Introduction
Gastric cancer is the fourth most commonly diagnosed
cancer type and is the second leading cause of cancer-associated
mortality worldwide (1). Numerous
molecules are responsible for gastric cancer tumorigenesis, and the
cause of carcinogenesis is complicated involving the dysregulation
of oncogenes and tumor suppressors (2). Gastrectomy remains the standard
treatment for gastric cancer clinically (3). The overall 5-year relative survival rate
for gastric cancer is ~28% and none of the current treatment
modalities are able to influence the overall survival rates
(4). The recurrence rate remains high
due to the absence of specific symptoms, thus rendering early
diagnosis of this fatal disease difficult (4), and the prognosis for patients with the
advanced stage remains poor. The outcome of patients with gastric
cancer is determined primarily by the presence or absence of
metastasis (5). Therefore, developing
novel prognostic factors and therapeutic strategies is essential,
and identification of the precise molecular mechanisms that
modulate malignant transformation is necessary.
As previously reported, Ras genes frequently undergo
activating mutations in several human and animal cancer types
(6–8).
However, the expression of Ras genes in gastric cancer and its
clinical values remain to be elucidated. Ras proteins are essential
in numerous signal transduction pathways that transfer information
from the extracellular environment to internal cellular
compartments (9). Thus,
mutation-induced deregulation of the Ras superfamily, and upstream
and downstream signaling components result in important alterations
to healthy cell growth control and differentiation, in addition,
they serve an important role in mechanisms of tumor formation
(8,10). In mammals, the Ras signal transduction
pathway is mediated by several effectors, including proteins with a
Ras-associated (RalGDS/AF-6) (RA)-domain, and the RA-domain is a
common feature of genes in the Ras association domain family
(RASSF) (9,11). However, the importance of RASSF
protein functions remains to be clarified and the Ras-binding
status of RASSF family members, particularly RASSF8, remain unknown
(12).
The complex interaction among different etiological
factors leads to genetic and epigenetic alterations of
proto-oncogenes, and tumor suppressor genes, which underlie the
pathogenesis of cancer. It has long been considered that
dysregulation of these genes results in abnormal function or
expression of oncogenic and tumor suppressor proteins (2). MicroRNAs (miRNAs) are small non-coding
RNA molecules that are generated within cells, which serve a role
in post-transcriptional gene regulation and in almost any cellular
biological function. Aberrant expression of miRNAs has been
identified in cancerous transformation and progression (13). miRNA has been demonstrated to be
involved in the pathogenesis of gastric cancer (14) and numerous other types of cancer
(15–17,19).
Furthermore, miRNA represses the expression of its target genes
through base pairing with endogenous mRNAs to perform its
biological functions, and miRNA genes have been characterized as
novel proto-oncogenes or tumor suppressor genes in gastric
carcinogenesis (20).
In the present study, the clinical significance of
microRNA-377/RASSF8 signaling axis was evaluated, in addition, the
expression and function of miRNA377/RASSF8 proteins were analyzed
in vitro. The correlation between miRNA377 and RASSF8
protein expression, and clinicopathological variables in cases of
gastric cancer were also investigated.
Materials and methods
Patients and samples
The present study was approved by the Medical Ethics
Committee of Shanghai Zhongshan Hospital, Fudan University
(Shanghai, China). Prior written informed consent was obtained from
each patient. A total of 10 patients with gastric cancer (6 male, 4
female; age range, 39–58 years) were included in the present study.
The specimens were handled and made anonymous according to the
ethical and legal standards. Paired tissue specimens (tumor and
adjacent normal mucosa) from 10 patients with gastric cancer were
obtained and histologically confirmed by a pathologist at Shanghai
Zhongshan Hospital. The samples were derived from patients who had
not received adjuvant treatment, including radiotherapy or
chemotherapy prior to surgery in order to eliminate potential
treatment-induced changes to gene expression profiles. Following
excision, tissue samples were immediately snap frozen in liquid
nitrogen and stored at −80°C until RNA extraction.
Immunohistochemistry and assessment of
RASSF8 expression
Immunohistochemical staining was performed for
RASSF8 to evaluate immunoreactivity in the aforementioned tissue
samples (21) using the Elivision™
Plus Two-Step system (Fuzhou Maixin Biotech Co., Ltd., Fuzhou,
China). Paraffin-embedded tissue sections (5-µm thick) were
prepared. Slides were deparaffinized, rehydrated and subjected to
microwave heat antigen retrieval in 0.01 M citrate buffer (pH 6.0)
for 20–25 min at 25°C. Following blocking of endogenous peroxidase
activity with 3% H2O2 for 15 min at 25°C, the
sections were incubated with a primary antibody directed against
RASSF8 (dilution, 1:300; cat. no. ab214324; Abcam, Cambridge, MA,
USA) overnight at 4°C. Following washing with PBS for 3 times (10
min each, 25°C), the specimens were incubated with F(ab')2-goat
anti-rabbit immunoglobulin G (IgG) (H+L) secondary antibody
(dilution, 1:200; cat. no. Q-11401MP; Thermo Fisher Scientific
Inc., Waltham, MA, USA) at 37°C for 1 h and subsequently washed
with PBS three times. Immunoreactivity was visualized using the
chromogen, 3,3′diaminobenzidine (Fuzhou Maixin Biotech Co., Ltd.).
Slides were then counterstained with 0.1% hematoxylin solution for
5 min at 25°C.
The immunostaining frequency for each tumor was
scored as follows: 0 (<10%); 1 (10–30%); 2 (31–60%); and 3
(>61%). Staining intensity was documented as: 0, no
immunostaining; 1, weak; 2, moderate and 3, strong. Total
immunostaining score results were calculated from the
multiplication of both parameters. Samples were scored totally as
follows: A scale of 0 (−, total immunostaining score, 0); 1+ (score
range, 1–2); 2+ (++, total immunostaining score, 3~4); and 3+ (+++,
total immunostaining score, 6~7). Immunostaining was assessed by an
experienced pathologist who was blinded to the clinical data of the
patients. RASSF8 expression was determined by assessing the
percentage and intensity of stained tumor cells. For RASSF8
protein, immunostainings were scored as strong (2++ and 3+++), weak
or negative (1+ and 0) according to the rate of labeled tumor cells
and the membrane staining intensity.
Cell culture
The human AGS, BGC-823, HGC-27, MKN-45 and SGC-7901
gastric cancer cell lines, GES-1 normal gastric cell line, and HEK
293T cells were purchased from the Cell Bank of Chinese Academy of
Science (Shanghai, China). AGS, BGC-823, MKN-45, SGC-7901 and HEK
293T cells were cultured in Dulbecco's modified Eagle's medium
(DMEM). HGC-27 and GES-1 were cultured in RPMI-1640. In each case,
the medium was supplemented with 10% fetal bovine serum (FBS) all
from Thermo Fisher Scientific, Inc. (Gibco) and 1% penicillin and
streptomycin (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). Cells
were maintained at 37°C in a humidified atmosphere consisting of 5%
CO2.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from cell lines or frozen
tissues using TRIzol reagent and miRNeasy mini kit (Invitrogen;
Thermo Fisher Scientific, Inc.). The RT-qPCR was performed using
All-in-One™ miRNA qRT-PCR Detection kit (GeneCopoeia Inc.,
Rockville, MD, USA) according to the manufacturer's protocol. The
iQ-5 system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) was
used to monitor the PCR in real-time. The PCR cycling profile was
as follows: Denaturation at 95°C for 2 min; followed by 40 cycles
of annealing at 95°C for 5 sec; and extension at 60°C for 35 sec.
