Introduction
Intrahepatic cholangiocarcinoma (IHCC) is the second
most common primary hepatic malignancy. The incidence rate and
cumulative mortality rate of IHCC have been steadily increasing
during past decades (1,2). Despite many advances having been made in
the diagnosis and treatment strategies, IHCC is still a great
challenge for doctors because of its heterogeneity and
aggressiveness. Due to its insidious development process, the vast
majority of IHCC patients are diagnosed at an advanced stage, with
no access to radical surgical resection. Moreover, the recurrence
rate of IHCC patients after surgical resection is high and advanced
IHCCs are for the most part unresponsive to systemic chemotherapy
and radiotherapy regimens (1). The
current overall 5-year survival rate range from 15 to 40%, and
patients with unresectable IHCC will typically die within 12–24
months of diagnosis (3). Therefore,
the molecular mechanisms of IHCC should be further elucidated,
which may shed new light on identifying more effective diagnostic
and prognostic markers and therapeutic targets.
Long non-coding RNAs (LncRNAs), defined as different
types of RNA polymerase II-transcribed molecules with size greater
than 200 nt in length, have been discovered to regulate cellular
biology at translational and posttranslational levels, the detailed
mechanisms include recruiting transcription factors or
chromatin-modifying complexes to their DNA targets, forming
heterogeneous nuclear ribonucleoprotein complexes, acting as decoys
to sequester RNA-binding proteins and microRNAs, or directly
interacting with RNAs and DNAs by base pairing (4). Recent literatures have indicated that
lncRNAs are involved in cancer initiation and progression, and
numerous lncRNAs have been implicated to be deregulated in cancers
and serve as essential regulators in many typical cancer biology
pathways (5). Thus, lncRNAs-based
therapeutics are considered as novel strategies in cancer treatment
in the future (6).
LncRNA colorectal neoplasia differentially expressed
(CRNDE), located on the long arm of chromosome 16 (16q12.2) of the
human genome, was originally identified as a lncRNA in human
colorectal cancer (7). The clinical
significance of lncRNA CRNDE has been well studied in multiple
tumors, such as colorectal cancer (7–9), renal
cell carcinoma (10), and breast
cancer (11). However, the potential
role of CRNDE in IHCC has not been investigated at present. This
study aims to detect the expression level and clinical significance
of CRNDE in IHCC, the potential mechanisms of CRNDE exerting its
function were also investigated.
Materials and methods
Clinical samples collection
A total of 118 IHCC tissues and paired adjacent
normal bile duct tissues were collected from primary IHCC patients
who underwent surgery at the Department of General Surgery, the
People's Hospital of Binzhou, Binzhou, Shandong Province, China.
Informed consents were obtained from all the participants. No
anti-cancer treatment was conducted for any patient prior to
surgery. Obtained IHCC tissues and paired adjacent normal bile duct
tissues were identified by pathological examination. This study
protocol was approved by the Ethics Committee of the People's
Hospital of Binzhou.
Cell culture and transfection
Normal human intrahepatic biliary epithelial cell
(HIBEpic) and four human IHCC cells (HuCCT1, RBE, HCCC9810 and
HUH28) were all purchased from the American Type Culture
Collection. These cell lines were routinely cultured in DMEM
(HIBEpic, HuCCT1, HUH28 and RBE) or RPMI-1640 (HCCC9810),
supplemented with 10% fetal bovine serum (FBS) (Sigma, St. Louis,
MO, USA), 100 U/ml penicillin sodium, and 100 mg/ml streptomycin
sulfate. All cells were cultured in a humidified atmosphere
containing 5% CO2 at 37°C.
HuCCT1 cells were transfected with siRNAs targeting
CRNDE (siCRNDE-1 and siCRNDE-2) or a scrambled negative control
(siNC) (GenePharma, Shanghai, China). HCCC9810 cells were
transfected with CRNDE overexpressing vector (pcDNA3.1-CRNDE) or
its control vector (pcDNA3.1-Vector) (GenePharma, China). The
transfection was conducted with Lipofectamine 2000 (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer's
protocol.
