Introduction
Breast cancer, a highly heterogeneous disease, is
one of the most commonly diagnosed types of cancer and the second
most common cause of cancer-associated mortality in women globally
(1,2).
Breast cancer metastasizes to the distant organs and an increased
number of patients with breast cancer with earlier stages survive
their disease for at least 5 years compared with patients diagnosed
with cancer metastasis (3). Detection
of breast cancer at an early stage is important to improve breast
cancer prognosis and reduce the mortality of this disease (4). At present, mammography and ultrasound
have been successfully used in the screening of early-stage breast
cancer (5). However, novel
non-invasive biomarkers are required to optimize individual
treatment.
Small non-coding microRNAs (miRNAs/miRs) are
epigenetic regulators that mediate specific cellular mRNA
degradation processes and inhibit translation to modulate gene
expression post-transcriptionally (6). Dysregulation of miRNA expression is
involved in the initiation and progression, including metastasis,
proliferation, chemoresistance, and recurrence of breast, prostate,
lung and colon cancer (5–8). Increasing evidence has indicated that
miRNAs may serve as tumor oncogenes or anti-oncogenes of types of
human cancer, including breast cancer (9–11). Several
studies have demonstrated that serum and plasma miRNAs (circulating
miRNAs) present potential as novel non-invasive biomarkers for the
early diagnosis of various types of cancer (12–14). In
breast cancer, studies have suggested that the various breast
cancer subtypes exhibit different molecular miRNA signatures
(14,15). miRNAs have been identified to be
stable in whole blood, plasma, serum, saliva and urine, and they
have been proposed as potentially accessible breast cancer
biomarkers for clinical use (10,14,16,17).
miRNA expression profiling of breast cancer has identified
signatures associated with diagnosis, staging, progression,
prognosis and response to treatment (10,14,15). For
example, several miRNA-based signatures have been identified with
notably high predictive values including a 3-miRNA (miR-199a,
miR-29c, and miR-424) signature with an area under the curve (AUC)
of 0.888 and a 7-miRNA panel with an AUC of 0.914 in sera samples
of patients with breast cancer (18,19).
miR-96 belongs to the same family as miR-183, and is
a well-recognized oncogenic miRNA in a variety of types of cancer,
including hepatocellular carcinoma, prostate cancer,
medulloblastoma, pancreatic cancer, colorectal carcinoma and breast
cancer (20–24). A previous study demonstrated that the
upregulation of miRNA-96 targeting Forkhead box protein (FOX)O3a
served a notable function in the pro-proliferation effect of breast
cancer and hepatoma cells (25,26). In
addition, miR-96 is overexpressed in papillary thyroid carcinoma
and prostate cancer cells, and functions as an oncogene through
repressing FOXO1 expression (27,28). In
breast cancer, the overexpression of miR-96 was also demonstrated
to induce the migration of breast cancer cells by downregulating
transcriptional factors FOXO3a, and FOXO1 (29). However, the expression of serum miR-96
in the metastasis and prognosis of breast cancer has not been fully
explored and its regulation mechanisms remain unclear. Metastasis
suppressor-1 (MTSS1) is a metastasis suppressor gene which was
first identified in non-metastatic bladder cancer cell lines
(30). Functional and mechanism
analysis suggested that MTSS1 protein may be associated with cancer
progression or tumor metastasis in a variety of organ sites
(31,32). Whether miR-96 targets MTSS1
dysregulation function in breast cancer metastasis remains
unknown.
In the present study, the expression of serum miR-96
was confirmed in healthy control, benign and malignant breast
cancer samples. Then, the effect of miR-96 expression on
chemotherapy prognosis was examined. The migration ability of
induced miR-96 overexpression in breast cancer cells was also
analyzed. Finally, the mechanism of direct targeting of MTSS1 by
miR-96 in breast cancer was explored. These results identified the
function and mechanism of miR-96 in the breast cancer progression,
and prognosis. Serum miR-96 may be used as a novel therapeutic and
prognostic marker in breast cancer.
Materials and methods
Cell culture and miR-96
transfection
The human breast cancer MCF-7 and MDA-MB-231 cell
lines were obtained from the Shanghai Institute of Biochemistry and
Cell Biology of the Shanghai Institutes of Biological Sciences,
Chinese Academy of Sciences (Shanghai, China). The cell lines were
maintained in plastic flasks as adherent monolayers in high glucose
Eagle's minimal essential medium (H-DMEM) supplemented with 10%
fetal bovine serum (FBS; Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and incubated at 37°C in a humidified
atmosphere supplemented with 5% CO2.
The cells were transfected with an miR-96 mimics and
inhibitors by using Lipofectamine™ 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). In order to induce overexpression of miR-96,
MCF-7 and MDA-MB-231 were treated with different concentrations of
miR-96 mimics and inhibitors (12.5, 25 and 50 nM) (Shanghai
GenePharma Biotechnology, Co., Ltd., Shanghai, China). The
sequences of the miR-96 mimics were designed as follows:
5′-UUUGGCACUAGCACAUUUUUGCU-3′ (sense) and
5′-CAAAAAUGUGCUAGUGCCAAAUU-3′ (antisense). The sequence of the
miR-96 inhibitors was designed as follows:
5′-AGCAAAAAUGUGCUAGUGCCAAA-3′. The cells were harvested 48 h
post-transfection, and total RNA was extracted for reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis. Cell lysates were prepared for western blot analysis.
Clinical tissue samples
Breast cancer tissues and adjacent control were
collected from 5 female patients (aged from 36 to 65 years, with an
average age 52.6 years) with breast cancer from the Nantong Tumor
Hospital (Nantong, China) who had undergone total or partial
mastectomy surgery between March 2015 and May 2016. The breast
cancer tissues and adjacent non-cancerous tissues were
histologically confirmed. All clinical procedures followed the
protocols approved by the Ethical Committee of Nantong Tumor
Hospital, and the methods were performed in accordance with the
approved guidelines. Written informed consent was obtained from all
patients prior to sample collection.
RNA extraction and miRNA RT-qPCR
assay
Blood samples were collected from 118 female
patients (aged from 18 to 76 years, with an average age 56 years)
with breast cancer from the Nantong Tumor Hospital (Nantong, China)
who had undergone total or partial mastectomy surgery between
December 2014 and May 2016. Informed written consent was provided
by all the patients. Each sample was centrifuged to collect sera at
1,500 × g, 4°C for 10 min and stored at −70°C for RNA extraction.
miRNA was isolated from 500 µl serum using miRNAeasy kit (Qiagen,
Inc., Valencia, CA, USA) according to the manufacturer's protocol.