The average cycle threshold (Cq), from triplicate assays, was used
for further calculations. Relative expression levels were
normalized to the control (GES-1, control cell line; normal gastric
mucosal tissue, control tissue). The endogenous U6 snRNA was chosen
as the internal control. The 2−ΔΔCq method (22) was used to quantify the relative amount
of miR-377/RASSF8, where
ΔΔCq=[(CqmiR-377/RASSF8)-CqU6RNA]-(Cqcontrol-CqU6RNA).
Lentivirus and lentivirus
infection
RASSF8 overexpression lentivirus was purchased from
Shanghai R&S Biotechnology Co., Ltd. (Shanghai, China). BGC-823
cells were seeded into 3.5-cm dishes (1×106 cells/dish)
24 h prior to lentivirus infection. The next day, lentivirus was
added into dishes with a multiplicity of infection of 4 to infect
cells. The infection efficiency was detected using fluorescence
microscopy analysis of green fluorescence protein 48 h after
infection, and was only accepted when the efficiency was
>90%.
miRNA mimics, plasmids and
transfection
The miR-377 mimic and negative control (miR-NC) were
purchased from Shanghai GenePharma, Co., Ltd. (Shanghai, China).
The transfection of miRNAs (50 nM) was performed using X-tremeGENE
(Roche Applied Science, Rotkreuz, Switzerland) according to the
manufacturer's protocol. The 3′-untranslated region (UTR) was
PCR-amplified from BGC-823 genomic DNA and cloned downstream of the
luciferase gene in the pGL vector (Promega Corporation, Madison,
WI, USA).
Western blot analysis
Total protein from cells was extracted according to
the protocol of the Whole Protein Extraction kit (KGP2100; Nanjing
KeyGen Biotech Co., Ltd., Nanjing, China). Equivalent quantities
(40 µg protein/lane) of protein were separated by 10% SDS-PAGE and
transferred to polyvinylidene difluoride membranes. Membranes were
blocked with 10% skimmed milk (10% bovine serum albumin (KGY00810,
Nanjing KeyGen Biotech Co., Ltd) for phosphorylated-protein), and
then incubated with the appropriate primary antibody overnight at
4°C. Membranes were subsequently washed with Tris-Buffered
saline-Tween 20 and incubated with the corresponding horseradish
peroxidase-conjugated AffiniPure goat anti-rabbit IgG (H+L)
secondary antibody (dilution, 1:200; cat. no. 111-035-003; Jackson
ImmunoResearch Inc., West Grove, PA, USA) at 25°C for 1 h. Bound
secondary antibody was visualized using an enhanced
chemiluminescence system (Pierce Biotechnology, Rockford, IL, USA).
The primary antibodies used were as follows: Anti-GAPDH (1:1,000;
cat. no. 2118; Cell Signal Technology, Inc., Danvers, MA, USA); and
anti-RASSF8 (1:2,000; cat. no. ab126110; Abcam). The results were
normalized to GAPDH to correct for loading.
Cell viability assay
A total of 1×104 BGC-823 cells/well were
plated in 96-well plates and cultured for 1–6 days in normal
conditions. Cell viability was assessed using Cell Counting kit-8
(Dojindo, Kumamoto, Japan). A total of 10 µl of cell counting assay
kit-8 (CCK-8) solution was added daily to each well. After
treatment with CCK-8 at 37°C for 2 h, the absorbance at 450 nm was
measured using a microplate reader.
Colony formation assay
The transfected cells were plated in 6-well plates
at a density of 1×103 cells/well and incubated in
culture medium (DMEM containing 10% FBS) for 10 days without any
disturbance. The cells were fixed with 70% (v/v) ethanol for 15 min
at 25°C and stained with 0.5% crystal violet for 1 h at 37°C.
Visible colonies were counted in 10 different fields and the mean
value was calculated.
Cell cycle assay
The cells were seeded into 6-well plates at a
density of 1×105 cells/well after transfection for 24 h,
and maintained in DMEM/RPMI-1640 containing 10% FBS. After being
cultured for 24, 4 h and 72 h, the cells were harvested and fixed
in 70% (v/v) ice-cold ethanol for 15 min at 4°C, then stored at
−20°C overnight. The fixed cells were washed twice with PBS (1 ml)
and stained with propidium iodide (PI) solution (Invitrogen; Thermo
Fisher Scientific, Inc.) (10 µg/ml) including RNase (0.5 mg/ml) for
30 min at 4°C in the dark. Flow cytometry was then performed using
a FACSCalibur cytometer (BD Biosciences, Franklin Lakes, NJ, USA)
to examine the cell cycle distribution. Experiments were performed
in triplicate.
Apoptosis assay
A total of 2×105lentivirus-infectedcells
BGC-823 cells/well were plated in 6-well plates, and then
transfected with miR-NC and miR-377 mimics for 48 h. The
transfected cells were harvested by brief trypsinization and
centrifugation (170 × g) for 5 min at 4°C, washed in ice-cold PBS
and resuspended in PBS containing Annexin V-fluorescein
isothiocyanate. PI (Thermo Fisher Scientific, Inc.) was then added,
and the cells were incubated for 30 min at 20°C. A total of
1×104 cells/sample were acquired using a FACScan flow
cytometer and the proportions of labeled cells were analyzed using
Paint-A-Gate software (BD Paint-A-Gate Pro for Windows; version
649728; BD Biosciences).
Invasion assay in vitro
The upper surface of a polycarbonate membrane in a
Transwell filter insert was coated with Matrigel (10 µg/µl; 30 µl).
After 30 min of incubation at 37°C, the Matrigel solidified and
served as the extracellular matrix for tumor cell invasion
analyses. The cells were harvested in 100 µl serum-free growth
medium and seeded into the upper compartment of the chamber.
BGC-823 cells or lentivirus-infected cells were seeded into the
upper chambers at the density of 8×105 cells in 100 µl
serum-free DMEM. The lower chambers were filled with 500 µl
RPMI-1640 containing 10% FBS. After 24 h of incubation,
non-invading cells on the top of the membrane were removed by
scraping. Invaded cells on the bottom of the membrane were fixed
with 4% paraformaldehyde for 15 min at 25°C, followed by staining
with 0.5% crystal violet for 20 min at 25°C. The invaded cells on
the membrane were then counted in three random microscopic
fields/well, using ImageJ software (k 1.45; National Institutes of
Health, Bethesda, MD, USA) (23).