RNA extraction and RT-qPCR
RNA extraction from cell lines and clinical tissues
was conducted with TRIzol reagent (Invitrogen, USA). To determine
the mRNA expression level of target genes, SYBR Green
fluorescent-based RT-qPCR was performed with an ABI PRISM 7900HT
sequence detection system. Relative mRNA expression levels were
calculated by the 2−ΔΔCT method. GAPDH was used as
internal control. Three independent experiments were conducted and
each reaction was performed in triplicate. The primers were as
follows: E-cadherin: 5′-CATTGCCACACATACACTCTCTTCT-3′ (forward);
5′-CGGTTACCGTGATCAAAATCTC-3′ (reverse); α-catenin:
5′-GAGCTGTCTACGCAAGTCCC-3′ (forward) and 5′-TTTCGGAGTACATGGGCAAT-3′
(reverse). Vimentin: 5′-GGAACAGCATGTCCAAATCG-3′ (forward);
5′-GCACCTGTCTCCGGTACTCA-3′ (reverse). N-cadherin:
5′-GGTGGAGGAGAAGAAGACCAG-3′ (forward) and 5′-GGCATCAGGCTCCACAGT-3′
(reverse).
Cell proliferation assay
The Cell Counting Kit-8 (CCK-8) (Dojindo Molecular
Technologies, Inc., Kumamoto, Japan) and colony formation assay
were employed to determine the proliferation of HuCCT1 and HCCC9810
cells in accordance with the manufacturer's instructions. Briefly,
HuCCT1 and HCCC9810 cells were seeded into 96-well plates. After
transfection, a total of 10 µl CCK-8 reagent was added to each well
at designated time points (24, 48, 72, 96 h). Then, cells were
incubated for another 3 h followed by measuring the absorbance at
450 nm using an enzyme-linked immunosorbent assay plate reader.
As for the colony formation assay, transfected
HuCCT1 and HCCC9810 cells were seeded in 6 cm plates and then
subjected to incubator for 7 days in an atmosphere of 5%
CO2 at 37°C. Formed colonies were counted with light
microscopy. All the experiments were performed in triplicate and
repeated at least three times.
Cell migration and invasion assay
Cell migration ability was assessed by Transwell
assay using 8 µm-pore-size chambers (BD, San Jose, CA, USA). Cells
were seeded in the upper chamber, which was filled with 250 µl
serum-free medium. The lower chamber was filled with 600 µl
complete medium as chemoattractant. Then, the chambers were
subjected to incubation at 37°C for 36 h. Then, cells were fixed
with 4% paraformaldehyde and stained with crystal violet. Finally,
cells invaded to the lower surface of the filter were counted in
five random fields using microscope. To measure the invasion
ability of IHCC cells, Matrigel assay was conducted as the
Transwell assay, except that each chamber was pre-coated with
matrigel (BD, USA) before cell seeding. All the experiments were
performed in triplicate and repeated at least three times.
Western blot analysis
Cells were harvested and rinsed with cold
phosphate-buffered saline. Then, the total proteins were extracted
with RIPA buffer (Thermo Scientific, Rockford, IL, USA)
supplemented with protease inhibitors. After protein
quantification, an equal amount of protein samples were size
fractionated by SDS-PAGE, and separated proteins were
electrotransferred onto PVDF membranes (Invitrogen, USA) in
transfer buffer. Membranes were then incubated overnight at 4°C
with primary antibody, followed by incubation with HRP-conjugated
secondary antibody. Antibody-antigen complexes were detected by
incubating with ECL detection reagent (Amersham Biosciences, Castle
Hill, Australia). ImageJ software was utilized to determine the
band intensity.
Statistical analysis
SPSS 17.0 software was utilized for statistical
analyses (SPSS, Chicago, IL, USA). Data was presented as mean ±
standard deviation (Mean ± SD). Statistically analysis was
performed using the Student's t-test for continuous variables and
Chi-square test for categorical variables. All tests were
two-tailed. By utilizing the mean CRNDE expression level as the
cutoff value, all involved patients were divided into the low CRNDE
expression group and the high CRNDE expression group. Subquently,
the Kaplan-Meier test was conducted to evaluate the survival data
and the log-rank test was performed to compare the cumulative
survival differencebetween the low CRNDE expression group and the
high CRNDE expression group. The statistical significance level was
set at 0.05.