RNA concentration was determined by NanoDrop ND1000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc., Pittsburgh, PA, USA). Following treatment with DNase (Life
Technologies), the RNA was eluted with 50 µl RNAse-free water.
Serum miR-96 expression was quantified using the
miScript SYBR Green PCR Kit (Qiagen GmbH, Hilden, Germany) and an
ABI Prism 7900HT Real Time PCR System. The thermocycling conditions
were as follows: Preheating at 95°C (for 5 min), followed by 40
cycles at 95°C (for 30 sec) and 60°C (for 45 sec). The primer for
miR-96 was forward, 5′-GCCCGCTTTGGCACTAGCACATT-3′ and reverse,
5′-GTGCAGGGTCCGAGGT-3′. U6 small nuclear RNA was used as an
internal control and the primer was forward,
5′-TGCGGGTGCTCGCTTCGGCAGC-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′. All the serum samples were analyzed in
triplicate. The relative expression of the miR-96 was calculated
using the comparative cycle threshold (2−ΔΔCq) method
and normalized to U6 (33).
Western blot analysis
Cultured cells were harvested and lysed in RIPA
buffer [10 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EGTA, 0.1% SDS]
supplemented with proteinase inhibitors (1 mM NaF, 1 mM Na3VO4, 1
mM PMSF, 1 mg/ml aprotinin and 1 mg/ml leupeptin; cat no. E211-02;
Vazyme, Piscataway, NJ, USA). Protein concentration was determined
using an BCA assay. Equal quantities of protein (20 µg) were loaded
and separated with 10% SDS-PAGE, transferred to methanol
pre-activated polyvinylidene fluoride membranes and blocked in 5%
(w/v) non-fat milk at room temperature for 1 h. Western blot
analysis was performed to detect the expression of various proteins
using the following primary antibodies: MTSS1 (1:1,000; cat no.
SC-101204; Santa Cruz Biotechnology Inc., Dallas, TX, USA),
E-cadherin (1:2,000; cat no. SC-7870; Santa Cruz Biotechnology
Inc.), N-cadherin (1:1,500; cat no. SC-1502; Santa Cruz
Biotechnology Inc.), vimentin (1:2,000; cat no. BS-1491; Biogot
Technology Co., Ltd., Nanjing, China) and GAPDH (1:3,000; cat no.
KC-5G4; Zhejiang Kangchen Biotech Co., Ltd., Wuhan, China).
Following incubation with the primary antibodies overnight at 4°C,
membranes were washed 3 times with tris-buffered saline containing
0.05% Tween-20 and incubated with horseradish peroxidase-conjugated
anti-rabbit (1:5,000; cat no. CW0103S; CWBIO Biotech Inc., Beijing,
China) or anti-mouse (1:5,000; cat no. CW0102S; CWBIO Biotech Inc.)
secondary antibodies at room temperature for 1 h. Proteins were
detected with an ECL enhanced chemiluminescence detection system
(GE Healthcare, Chicago, IL, USA).
Wound healing assay
MDA-MB-231, and MCF-7 cells (2×105) were
seeded in six-well plates and cultured in H-DMEM with 10% FBS at
37°C to reach 95% confluence. A wound, 0.35 mm in width, was
generated by scraping with a 10 µl pipette tip. Images of the cells
in the wounded monolayer were captured at 24, 48 and 72 h, and cell
migration was assessed by measuring the gap sizes at five fields
under a light microscope (Ti; Nikon Corporation, Tokyo, Japan) at
magnification of ×100.
Transwell migration assay
A Transwell migration assay was performed using a
specialized chamber pre-coated with a thin layer of basement
membrane matrix (ECMatrix) (Merck KGaA, Darmstadt, Germany). Medium
containing 10% FBS was placed in the lower chambers to act as a
chemoattractant. Cells (5×105) in a 300 µl serum-free
medium were placed in the upper chambers and incubated at 37°C for
24, 48 and 72 h. Invasive cells on the lower surface of the
membrane, which had migrated through the polycarbonate membrane,
were stained with 10% hematoxylin at room temperature for 2 min and
counted under a light microscope (Ti; Nikon Corporation) in five
selected fields at magnification of ×200.
Immunohistochemistry
Breast cancer tissue slides were fixed in 4%
formaldehyde solution at room temperature overnight. These slides
were incubated with MTSS1, E-cad, N-cad and vimentin antibodies
overnight at 4°C according to the manufacturer's protocol. The
signals were visualized with 3,3′-diminobenzidine (Wuhan Boster
Biological Technology, Ltd, Wuhan, China) and counterstained with
10% hematoxylin at room temperature for 2 min. The morphological
sections were evaluated and imaged with high-power light microscopy
(Nikon Corporation, Tokyo, Japan) at magnification of ×200.
Prediction of miR-96 target
Targets of miR-96 were predicted using online
TargetScan software with a search term of has-mir-96 (Release 3.1:
October 2016, URL http://www.targetscan.org/mamm_31/).
Statistical analysis
Statistical analysis was performed using SPSS
software (version 22.0; IBM Corp., Armonk, NY, USA). Data are
expressed as the mean ± standard deviation. Differences of miR-96
expression in serum samples and cancer cells between two groups
were analyzed using a Student's t-test (two-tailed). Differences
among three groups were compared using one-way analysis of variance
with Bonferroni post-hoc test. P<0.05 was considered to indicate
a statistically significant difference.