Invasion studies were repeated three times.
Databases for prediction of the
targeting relationship
TargetScan (www.targetscan.org/vert_71) and Starbase (starbase.sysu.edu.cn) were used to predict the
targeting association between miR-377 and RASSF8.
Luciferase reporter assays
A total of 1×105 HEK 293 T cells/well
were seeded into 24-well culture plates in RPMI-1640 containing 10%
FBS without antibiotics. After 24 h, the cells were cotransfected
with miR-NC, miR-377 mimics and the 3′-UTR of RASSF8 responsive
firefly luciferase reporter plasmid and Renilla luciferase
plasmid (R&S Biotechnology Co., Ltd., Shanghai, China) and
transfection reagent (POLO deliverer TM 3000 Transfection Reagent
POLO3000, CT001; R&S Biotechnology Co., Ltd.) according to the
manufacturer's protocol. Cells were incubated for 24 h under normal
conditions, then cell lysates were prepared and luciferase
activities were measured using the Dual-Luciferase Reporter Assay
system (Promega Corporation). Firefly luciferase activity was
normalized to the activity of Renilla luciferase.
Statistical analysis
Results are presented as the means ± standard
deviation of three independent samples. A comparison of the level
of RASSF/miR-377 expression between gastric cancer and adjacent
normal tissue was performed using the Wilcoxon signed-rank test.
Significant differences in the mean values were evaluated using the
Student's unpaired t-test. Where multiple comparisons were
required, analysis was performed using one-way analysis of variance
with Bonferroni correction. P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of RASSF8 and miR-377 in
human gastric cancer tissues and cell lines
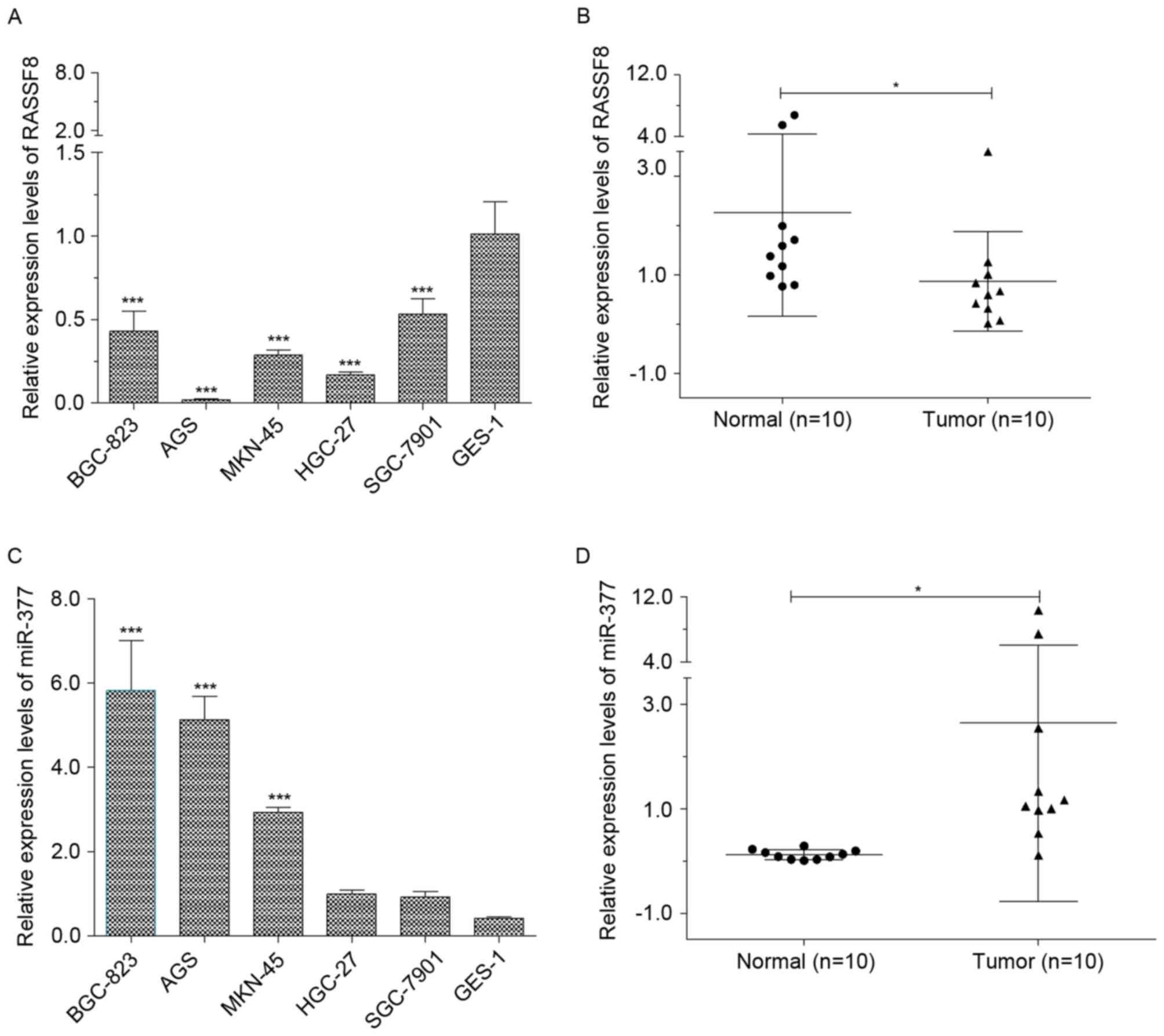
The overall expression levels of RASSF8 and miR-377
human gastric cancer tissues, and cell lines were determined using
RT-qPCR. As presented in Fig. 1A,
levels of RASSF8 in the human GES-1 normal cells were significantly
higher compared with the levels of RASSF8 in BGC-823, AGS, MKN-45,
HGC-27 and SGC-7901. In addition, RASSF8 expression in tumor
tissues demonstrated significantly attenuated levels compared with
the corresponding normal tissues, with a mean1.6-fold decrease
(Fig. 1B). Furthermore, levels of
miR-377 in the BGC-823, AGS and MKN-45 cell lines were
significantly higher compared with that in the normal GES-1 cell
line (P<0.01; Fig. 1C), which was
consistent with a previous report (24). However, no significant differences
were identified among the levels of miR-377 in the HGC-27, SGC-7901
and normal GES-1 cell lines in the present study. Furthermore, a
significantly increased expression level of miR-377 between the
tumor and normal groups was identified, with a mean 20.4-fold
increase (P<0.05; Fig. 1D).