Results
LncRNA CRNDE is notably overexpressed
in IHCC and significantly correlates with IHCC clinical
progression
The expression of lncRNA CRNDE was evaluated in IHCC
cell lines and clinical specimens. The result of RT-qPCR assay
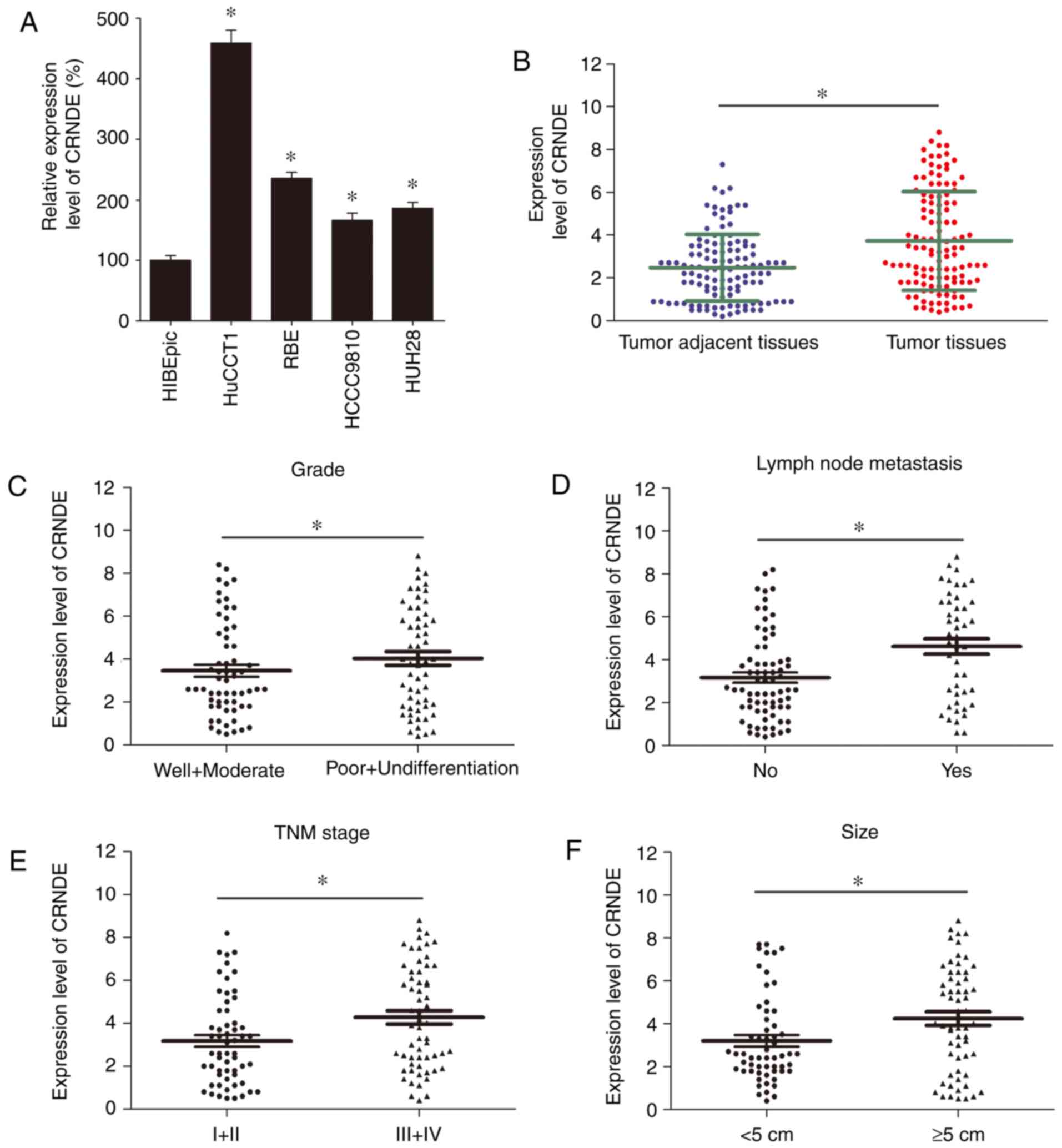
showed that CRNDE was remarkably up-regulated in IHCC cell lines
compared with HIBEpics (Fig. 1A).
Further investigation of CRNDE expression in 118 IHCC clinical
specimens revealed that tumor tissues displayed much higher
expression level relative to tumor adjacent tissues (Fig. 1B). Besides, the expression levels of
CRNDE in IHCC patients with poor and undifferentiated grade, lymph
node metastasis, tumor-nodes-metastasis (TNM) stage III and stage
IV and tumor size no less than 5 cm were much higher than those
with well and moderate grade, without lymph node metastasis, TNM
stage I and stage II and tumor size less than 5 cm, respectively
(Fig. 1C-F).