Results
Dysregulated serum miR-96 in patients
with breast cancer
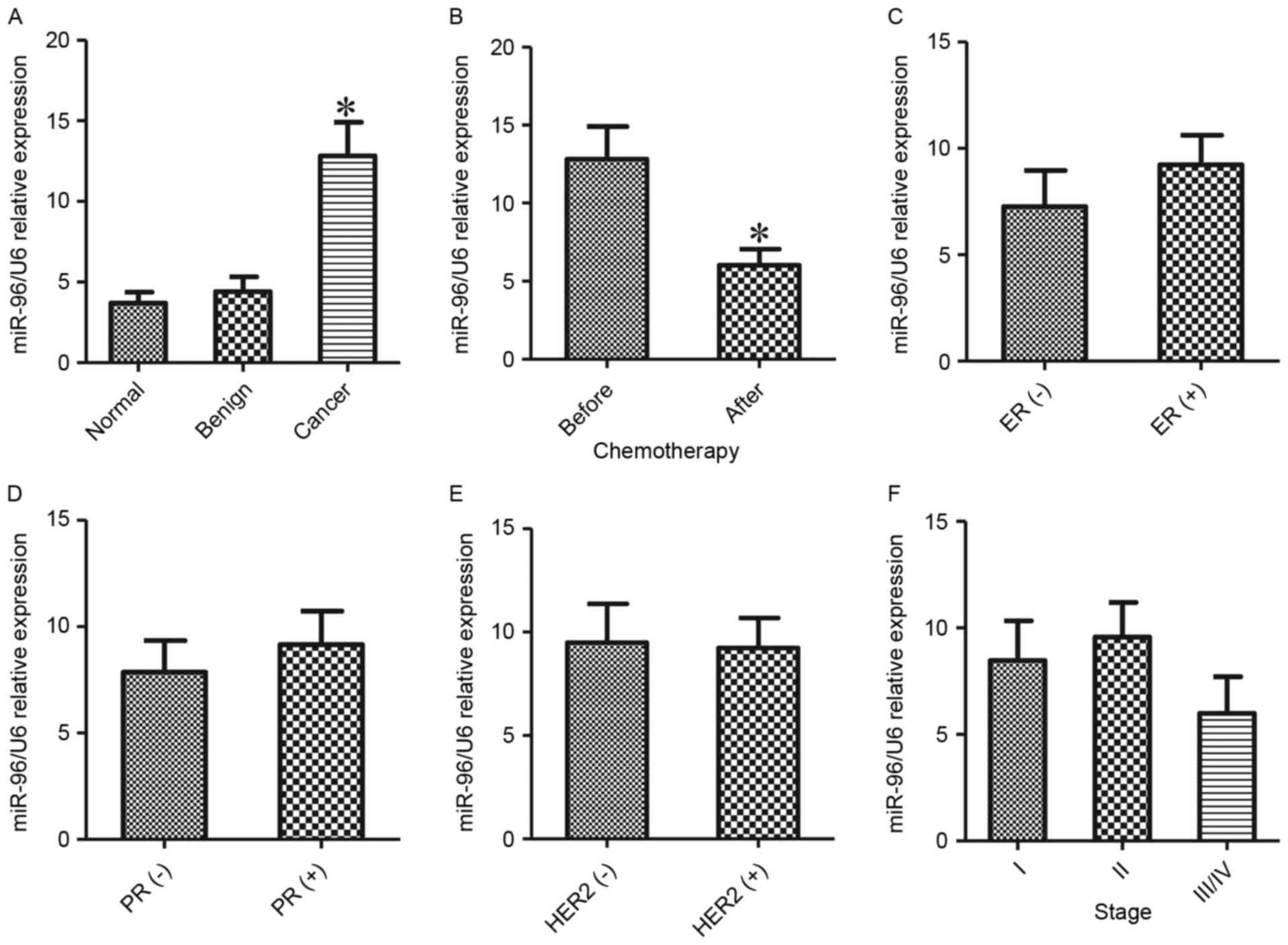
To verify the role of miR-96 in breast cancer, sera
samples were collected from patients with breast cancer (n=44),
benign breast tumors (n=18) and health controls (n=18). RT-qPCR
assays were used to detect the level of miR-96 in different groups.
The results indicated that the level of miR-96 was significantly
elevated in breast cancer samples compared with benign breast
tumors and health controls (P<0.05; Fig. 1A). Subsequently, miR-96 expression was
compared between the patients with breast cancer with (n=26) or
without chemotherapy (n=18). Notably, higher expression of miR-96
was detected in the blood samples from the patients with breast
cancer without chemotherapy (P<0.05; Fig. 1B) compared with the patients who has
undergone chemotherapy. The data indicate that miR-96 may be used
as biomarker for breast cancer diagnosis and therapeutic
outcomes.
To additionally explore the association between the
expression levels of miR-96 and prognosis of patients with breast
cancer, the miR-96 expression in different breast cancers was
compared based on their clinical features, surface markers and
clinical stages. As indicated in Fig. 1C
and D, the expression of miR-96 was almost equivalent, even
slightly increased in cases of estrogen receptor (ER)+ and
progesterone receptor (PR)+ breast cancer compared with cases of
ER- and PR-cancer. No associations between the miR-96 level and the
human epidermal growth factor 2 (HER2)/neu receptor (P>0.05;
Fig. 1E) or between the levels of
miR-96 and stages of breast cancer (P>0.05; Fig. 1F) were observed.