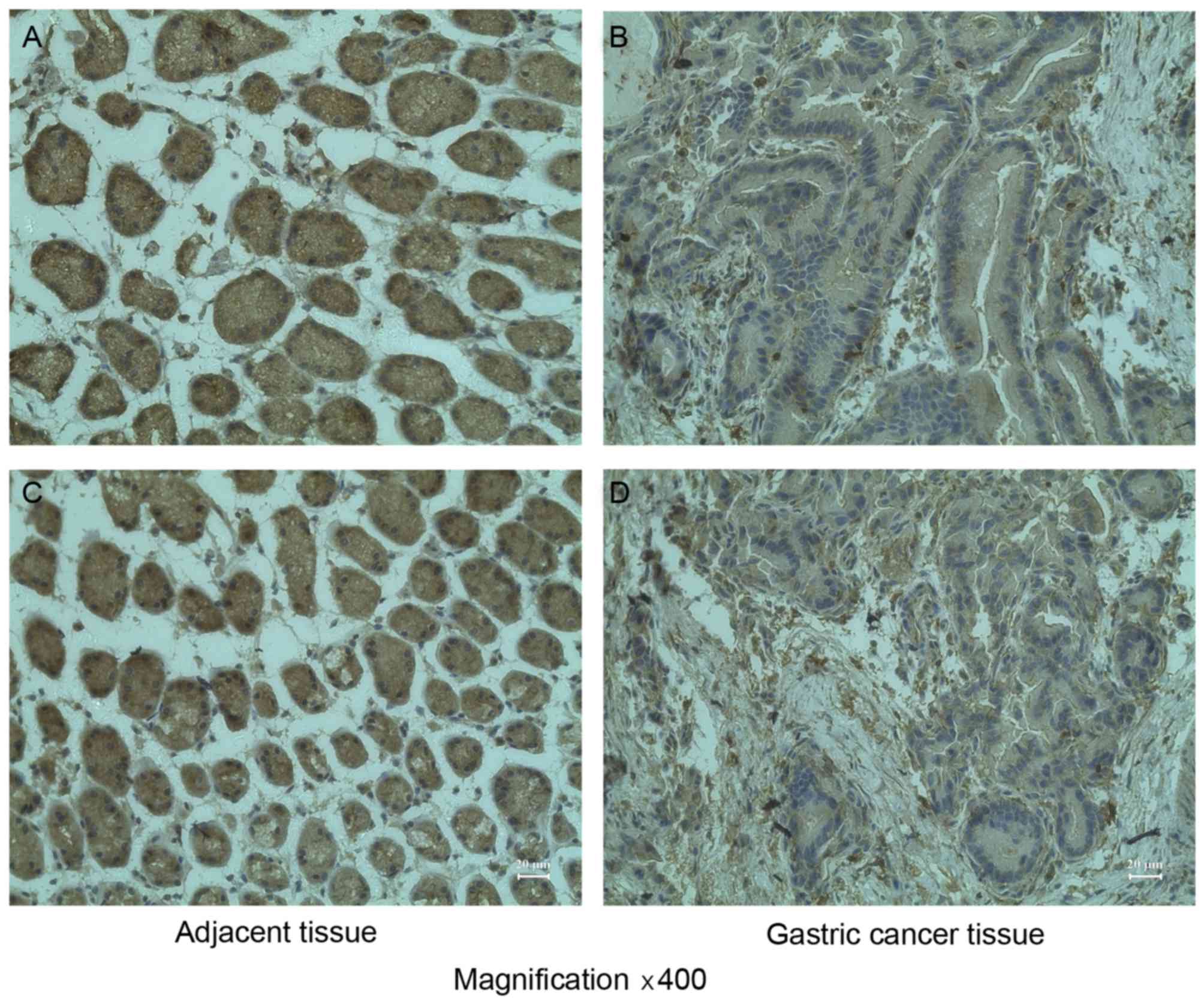
RASSF8 was immunohistochemically stained in the
tissue sections of gastric cancer and their corresponding adjacent
non-cancerous mucosa. It was demonstrated that the RASSF8 protein
was abundantly expressed in the upper glandular layer of the
superficial epithelium, while expression of RASSF8 protein was
significantly downregulated in gastric cancer tissue compared with
normal gastric mucosa (Table I;
Fig. 2; P<0.05).
 | Table I.RASSF8 expression detected by
immunohistochemistry in gastric tissues. |
Table I.
RASSF8 expression detected by
immunohistochemistry in gastric tissues.
|
|
| RASSF8 expression, n
(%) |
|---|
|
|
|
|
|---|
| Histological
type | Number of
patients | − | + | ++ | +++ |
|---|
| Normal gastric
mucosa | 10 | 0 (0) | 0 (0) | 1 (10) | 9
(90) |
| Gastric cancer | 10 | 0 (0) | 8
(80) | 2 (20) | 0 (0) |
RASSF8 attenuates gastric cancer cell
proliferation
To examine the role of RASSF8/miR-377 in
tumorigenesis of gastric cancer, the effect of RASSF8
overexpression and miR-377 on the proliferation of BGC-823 gastric
cancer cell lines was examined. Following infection with RASSF8
overexpression lentivirus the cells were transfected with miR-377
mimic or the miR377-NC. The RT-qPCR results demonstrated that
RASSF8 was significantly increased in the cells transfected with
RASSF8 overexpression lentivirus (Fig.
3A). The CCK-8 assay demonstrated that the overexpression of
RASSF8 significantly suppressed the proliferation of BGC-823 cells,
whereas miR-377 attenuated the effect of RASSF8 on cell
proliferation (Fig. 3B). The
proliferation of the RASSF8 overexpression with miR377 group
remained significantly lower compared with that of the negative
control group, in which cells were transfected by negative control
miRNA. The colony formation assay was performed to further confirm
the effect of RASSF8/miR-377 on gastric cancer cell proliferation,
and the data indicated that the overexpression of RASSF8
significantly decreased colony numbers in BGC-823 cell cultures,
which was not reversed by treatment with miR-377, which confirmed
the results of CCK-8 assay (Fig. 3C).
Furthermore, the cell cycle distribution was assessed by flow
cytometry. As presented in Fig. 3D,
the overexpression ofRASSF8decreased the proportion of cells in the
G1phase at 24, 48 and 72 h, compared with the control
(BGC-823 cells without transfection) or negative control.
Additionally, overexpression of RASSF8 increased the proportion of
cells in the S phase at 24 and 48 h, compared with the control or
negative control, and at 72 h, compared with the control, as well
slightly decreased at 72 h compared with the negative control and
led to cell cycle arrests in BGC-823 cells. Similarly, miR-377
attenuated rather than reversed the effect of RASSF8 on cell cycle.
Collectively, these results demonstrated that RASSF8 inhibits
gastric cancer cell growth.