Furthermore, with mean CRNDE expression level
serving as the cutoff value, all IHCC patients were divided into
the high CRNDE expression group (n=51) and the low CRNDE expression
group (n=67). Statistical analysis between CRNDE expression and
IHCC clinicopathological parameters found that high CRNDE
expression was obviously correlated with poor and undifferentiated
grade (P=0.046), lager tumor size (P=0.001), lymph node metastasis
(P=0.001), and advanced TNM stage (P=0.024). (Table I) These findings suggested that CRNDE
overexpression could promote the clinical progression of IHCC.
 | Table I.Correlation between lncRNA CRNDE
expression and IHCC clinicopathological characteristics. |
Table I.
Correlation between lncRNA CRNDE
expression and IHCC clinicopathological characteristics.
| Parameters | No. of patients | CRNDE high/low | P-value |
|---|
| Age |
|
| 0.276 |
| <60
years | 58 | 28/30 |
|
| ≥60
years | 60 | 23/37 |
|
| Gender |
|
| 0.306 |
| Male | 80 | 32/48 |
|
|
Female | 38 | 19/19 |
|
| TBIL |
|
| 0.453 |
| ≥20.4
µmol/l | 81 | 33/47 |
|
| <20.4
µmol/l | 37 | 18/19 |
|
| AFP |
|
| 0.374 |
| ≥20
ng/ml | 77 | 31/46 |
|
| <20
ng/ml | 41 | 20/21 |
|
| CEA |
|
| 0.553 |
| ≥5
µg/l | 45 | 21/24 |
|
| <5
µg/l | 73 | 30/43 |
|
| CA19-9 |
|
| 0.172 |
| <100
U/ml | 54 | 27/27 |
|
| ≥100
U/ml | 64 | 24/40 |
|
| Cirrhosis |
|
| 0.298 |
| No | 56 | 27/29 |
|
| Yes | 62 | 24/38 |
|
| Satellitosis |
|
| 0.499 |
| Yes | 31 | 15/16 |
|
| No | 87 | 36/51 |
|
| Perineural
invasion |
|
| 0.187 |
| Yes | 43 | 22/21 |
|
| No | 75 | 29/46 |
|
| Lymphovascular
invasion |
|
| 0.742 |
| No | 42 | 19/23 |
|
| Yes | 76 | 32/44 |
|
| Grade |
|
| 0.046 |
|
Well+moderate | 61 | 21/40 |
|
|
Poor+undifferentiation | 57 | 30/27 |
|
| Size |
|
| 0.001 |
| <5
cm | 58 | 16/42 |
|
| ≥5
cm | 60 | 35/25 |