Effects of miR-96 on breast cancer
cell migration
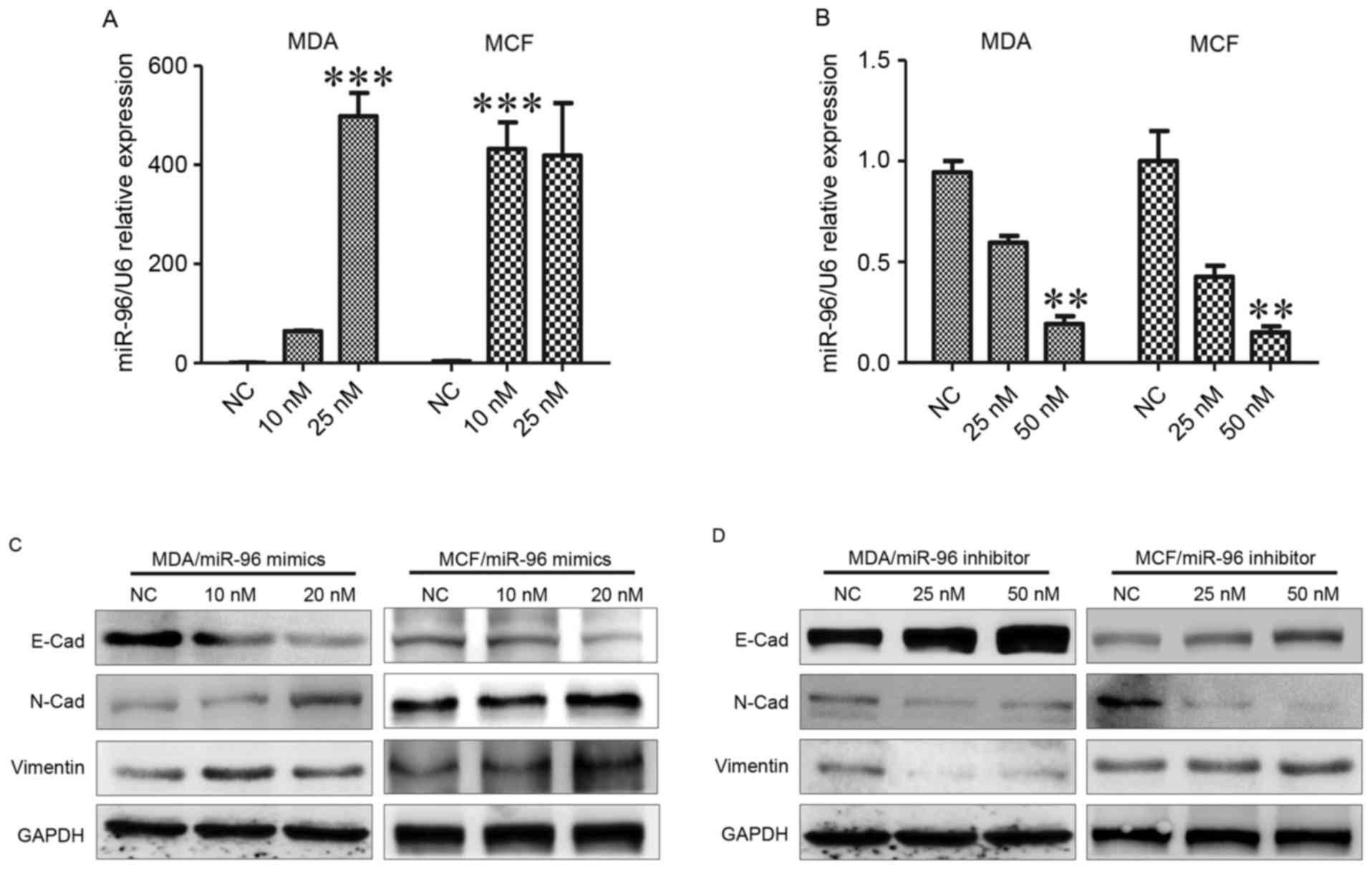
Epithelial-to-mesenchymal transition (EMT) is a
critical process in the progression of breast cancer. The present
study investigated the effects of miR-96 manipulation on
EMT-associated proteins expression in breast cancer cell lines.
Downregulation of epithelial markers and upregulation of
mesenchymal markers serves an important role in EMT. Synthetic
mimics and inhibitors were employed to increase and suppress
endogenous miR-96 expression in MDA-MB-231, and MCF-7 cells
(P<0.001; Fig. 2A and B). It was
demonstrated that miR-96 overexpression in MDA-MB-231 and MCF-7
cell lines resulted in the downregulation of epithelial marker
E-cad and the upregulation of the mesenchymal markers N-cad and
vimentin (Fig. 2C). It was
additionally identified that miR-96 knockdown promoted E-cad
expression and inhibited N-cad expression. Vimentin downregulation
was observed in MDA-MB-231 cells, but not in MCF-7 cells (Fig. 2D). These data reinforce that
MDA-MB-231 and MCF-7 may represent different types of breast
cancer.
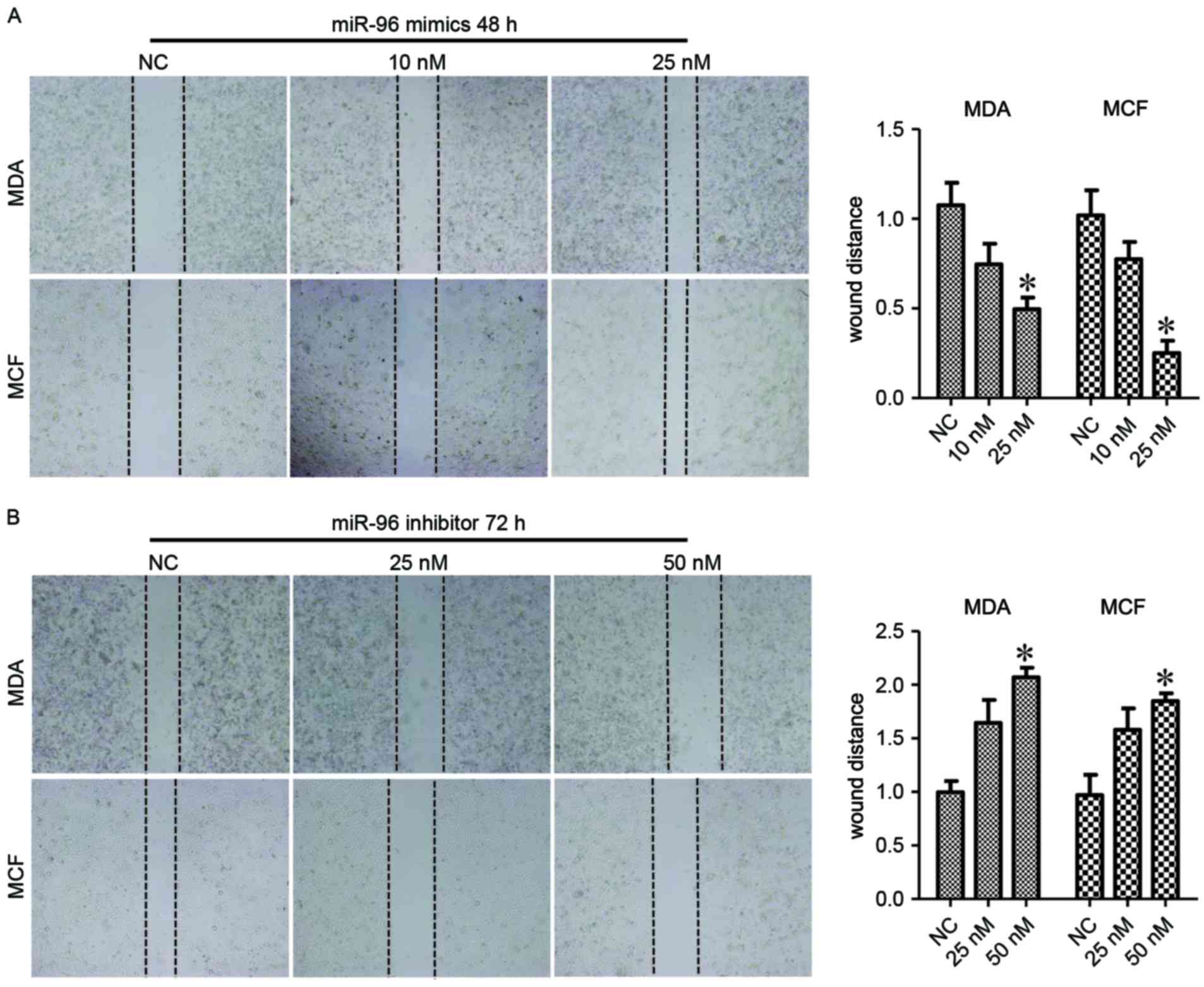
Next, the biological effects of miR-96 in migration
of breast cancer cells was investigated. The results of the wound
healing assay indicated that the migration of MDA-MB-231 and MCF-7
cells were enhanced by 25 nM miR-96 mimics (P<0.05; Fig. 3A) at 48 h post-treatment.
