, Rhee CK,
Kim J, Kim SJ, Lee SH, Lee KY and Yoon HK: Simultaneous diagnostic
platform of genotyping EGFR, KRAS, and ALK in 510 Korean patients
with non-small-cell lung cancer highlights significantly higher ALK
rearrangement rate in advanced stage. J Surg Oncol. 110:245–251.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tokumo M, Toyooka S, Kiura K, Shigematsu
H, Tomii K, Aoe M, Ichimura K, Tsuda T, Yano M, Tsukuda K, et al:
The relationship between epidermal growth factor receptor mutations
and clinicopathologic features in non-small cell lung cancers. Clin
Cancer Res. 11:1167–1173. 2005.PubMed/NCBI
|
|
20
|
Sahoo R, Harini V, Babu V, Okaly Patil GV,
Rao S, Nargund A, Venkataswamy E, Rao R and Kumar BS: Screening for
EGFR mutations in lung cancer, a report from India. Lung Cancer.
73:316–319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xie G, Xie F, Wu P, Yuan X, Ma Y, Xu Y, Li
L, Xu L, Yang M and Shen L: The mutation rates of EGFR in non-small
cell lung cancer and KRAS in colorectal cancer of Chinese patients
as detected by pyrosequencing using a novel dispensation order. J
Exp Clin Cancer Res. 34:632015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ullrich A and Schlessinger J: Signal
transduction by receptors with tyrosine kinase activity. Cell.
61:203–212. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sequist LV, Bell DW, Lynch TJ and Haber
DA: Molecular predictors of response to epidermal growth factor
receptor antagonists in non-small-cell lung cancer. J Clin Oncol.
25:587–595. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oxnard GR, Arcila ME, Chmielecki J,
Ladanyi M, Miller VA and Pao W: New strategies in overcoming
acquired resistance to epidermal growth factor receptor tyrosine
kinase inhibitors in lung cancer. Clin Cancer Res. 17:5530–5537.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ohashi K, Sequist LV, Arcila ME, Moran T,
Chmielecki J, Lin YL, Pan Y, Wang L, de Stanchina E, Shien K, et
al: Lung cancers with acquired resistance to EGFR inhibitors
occasionally harbor BRAF gene mutations but lack mutations in KRAS,
NRAS, or MEK1. Proc Natl Acad Sci USA. 109:E2127–E2133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Engelman JA and Jänne PA: Mechanisms of
acquired resistance to epidermal growth factor receptor tyrosine
kinase inhibitors in non-small cell lung cancer. Clin Cancer Res.
14:2895–2899. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Engelman JA, Mukohara T, Zejnullahu K,
Lifshits E, Borrás AM, Gale CM, Naumov GN, Yeap BY, Jarrell E, Sun
J, et al: Allelic dilution obscures detection of a biologically
significant resistance mutation in EGFR-amplified lung cancer. J
Clin Invest. 116:2695–2706. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tam IY, Leung EL, Tin VP, Chua DT, Sihoe
AD, Cheng LC, Chung LP and Wong MP: Double EGFR mutants containing
rare EGFR mutant types show reduced in vitro response to gefitinib
compared with common activating missense mutations. Mol Cancer
Ther. 8:2142–2151. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stamos J, Sliwkowski MX and Eigenbrot C:
Structure of the epidermal growth factor receptor kinase domain
alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol
Chem. 277:46265–46272. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Michalczyk A, Klüter S, Rode HB, Simard
JR, Grütter C, Rabiller M and Rauh D: Structural insights into how
irreversible inhibitors can overcome drug resistance in EGFR.
Bioorg Med Chem. 16:3482–3488. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cross DA, Ashton SE, Ghiorghiu S, Eberlein
C, Nebhan CA, Spitzler PJ, Orme JP, Finlay MR, Ward RA, Mellor MJ,
et al: AZD9291, an irreversible EGFR TKI, overcomes T790M-mediated
resistance to EGFR inhibitors in lung cancer. Cancer Discov.
4:1046–1061. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu B, Bernard B and Wu JH: Impact of EGFR
point mutations on the sensitivity to gefitinib: Insights from
comparative structural analyses and molecular dynamics simulations.
Proteins. 65:331–346. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Balius TE and Rizzo RC: Quantitative
prediction of fold resistance for inhibitors of EGFR. Biochemistry.
48:8435–8448. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Martínez-Jiménez F, Overington JP,
Al-Lazikani B and Marti-Renom MA: Rationally designed drug blending
as a mechanism to overcome drug resistance in cancer: An
application in EGFR. http://sgt.cnag.cat/www/presentations/files/slides/20150709_Fran_Cancer_poster.pdfMarch
27–2017
|