|
| T stage |
|
| 0.050 |
|
T1+T2 | 85 | 32/53 |
|
|
T3+T4 | 33 | 19/14 |
|
| N stage |
|
| 0.001 |
| N0 | 73 | 23/50 |
|
| N1 | 45 | 28/17 |
|
| M stage |
|
| 0.114 |
| M0 | 111 | 46/75 |
|
| M1 | 7 | 5/2 |
|
| TNM stage |
|
| 0.024 |
|
I+II | 58 | 19/39 |
|
|
III+IV | 60 | 32/28 |
|
High CRNDE expression is an
independent risk factor of IHCC poor prognosis
For further verifying the clinical significance of
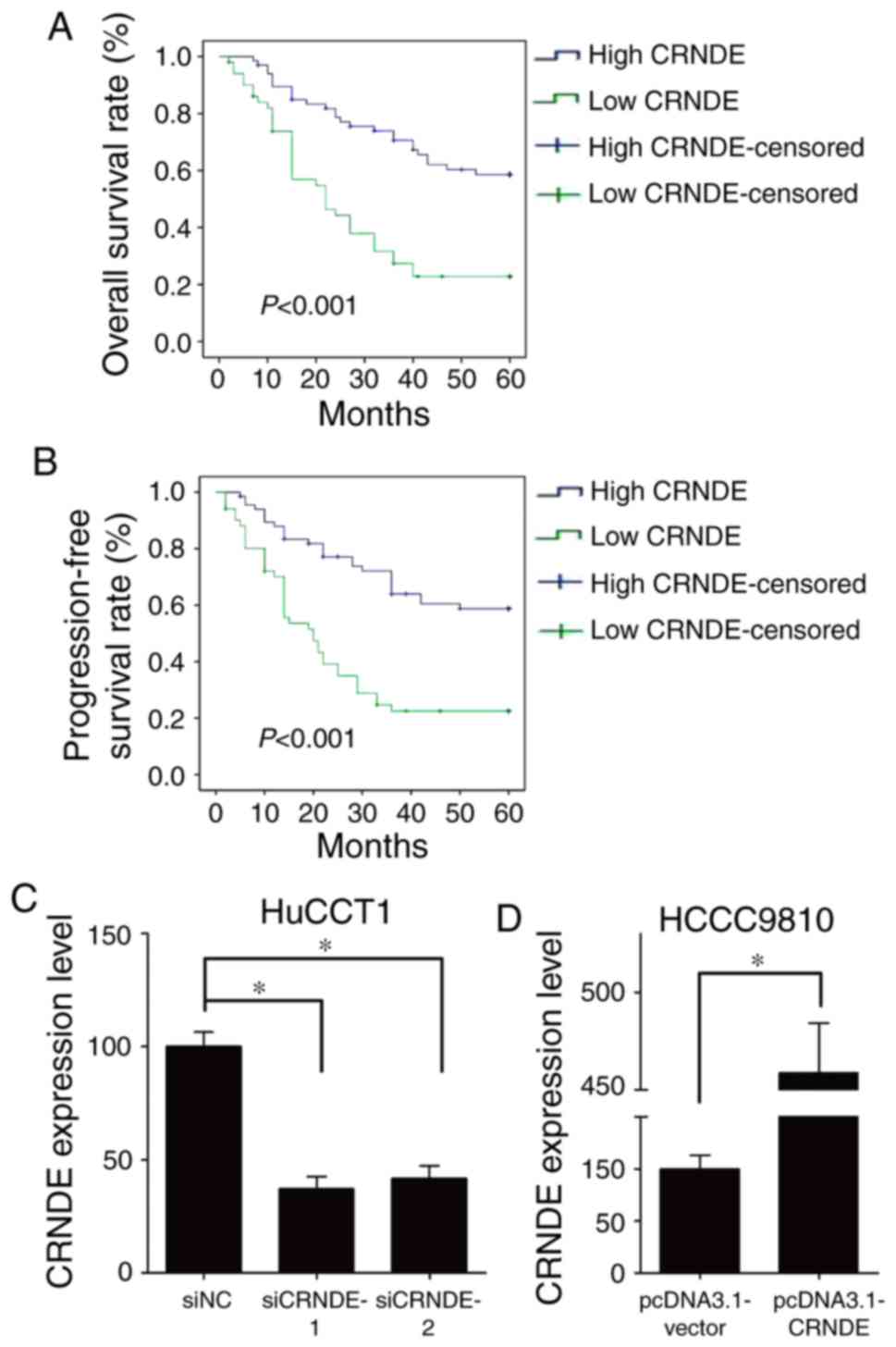
high CRNDE expression in IHCC, the prognostic value of CRNDE was
evaluated. Intriguingly, high CRNDE expression was discovered to
correlate with poorer overall survival (OS) rate (P<0.001),
(Fig. 2A) and shorter
progression-free survival (PFS) period (P<0.001) (Fig. 2B).
In addition, the risk factors of IHCC OS and PFS
were statistically analyzed by univariate COX regression analysis
and multivariate COX regression analysis. Univariate analysis
revealed 5 risk factors for worse OS (Table II) and 6 risk factors for poor PFS
(Table III) in IHCC patients,
respectively. By further analyzing these factors with multivariate
analysis, lymphovascular invasion (HR=0.530, 95% CI=0.313–1.224,
P=0.018), advanced TNM stage (HR=3.695, 95% CI=1.357–10.060,
P=0.011) and high CRNDE expression (HR=1.309, 95% CI=1.160–1.478,
P<0.001) were highlighted as independent risk factors of poor OS
in IHCC. (Table II) Interestingly,
these three factors, lymphovascular invasion (HR=0.432, 95%
CI=0.255–0.734, P=0.002), advanced TNM stage (HR=2.484, 95%
CI=1.104–5.586, P=0.028) and high CRNDE expression (HR=1.305, 95%
CI=1.168–1.459, P<0.001), were also identified as independent
risk factors of shorter PFS period. (Table III).
 | Table II.Univariate and multivariate analysis
of clinicopathologic features for overall survival of IHCC
patients. |
Table II.