Consistently, miR-96 inhibitors treatment suppressed the migration
of MDA-MB-231 and MCF-7 cells at 72 h (P<0.05; Fig. 3B). The Transwell migration assay also
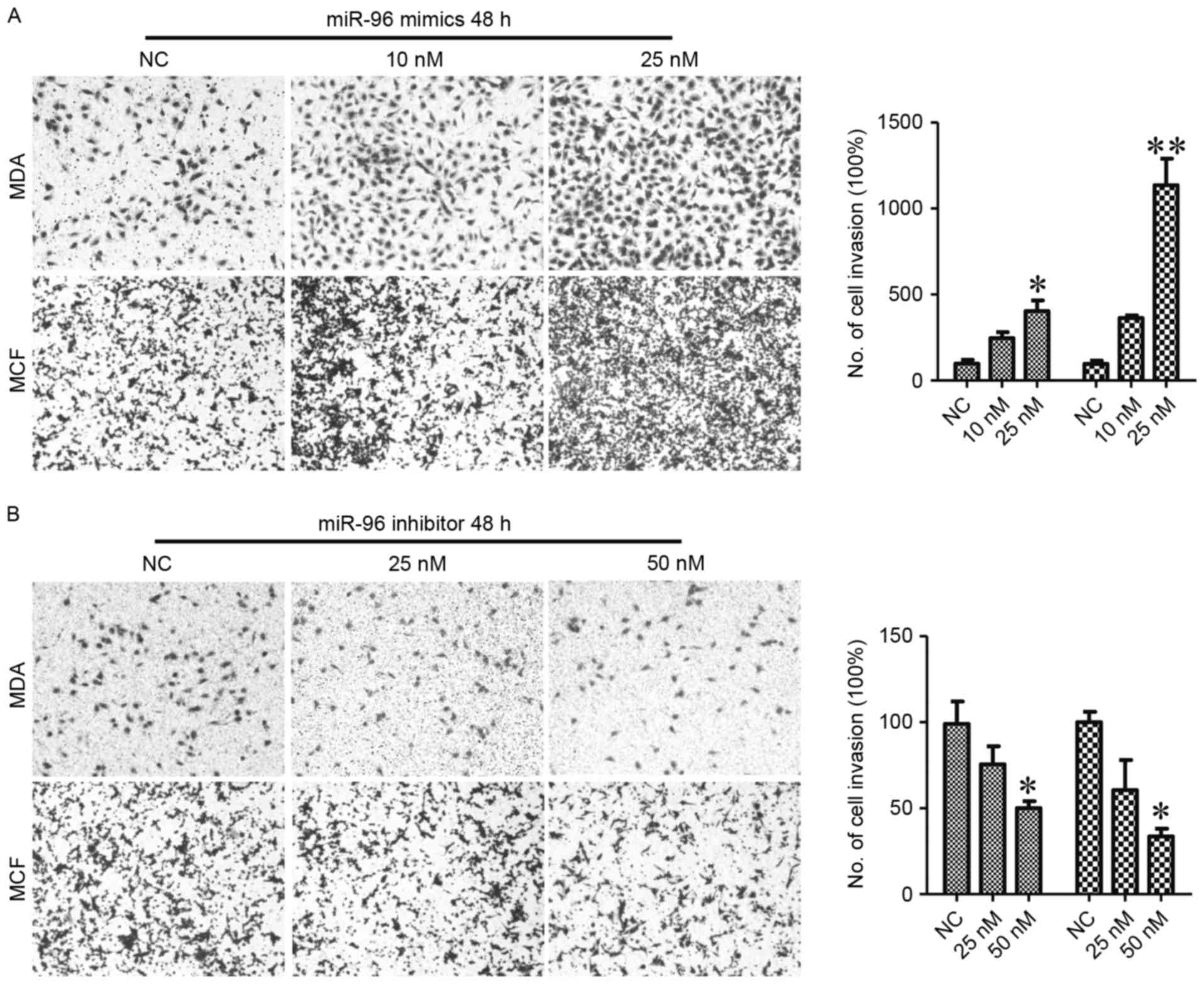
revealed that miR-96 upregulation significantly promoted the
migration of breast cancer cells (P<0.01; Fig. 4A). miR-96 suppression also decreased
the migration of breast cancer cells (P<0.05; Fig. 4B). These data indicated that miR-96
serves an important role in the maintenance of malignant phenotypes
of breast cancer cells.