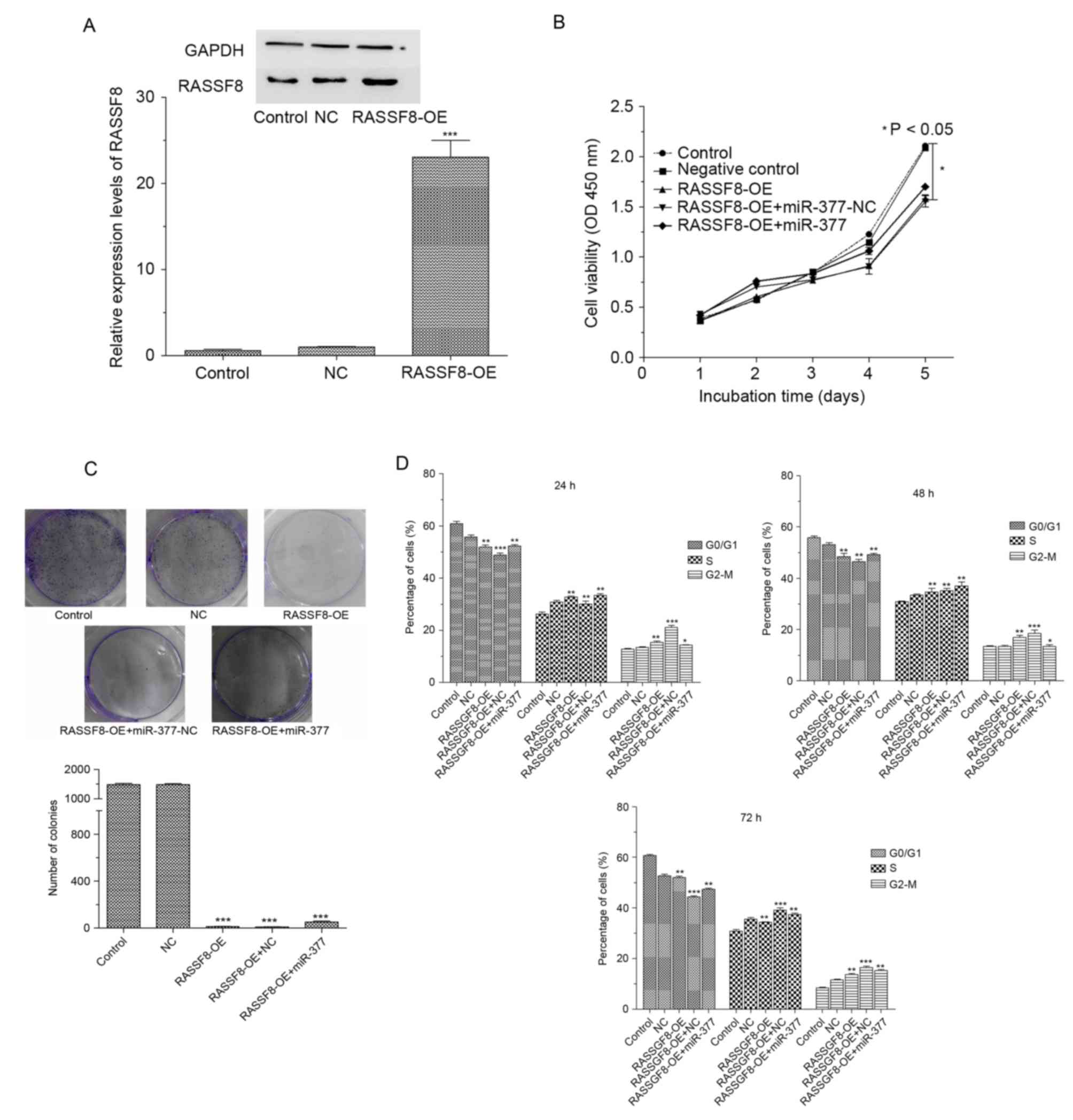 | Figure 3.RASSF8 inhibits cell growth in BGC-823
cells. (A) Western blot analyses of RASSF8 levels in control, NC
and RASSF8-OE transfected BGC-823 cells. (B) Effects of RASSF8 and
miR-377 on tumor cell proliferation at 0, 1, 2, 3, 4 and 5 day
using the CCK8 assay. (C) Representative quantification of colony
formation in BGC-827 cells transfected with control, negative
control, RASSF8-OE and RASSF8 overexpression with miR-377
(RASSF8-OE+ miR-377). (D) Cell cycle profiles of BGC-823 cells
transfected with control, negative control, RASSF8-OE and
RASSF8-OE+ miR-377. Data are presented as the mean ± standard
deviation of three independent experiments. *P<0.05, **P<0.01
and ***P<0.001 vs. control. RASSF8, Ras association domain
family 8; miR, microRNA; OE, overexpression; NC, negative
control. |
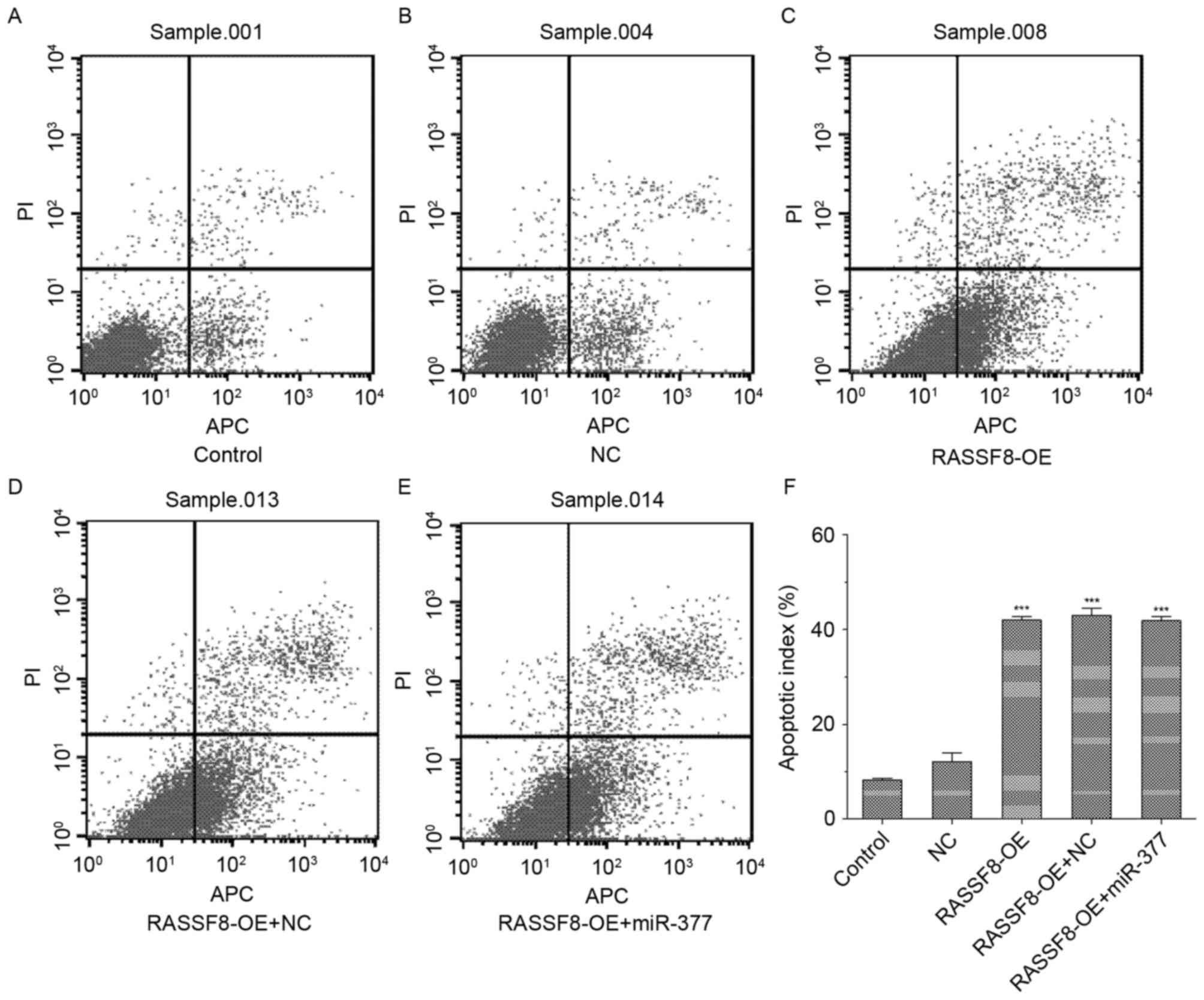
RASSF8 promotes gastric cancer cell
apoptosis
As presented in Fig.