Univariate and multivariate analysis
of clinicopathologic features for overall survival of IHCC
patients.
|
| Univariate
analysis |
| Multivariate
analysis |
|
|---|
|
|
|
|
|
|
|---|
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age ≥60 years vs.
<60 years | 1.125 | 0.685–1.847 | 0.643 |
|
|
|
| Gender female vs.
male | 0.627 | 0.355–1.108 | 0.108 |
|
|
|
| TBIL ≥20.4 µmol/l
vs. <20.4 µmol/l | 1.362 | 0.818–2.266 | 0.235 |
|
|
|
| AFP ≥20 ng/ml vs.
<20 ng/ml | 1.659 | 1.002–2.748 | 0.049 | 1.193 | 0.682–2.086 | 0.536 |
| CEA ≥5 µg/l vs.
<5 µg/l | 1.333 | 0.809–2.196 | 0.259 |
|
|
|
| CA19-9 <100 U/ml
vs. ≥100 U/ml | 0.836 | 0.510–1.372 | 0.479 |
|
|
|
| Cirrhosis yes vs.
no | 1.102 | 0.672–1.808 | 0.701 |
|
|
|
| Satellitosis yes
vs. no | 1.369 | 0.799–2.346 | 0.254 |
|
|
|
| Perineural invasion
yes vs. no | 1.182 | 0.708–1.976 | 0.522 |
|
|
|
| Lymphovascular
invasion no vs. yes | 0.462 | 0.281–0.760 | 0.002 | 0.530 | 0.313–1.224 | 0.018 |
| Grade
poor+undifferentiated vs. well+moderate | 1.629 | 0.984–2.696 | 0.058 |
|
|
|
| Size ≥5 cm vs.
<5 cm | 1.426 | 0.867–2.347 | 0.162 |
|
|
|
| T stage (T3+T4) vs.
(T1+T2) | 1.444 | 1.030–2.024 | 0.033 | 0.621 | 0.315–1.224 | 0.169 |
| N stage N1 vs.
N0 | 2.982 | 1.798–4.945 | <0.001 | 1.042 | 0.473–2.295 | 0.918 |
| M stage M1 vs.
M0 | 2.947 | 1.054–8.243 | 0.039 | 1.414 | 0.468–4.272 | 0.539 |
| TNM stage (III+IV)
vs. (I+II) | 3.708 | 2.126–6.468 | <0.001 | 3.695 | 1.357–10.060 | 0.011 |
| CRNDE high vs.
low | 1.365 | 1.221–1.525 | <0.001 | 1.309 | 1.160–1.478 | <0.001 |
 | Table III.Univariate and multivariate analysis
of clinicopathologic features for progrerssion-free survival of
IHCC patients. |
Table III.
Univariate and multivariate analysis
of clinicopathologic features for progrerssion-free survival of
IHCC patients.
|
| Univariate
analysis |
| Multivariate
analysis |
|
|---|
|
|
|
|
|
|
|---|
| Parameters | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age ≥60 years vs.
<60 years | 1.152 | 0.704–1.885 | 0.574 |
|
|
|
| Gender female vs.
male | 0.676 | 0.388–1.180 | 0.168 |
|
|
|
| TBIL ≥20.4 µmol/l
vs. <20.4 µmol/l | 1.227 | 0.736–2.045 | 0.434 |
|
|
|
| AFP ≥20 ng/ml vs.
<20 ng/ml | 1.605 | 0.972–2.648 | 0.064 |
|
|
|
| CEA ≥5 µg/l vs.
<5 µg/l | 1.274 | 0.776–2.093 | 0.338 |
|
|
|
| CA19-9 <100 U/ml
vs. ≥100 U/ml | 0.810 | 0.496–1.323 | 0.399 |
|
|
|
| Cirrhosis yes vs.
no | 1.134 | 0.693–1.853 | 0.617 |
|
|
|
| Satellitosis yes
vs. no | 1.465 | 0.863–2.478 | 0.158 |
|
|
|
| Perineural invasion
yes vs. no | 1.206 | 0.727–2.002 | 0.469 |
|
|
|
| Lymphovascular
invasion no vs. yes | 0.430 | 0.262–0.704 | 0.001 | 0.432 | 0.255–0.734 | 0.002 |
| Grade
poor+undifferentiated vs. well+moderate | 1.679 | 1.107–2.770 | 0.043 | 0.792 | 0.436–1.442 | 0.446 |
| Size ≥5 cm vs.