miR-96 suppresses MTTS1 expression in
breast cancer cells
To verify the factors mediating the effect of miR-96
on breast cancer, the target of miR-96 were predicted by using
online database TargetScan (Release 3.1: October 2016). A total of
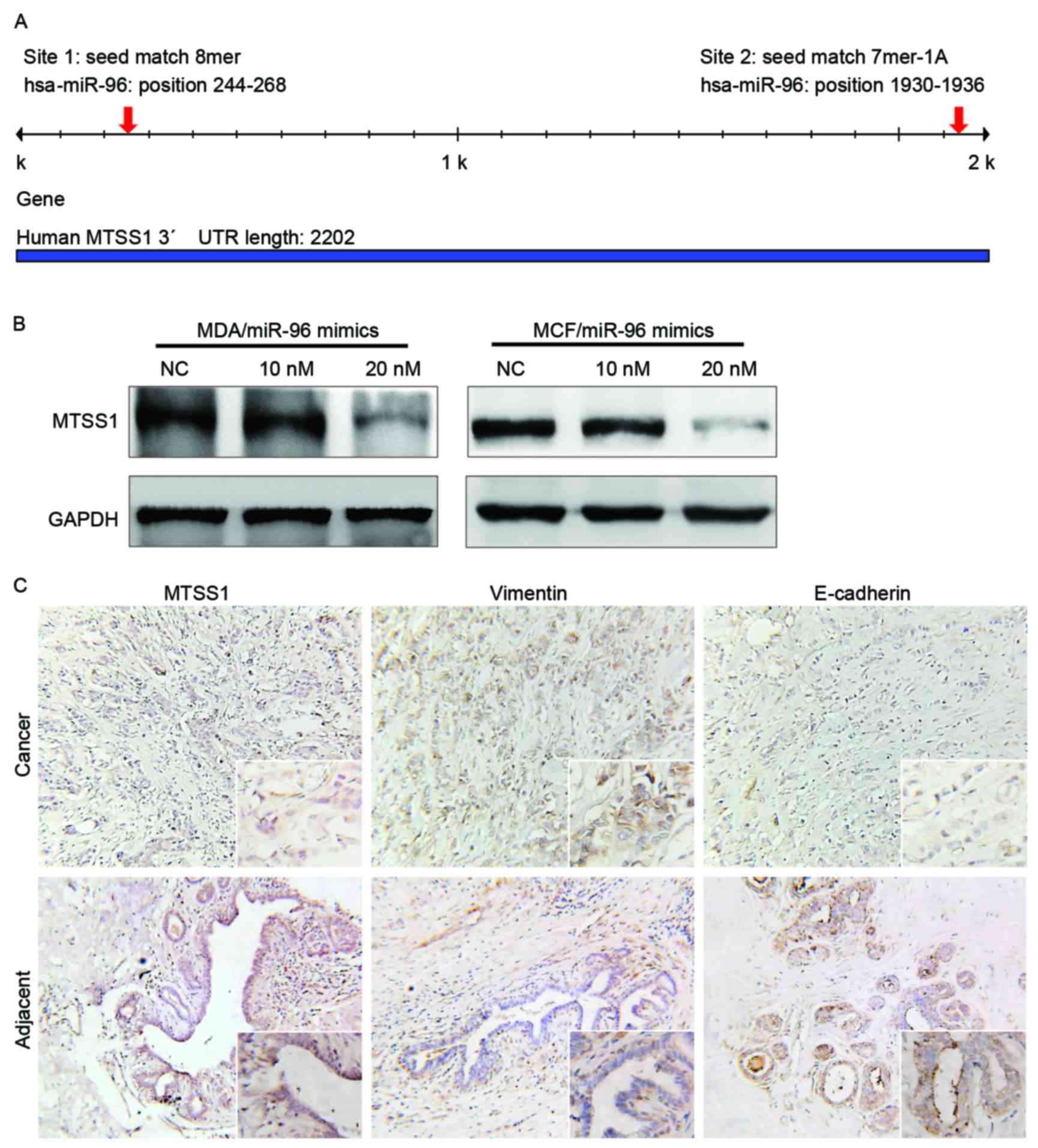
2 conserved miR-96 binding sites were identified in the
3′untranslated region of the human MTSS1 gene (Fig. 5A). To additionally confirm whether the
MTSS1 gene was a target of miR-96, MTSS1 expression was
investigated in breast cancer tissues and miR-96-manipulated cell
lines. The results of the western blot analysis indicated that
treatment with miR-96 mimics suppressed MTSS1 expression and miR-96
inhibitors increased MTSS1 expression in breast cancer cells
(Fig. 5B). The results of
immunohistochemistry also revealed that MTSS1 was downregulated in
breast cancer samples compared with noncancerous tissues (Fig. 5C). In contrast, E-cad expression was
upregulated in noncancerous tissues and reduced in breast cancer
samples (Fig. 5C).
Discussion
miR-96 is highly expressed in various types of
cancers, including breast cancer, hepatocellular carcinoma,
prostate carcinoma and non-small cell lung cancer (20–24). The
role of miR-96 in the prognosis of breast cancer remains unknown.
In the present study, it was identified that serum miR-96 levels
were significantly increased in breast cancer cells compared with
the benign and healthy controls, and miR-96 expression decreased
markedly in patients with breast cancer undergoing chemotherapy.
The expression of miR-96 was almost equivalent in ER+, PR+ and HER+
types of cancer compared with in ER- and PR- and HER-types of
cancer. miR-96 expression was also not changed between different
stages of breast cancer. Previously, Li et al demonstrated
that miR-96 was decreased in ER+ and PR+ breast cancer and
increased in HER2-enriched breast cancer (34). In the present study, 44 breast cancer
samples were collected to investigate miR-96 expression in
different types of breast cancer. Additional samples of breast
cancer should be examined to comprehensively elucidate miR-96
expression in breast cancer.
Previous studies have demonstrated that miR-96 may
increase cancer cell proliferation and migration in prostate cancer
and breast cancer (24,34). The data from the present study support
a proto-oncogenic miRNA role for miR-96 in breast cancer cell
lines, as overexpression of miR-96 by mimics in MCF-7 and
MDA-MB-231 cell lines induced cell migration. The migration results
of MCF-7 were similar to those demonstrated by Li et al
(34), who also identified that
upregulation of miR-96 promoted migration of the breast cancer
MCF-7 and T47D cell lines. The present study indicated that
downregulation of miR-96 by inhibitors in MCF-7 and MDA-MB-231 cell
lines also decreased cell migration. Xu et al (24) also suggested that the invasiveness of
prostate cancer cells was partially suppressed by miR-96 inhibitor
treatment. Furthermore, the data of the present study revealed that
the expression of epithelial marker E-cad was decreased, and
mesenchymal markers N-cad and vimentin were induced in
miR-96-overexpressed breast cancer cells.