4, RASSF8 overexpression significantly increased the rate of
apoptotic cells compared with the control (P<0.01). Furthermore,
no significant differences were identified between the RASSF8 and
RASSF8 combined with miR-377 groups regarding the rate of apoptotic
cells.
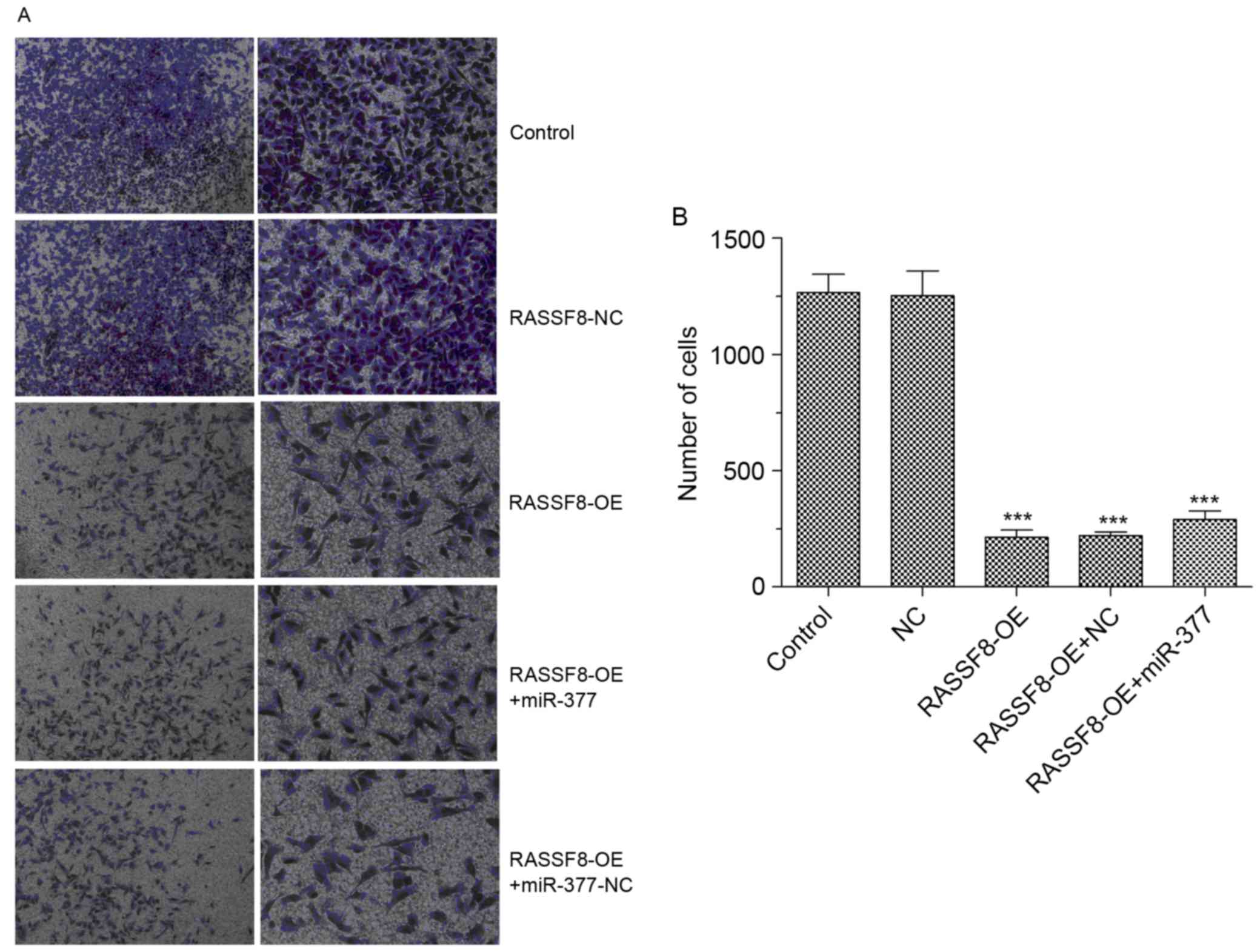
RASSF8 inhibits the invasion
capability of BGC-823 cells
To investigate the effect of RASSF8
ontheinvasivecapability of gastric cancer cells, Transwell assays
were performed in BGC-823 cells. Fig.
5A demonstrates the changes in theinvasive capacity of BGC-823
cells. BGC-823 cells infected with RASSF8 overexpression lentivirus
revealed significantly decreased invasive capabilities compared
with NC lentivirus infected-BGC-823 cells, with an 80% reduced
ability to invade through Matrigel membranes (Fig. 5B). Compared with the RASSF8
overexpression group, no significant differences were observed in
the invasive capabilities of BGC-823 cells transfected with RASSF8
and miR-377. Collectively, these results suggest that RASSF8
decreased BGC-823 cell invasion capability and miR-377 could not
reverse the effect of RASSF8 on BGC-823 cell invasion.
miR-377 directly targets RASSF8
To gain insight into the biological implications of
RASSF8/miR-377 on gastric cancer tumorigenesis, TargetScan, miRanda
and DIANA were used to identify putative human protein-coding gene
targets of miR-377. The tumor suppressor gene RASSF8 was predicted
to have miR-377-binding elements in its 3′-UTRs with high fidelity
scores (Fig. 6A). To assess whether
the predicted miR-377-binding sites in the 3′-UTR of the target
gene was responsible for miR-377 regulation, the 3′-UTR regions
downstream of a luciferase reporter gene were cloned and
transfected into the BGC-823 control, RASSF8 overexpression with
miR-377, and RASSF8 overexpression with miR-377-NC groups. The
luciferase activity of RASSF8-overexpressed cells was significantly
increased compared with the control. miR-377 significantly
suppressed the luciferase activity of RASSF8-overexpressed cells,
which was not observed following transfection with miR-377-NC
(Fig. 6B). However, miR-377
attenuated but not reversed the luciferase activity of cells
transfected with RASSF8 overexpression compared with control, which
suggested that miR-377 could decreased but not inhibit the effect
of overexpression of RASSF8 compared with control or negative
control.
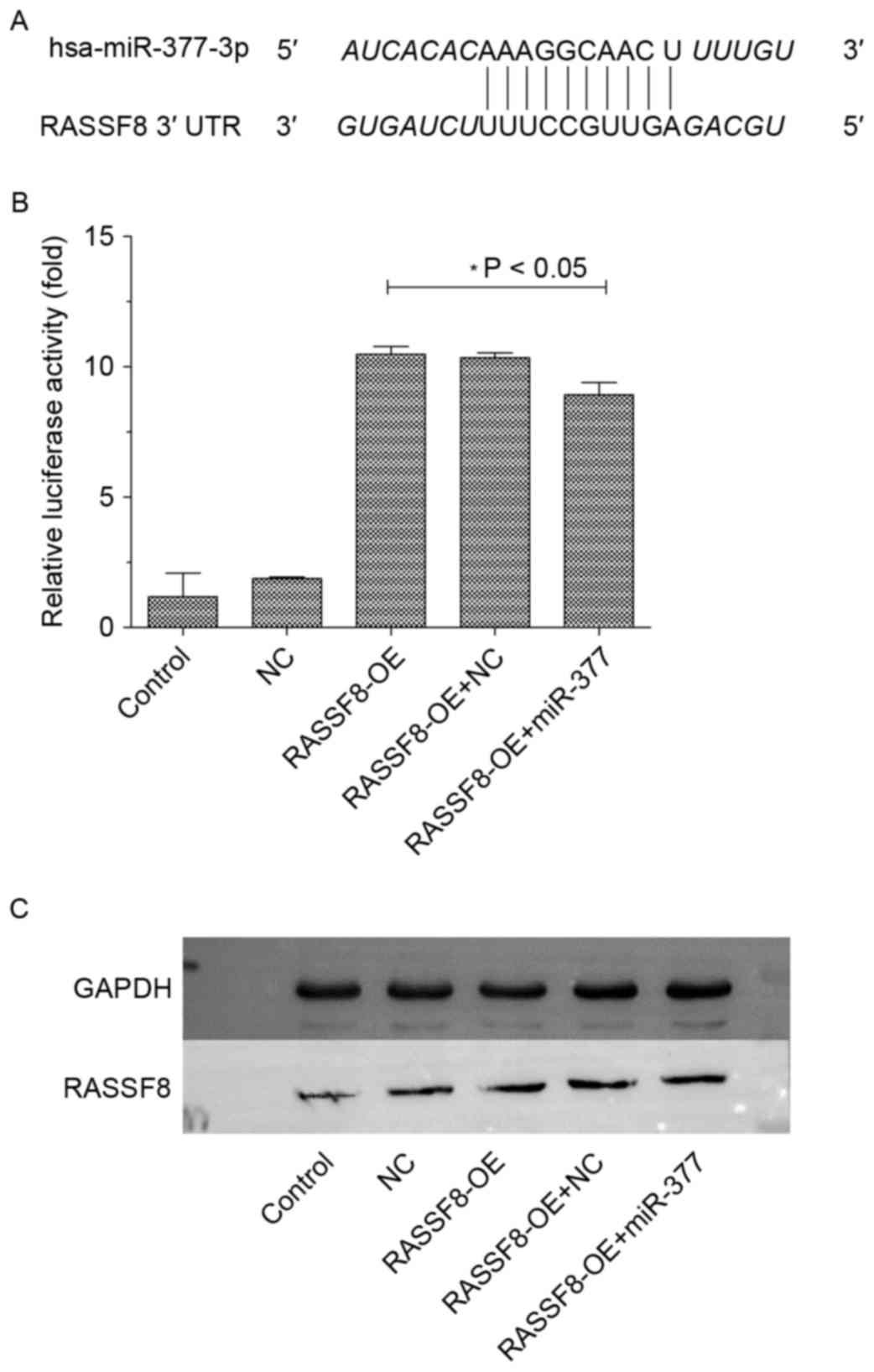 | Figure 6.RASSF8 is direct target of miR377. (A)
Sequences of the putative miR-377 binding sites in the 3′-UTR of
RASSF8. (B) Luciferase assay of BGC-823 cells transfected with
hRluc-RASSF8-3′UTR and control, negative control, RASSF8-OE,
RASSF8-OE+ miR-377, respectively. (C) Western blot analysis of
RASSF8 in BGC-823 cells transfected with control, negative control,
RASSF8-OE or RASSF8-OE+ miR-377. Data are presented as the mean ±
standard deviation of three independent experiments. RASSF8, Ras
association domain family 8; miR, microRNA; OE, overexpression; NC,
negative control; UTR, untranslated region. |
Discussion
In the current study, the expression of
RASSF8/miR-377 was investigated in gastric cancer cell lines and
tissue specimens. RASSF8 expression was identified to be
significantly upregulated in normal gastric mucosa adjacent tissue
compared with that in gastric cancer tissue, indicating that the
loss of RASSF8 expression may contribute to gastric carcinogenesis.
The current immunohistochemical data also revealed that RASSF8
protein was absent in the gastric cancer tissue and expressed in
normal adjacent gastric mucosa. However, miR-377 was significantly
upregulated in gastric cancer tissues compared with in their normal
adjacent mucosa, indicating that the upregulation of miR-377
expression may contribute to gastric carcinogenesis, which was
consistent with a previous report (24).
To further investigate the significance of
RASSF8/miR-377 in gastric cancer, RASSF8 was successfully cloned
and transfected into gastric cancer BGC-823 cells that
overexpressed RASSF8 protein, and investigated the effect of miR377
on RASSF8. Cell growth assays on BGC-823 cells were performed to
assess the effect of differential RASSF8/miR-377 expression on cell
viability. The data of the colony formation assay in BGC-823 cells
further confirmed the effect of RASSF8/miR377 on cell growth. These
results indicated that the upregulation of RASSF8 expression
significantly suppressed cell growth. The effect of RASSF8 on
cancer cell growth was attenuated, but not reversed by miR-377.
When assessing the cell cycle of BGC-823 cells by flow cytometry,
it was identified that RASSF8 overexpression resulted in a
population with more cells in the S/G2/M-phase, and
fewer cells in the G1-phase. However, cells transfected
with RASSF8 overexpression lentivirus and miR-377-mimic yielded a
population with more cells in the S-phase and fewer cells in
G2/M-phase compared with RASSF8 overexpression cells.