MTSS1 is known to be a metastasis suppressor, and to
suppress proliferation and EMT in non-small cell lung cancer,
hepatitis B-associated hepatocellular carcinoma and bladder
urothelial carcinoma cells, prostate carcinoma cells, chronic
myeloid leukemia and the tongue squamous cellular carcinoma Tca8113
cell line (24,31,32,35). Loss
of MTSS1 facilitates the progression of prostate and breast
cancers. Similar to other types of cancer, MTSS1 has also been
suggested to demonstrate prognostic value and anti-metastatic
effects in breast cancer (36–40). The
immunohistochemistry results of the present study indicated that
invasive breast cancer tumors exhibited decreased expression of
MTSS1 compared with paracancerous tissue, which additionally
confirmed that MTSS1 is a tumor suppressor in breast cancer.
The regulation of MTSS1 is also of interest for the
study of prostate cancer biology. Downregulation of MTSS1
expression contributes to the growth, development, and metastasis
of breast and prostate cancer (36–38). Zhong
et al (38) demonstrated that
Skp, Cullin, F-box containing complex β-transducin
repeat-containing protein, a E3 ubiquitin ligase complex with a
function in different types of cancer including breast or prostate
cancer cells, inhibited MTSS1 expression in a
ubiquitination-dependent fashion. miR-15 and miR-182-5p were also
identified to participate in the regulation of MTSS1 transcription
in prostate cancer cells (37,41).
However, it is necessary to investigate the mechanisms involved
with MTSS1 deregulation in breast cancer. The results of the
present study revealed that miR-96 may downregulate MTSS expression
in breast cancer cells.
In conclusion, miR-96 was indicated to be associated
with the prognosis of patients with breast cancer, and may suppress
migration and invasiveness of breast cancer cells by downregulating
MTSS1 expression. The present study implied that miR-96 may be a
useful therapeutic target and prognostic marker for breast cancer
treatment.
Acknowledgements
The present study was supported by the Project of
Nantong Science and Technology bureau guiding science and
technology (grant no. HS149134).
References
|
1
|
Huang X, Li X and Xie X, Ye F, Chen B,
Song C, Tang H and Xie X: High expressions of LDHA and AMPK as
prognostic biomarkers for breast cancer. Breast. 30:39–46. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kakushadze Z, Raghubanshi R and Yu W:
Estimating Cost Savings from Early Cancer Diagnosis. SSRN
Electronic J. 2017. View Article : Google Scholar
|
|
4
|
Schootman M, Fuortes L and Aft R:
Prognosis of metachronous contralateral breast cancer according to
stage at diagnosis: The importance of early detection. Breast
Cancer Res Treat. 99:91–95. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bucchi L, Belli P, Benelli E, Bernardi D,
Brancato B, Calabrese M, Carbonaro LA, Caumo F, Cavallo-Marincola
B, Clauser P, et al: Recommendations for breast imaging follow-up
of women with a previous history of breast cancer: Position paper
from the Italian Group for Mammography Screening (GISMa) and the
Italian College of Breast Radiologists (ICBR) by SIRM. Radiol Med.
121:891–896. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Svoronos A, Engelman D and Slack F:
OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer.
Cancer Res. 76:3666–3670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alhasan AH, Scott AW, Wu JJ, Feng G, Meeks
JJ, Thaxton CS and Mirkin CA: Circulating microRNA signature for
the diagnosis of very high-risk prostate cancer. Proc Natl Acad Sci
USA. 113:pp. 10655–10660. 2016; View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kang J, Kim W, Lee S, Kwon D, Chun J, Son
B, Kim E, Lee JM, Youn H and Youn B: TFAP2C promotes lung
tumorigenesis and aggressiveness through miR-183- and
miR-33a-mediated cell cycle regulation. Oncogene. 36:1585–1596.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pichler M, Stiegelbauer V,
Vychytilova-Faltejskova P, Ivan C, Ling H, Winter E, Zhang X,
Goblirsch M, Wulf-Goldenberg A, Ohtsuka M, et al: Genome-wide
microRNA analysis identifies miR-188-3p as novel prognostic marker
and molecular factor involved in colorectal carcinogenesis. Clin
Cancer Res. 23:1323–1333. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hannafon BN, Trigoso YD, Calloway CL, Zhao
YD, Lum DH, Welm AL, Zhao ZJ, Blick KE, Dooley WC and Ding WQ:
Plasma exosome microRNAs are indicative of breast cancer. Breast
Cancer Res. 18:902016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu M, Yang R, Urrehman U, Ye C, Yan X,
Cui S, Hong Y, Gu Y, Liu Y, Zhao C, et al: miR-19b suppresses PTPRG
to promote breast tumorigenesis. Oncotarget. 7:64100–64108.
2016.PubMed/NCBI
|
|
12
|
Li C, Li J, Cai Q, Qiu QQ, Yan M, Liu BY
and Zhu ZG: miRNA-199a-3p: A potential circulating diagnostic
biomarker for early gastric cancer. J Surg Oncol. 108:89–92. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qi Y, Cui L, Ge Y, Shi Z, Zhao K, Guo X,
Yang D, Yu H, Cui L, Shan Y, et al: Altered serum microRNAs as
biomarkers for the early diagnosis of pulmonary tuberculosis
infection. BMC Infect Dis. 12:3842012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Adhami M, Haghdoost AA, Sadeghi B and
Malekpour Afshar R: Candidate miRNAs in human breast cancer
biomarkers: A systematic review. Breast Cancer. Nov 3–2017.(Epub
ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ohzawa H, Miki A, Teratani T, Shiba S,
Sakuma Y, Nishimura W, Noda Y, Fukushima N, Fujii H, Hozumi Y, et
al: Usefulness of miRNA profiles for predicting pathological
responses to neoadjuvant chemotherapy in patients with human
epidermal growth factor receptor 2-positive breast cancer. Oncol
Lett. 13:1731–1740. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hemmatzadeh M, Mohammadi H, Jadidi-Niaragh
F, Asghari F and Yousefi M: The role of oncomirs in the
pathogenesis and treatment of breast cancer. Biomed Pharmacother.
78:129–139. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mar-Aguilar F, Rodríguez-Padilla C and
Reséndez-Pérez D: Use of serum-circulating miRNA profiling for the
identification of breast cancer biomarkers. Methods Mol Biol.