The following apoptosis data demonstrated that RASSF8 may induce
apoptosis in gastric cancer cells while miR-377 may inhibit
apoptosis. The results of the present study indicated that RASSF8
may suppress gastric cancer cell proliferation by arresting cells
in the S/G2/M-phase, whereas miR-377 increased the
percentage of cells in the S-phase, thus promoting gastric cancer
cell growth. Furthermore, miR-377 significantly increased the
percentage of cells in the G2/M-phase and decreased that
in the S-phase correspondingly at 72 h, which confirmed the effect
and mechanism of miR377 in gastric cancer cells proliferation. The
data of the Transwell assay in BGC-823 cells suggested that RASSF8
significantly inhibited BGC-823 cell invasion capability, which
could not be reversed bymiR377. As previously reported, RASSF8 is
essential for maintaining adherent junction function, and is
involved in regulating the migration of epithelial cells and
inhibiting cell proliferation (25,23), the
results of the present study suggested that RASSF8 can inhibit
BGC-823 cell invasion capability and suppress gastric cancer cell
proliferation.
To investigate the association between RASSF8 and
miR-377 in gastric cancer, a luciferase-reporter system was
developed to confirm whether miR-377 can regulate RASSF8. The
results suggested that miR-377 could target to RASSF8. In previous
reports, miR-377 was identified to be overexpressed in tumor tissue
and cell lines, in addition to being associated with tumorigenesis,
and poor prognosis (3,13), which is consistent with the
observations of the present study whereby BGC-823 cells exhibit
increased miR-377 expression. Furthermore, miR-377 has been
identified to increase the proliferation ability and cell invasion
capability and decrease the apoptosis of gastric cancer cells
(24). Notably, in the present
studymiR-377 was not able to reverse the effect of RASSF8 on
proliferation, apoptosis and cell invasion of BGC-823 cells.
In conclusion, the results of present study
demonstrated that RASSF8 was overexpressed in normal adjacent
gastric mucosa compared with gastric cancer tissue. However,
miR-377 was overexpressed in gastric cancer tissue and cell lines.
RASSF8 overexpression inhibited proliferation ability, apoptosis
and invasion capability of BGC-823 cells. It was suggested that
miR-377 directly targets RASSF8 through binding to its 3′-UTR, thus
increasing the cell proliferative and invasive capabilities, while
decreasing the apoptosis of gastric cancer cells. miR-377
attenuated, but did not reverse the effect of RASSF8 on
proliferation, apoptosis and cell invasion in BGC-823 cells. Thus,
the RASSF8 gene may represent a novel molecular target involved in
gastric cancer development and may offer a promising therapeutic
approach for the treatment of patients with gastric cancer.
References
|
1
|
Gibson CJ, Britton KA, Miller AL and
Loscalzo J: Clinical problem-solving. Out of the blue. N Engl J
Med. 370:1742–1748. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J
and Sung JJ: MicroRNA dysregulation in gastric cancer: A new player
enters the game. Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Catalano V, Labianca R, Beretta GD, Gatta
G, de Braud F and Van Cutsem E: Gastric cancer. Crit Rev Oncol
Hematol. 71:127–164. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gomceli I, Demiriz B and Tez M: Gastric
carcinogenesis. World J Gastroenterol. 18:5164–5170.
2012.PubMed/NCBI
|
|
5
|
Beckman JD, Chen C, Nguyen J, Thayanithy
V, Subramanian S, Steer CJ and Vercellotti GM: Regulation of heme
oxygenase-1 protein expression by miR-377 in combination with
miR-217. J Biol Chem. 286:3194–3202. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mascaux C, Iannino N, Martin B, Paesmans
M, Berghmans T, Dusart M, Haller A, Lothaire P, Meert AP, Noel S,
et al: The role of RAS oncogene in survival of patients with lung
cancer: A systematic review of the literature with meta-analysis.
Br J Cancer. 92:131–139. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ellis CA and Clark G: The importance of
being K-Ras. Cell Signal. 12:425–434. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Falvella FS, Manenti G, Spinola M,
Pignatiello C, Conti B, Pastorino U and Dragani TA: Identification
of RASSF8 as a candidate lung tumor suppressor gene. Oncogene.
25:3934–3938. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Malumbres M and Barbacid M: RAS oncogenes:
The first 30 years. Nat Rev Cancer. 3:459–465. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Downward J: Targeting RAS signalling
pathways in cancer therapy. Nat Rev Cancer. 3:11–22. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wohlgemuth S, Kiel C, Krämer A, Serrano L,
Wittinghofer F and Herrmann C: Recognizing and defining true Ras
binding domains I: Biochemical analysis. J Mol Biol. 348:741–758.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sherwood V, Manbodh R, Sheppard C and
Chalmers AD: RASSF7 is a member of a new family of RAS association
domain-containing proteins and is required for completing mitosis.
Mol Biol Cell. 19:1772–1782. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zehavi L, Schayek H, Jacob-Hirsch J, Sidi
Y, Leibowitz-Amit R and Avni D: MiR-377 targets E2F3 and alters the
NF-kB signaling pathway through MAP3K7 in malignant melanoma. Mol
Cancer. 14:682015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ueda T, Volinia S, Okumura H, Shimizu M,
Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: A microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rachagani S, Kumar S and Batra SK:
MicroRNA in pancreatic cancer: Pathological, diagnostic and
therapeutic implications. Cancer Lett. 292:8–16. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang QZ, Xu W, Habib N and Xu R: Potential
uses of microRNA in lung cancer diagnosis, prognosis, and therapy.
Curr Cancer Drug Targets. 9:572–594. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Balch C, Fang F, Matei DE, Huang TH and
Nephew KP: Minireview: Epigenetic changes in ovarian cancer.
Endocrinology. 150:4003–4011. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Faber C, Kirchner T and Hlubek F: The
impact of microRNAs on colorectal cancer. Virchows Arch.
454:359–367. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mott JL: MicroRNAs involved in tumor
suppressor and oncogene pathways: Implications for hepatobiliary
neoplasia. Hepatology. 50:630–637. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu C, Guo X, Wang W, Wang Y, Shan Y, Zhang
B, Song W, Ma S, Ge J, Deng H and Zhu M:
N-Acetylgalactosaminyltransferase-14 as a potential biomarker for
breast cancer by immunohistochemistry. BMC Cancer. 10:1232010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lang Y, Xu S, Ma J, Wu J, Jin S, Cao S and
Yu Y: MicroRNA-429 induces tumorigenesis of human non-small cell
lung cancer cells and targets multiple tumor suppressor genes.
Biochem Biophys Res Commun. 450:154–159. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wen X, Wu JQ, Peng W, Feng JF and Tang JH:
MicroRNA-377 predicts poor clinical outcome of gastric cancer and
induces tumorigenesis by targeting multiple tumor-suppressor genes.
Oncol Rep. 34:203–210. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lock FE, Underhill-Day N, Dunwell T,
Matallanas D, Cooper W, Hesson L, Recino A, Ward A, Pavlova T,
Zabarovsky E, et al: The RASSF8 candidate tumor suppressor inhibits
cell growth and regulates the Wnt and NF-kappaB signaling pathways.
Oncogene. 29:4307–4316. 2010. View Article : Google Scholar : PubMed/NCBI
|