1165:71–80. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang L, Xu Y, Jin X, Wang Z, Wu Y, Zhao
D, Chen G, Li D, Wang X, Cao H, et al: A circulating miRNA
signature as a diagnostic biomarker for non-invasive early
detection of breast cancer. Breast Cancer Res Treat. 154:423–434.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huo D, Clayton WM, Yoshimatsu TF, Chen J
and Olopade OI: Identification of a circulating MicroRNA signature
to distinguish recurrence in breast cancer patients. Oncotarget.
7:55231–55248. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cheung CC, Lun SW, Chung GT, Chow C, Lo C,
Choy KW and Lo KW: MicroRNA-183 suppresses cancer stem-like cell
properties in EBV-associated nasopharyngeal carcinoma. BMC Cancer.
16:4952016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gokhale A, Kunder R, Goel A, Sarin R,
Moiyadi A, Shenoy A, Mamidipally C, Noronha S, Kannan S and Shirsat
NV: Distinctive microRNA signature of medulloblastomas associated
with the WNT signaling pathway. J Cancer Res Ther. 6:521–529. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Leung WK, He M, Chan AW, Law PT and Wong
N: Wnt/β-Catenin activates miR-183/96/182 expression in
hepatocellular carcinoma that promotes cell invasion. Cancer Lett.
362:97–105. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rapti S, Kontos CK, Papadopoulos IN and
Scorilas A: High miR-96 levels in colorectal adenocarcinoma predict
poor prognosis, particularly in patients without distant metastasis
at the time of initial diagnosis. Tumour Biol. 37:11815–11824.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu L, Zhong J, Guo B, Zhu Q, Liang H, Wen
N, Yun W and Zhang L: miR-96 promotes the growth of prostate
carcinoma cells by suppressing MTSS1. Tumour Biol. 37:12023–12032.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin H, Dai T, Xiong H, Zhao X, Chen X, Yu
C, Li J, Wang X and Song L: Unregulated miR-96 induces cell
proliferation in human breast cancer by downregulating
transcriptional factor FOXO3a. PLoS One. 5:e157972010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu D, He X, Chang Y, Xu C, Jiang X, Sun S
and Lin J: Inhibition of miR-96 expression reduces cell
proliferation and clonogenicity of HepG2 hepatoma cells. Oncol Rep.
29:653–661. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Haflidadottir BS, Larne O, Martin M,
Persson M, Edsjö A, Bjartell A and Ceder Y: Upregulation of miR-96
enhances cellular proliferation of prostate cancer cells through
FOXO1. PLoS One. 8:e724002013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Song HM, Luo Y, Li DF, Wei CK, Hua KY,
Song JL, Xu H, Maskey N and Fang L: MicroRNA-96 plays an oncogenic
role by targeting FOXO1 and regulating AKT/FOXO1/Bim pathway in
papillary thyroid carcinoma cells. Int J Clin Exp Pathol.
8:9889–9900. 2015.PubMed/NCBI
|
|
29
|
Gao F and Wang W: MicroRNA-96 promotes the
proliferation of colorectal cancer cells and targets tumor protein
p53 inducible nuclear protein 1, forkhead box protein O1 (FOXO1)
and FOXO3a. Mol Med Rep. 11:1200–1206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee YG, Macoska JA, Korenchuk S and Pienta
KJ: MIM, a potential metastasis suppressor gene in bladder cancer.
Neoplasia. 4:291–294. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Huang XY, Huang ZL, Xu B, Chen Z, Re TJ,
Zheng Q, Tang ZY and Huang XY: Elevated MTSS1 expression associated
with metastasis and poor prognosis of residual hepatitis B-related
hepatocellular carcinoma. J Exp Clin Cancer Res. 35:852016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li XD, Zhang JX, Jiang LJ, Wang FW, Liu
LL, Liao YJ, Jin XH, Chen WH, Chen X, Guo SJ, et al: Overexpression
of maelstrom promotes bladder urothelial carcinoma cell
aggressiveness by epigenetically downregulating MTSS1 through
DNMT3B. Oncogene. 35:6281–6292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li P, Sheng C, Huang L, Zhang H, Huang L,
Cheng Z and Zhu Q: miR-183/-96/-182 cluster is up-regulated in most
breast cancers and increases cell proliferation and migration.
Breast Cancer Res. 16:4732014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guo Y, Ren M, Shang C, Zhu L and Zhong M:
MTSS1 gene regulated by miR-96 inhibits cell proliferation and
metastasis in tongue squamous cellular carcinoma Tca8113 cell line.
Int J Clin Exp Med. 8:15441–15449. 2015.PubMed/NCBI
|
|
36
|
Giacobbe A, Compagnone M,
Bongiorno-Borbone L, Antonov A, Markert EK, Zhou JH,
Annicchiarico-Petruzzelli M, Melino G and Peschiaroli A: p63
controls cell migration and invasion by transcriptional regulation
of MTSS1. Oncogene. 35:1602–1608. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kedmi M, Ben-Chetrit N, Körner C, Mancini
M, Ben-Moshe NB, Lauriola M, Lavi S, Biagioni F, Carvalho S,
Cohen-Dvashi H, et al: EGF induces microRNAs that target
suppressors of cell migration: miR-15b targets MTSS1 in breast
cancer. Sci Signal. 8:ra292015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhong J, Shaik S, Wan L, Tron AE, Wang Z,
Sun L, Inuzuka H and Wei W: SCF β-TRCP targets MTSS1 for
ubiquitination-mediated destruction to regulate cancer cell
proliferation and migration. Oncotarget. 4:2339–2353. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hicks DG, Yoder BJ, Short S, Tarr S,
Prescott N, Crowe JP, Dawson AE, Budd GT, Sizemore S, Cicek M, et
al: Loss of breast cancer metastasis suppressor 1 protein
expression predicts reduced disease-free survival in subsets of
breast cancer patients. Clin Cancer Res. 12:6702–6708. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Parr C and Jiang WG: Metastasis suppressor
1 (MTSS1) demonstrates prognostic value and anti-metastatic
properties in breast cancer. Eur J Cancer. 45:1673–1683. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hirata H, Ueno K, Shahryari V, Deng G,
Tanaka Y, Tabatabai ZL, Hinoda Y and Dahiya R: MicroRNA-182-5p
promotes cell invasion and proliferation by down regulating FOXF2,
RECK and MTSS1 genes in human prostate cancer. PLoS One.
8:e555022013. View Article : Google Scholar : PubMed/NCBI
|