<5 cm | 1.474 | 0.898–2.418 | 0.125 |
|
|
|
| T stage (T3+T4) vs.
(T1+T2) | 1.275 | 0.881–1.845 | 0.198 |
|
|
|
| N stage N1 vs.
N0 | 2.800 | 1.698–4.617 | <0.001 | 1.294 | 0.646–2.590 | 0.467 |
| M stage M1 vs.
M0 | 2.896 | 1.039–8.084 | 0.042 | 1.311 | 0.437–3.928 | 0.629 |
| TNM stage (III+IV)
vs. (I+II) | 3.438 | 1.994–5.928 | <0.001 | 2.484 | 1.104–5.586 | 0.028 |
| CRNDE high vs.
low | 1.366 | 1.224–1.525 | <0.001 | 1.305 | 1.168–1.459 | <0.001 |
Overexpression of lncRNA CRNDE could
promote the proliferation of IHCC cells
To detect the functional role of CRNDE in IHCC
cells, the expression of CRNDE was silenced with siRNAs in HuCCT1
cells (Fig. 2C), and ectopic
overexpressed in HCCC9810 cells (Fig.
2D), respectively. Then, the effect of CRNDE on IHCC
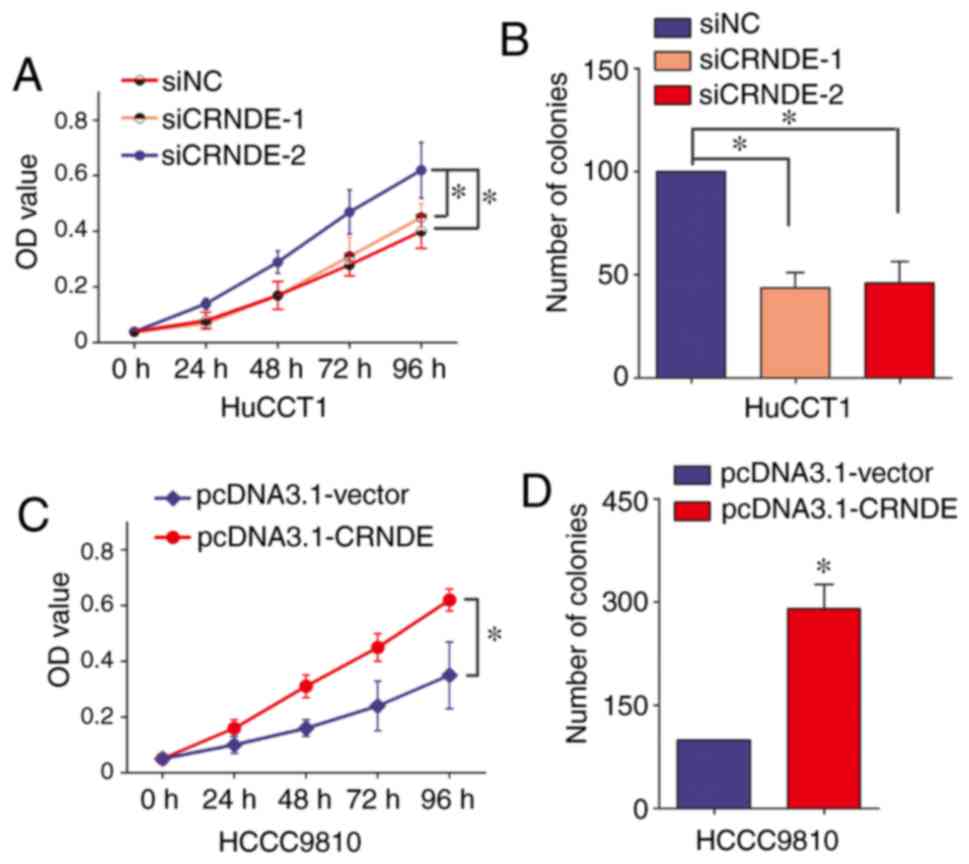
proliferation was evaluated with CCK-8 assay and colony formation
assay. CRNDE silencing obviously inhibited the proliferation of
HuCCT1 cells on CCK-8 assay and colony formation assay (Fig. 3A and 3B). Consistently, the
proliferation ability of HCCC9810 cells was notably increased with
CRNDE ectopic overexpression (Fig. 3C and
D). It was concluded that CRNDE could promote the proliferation
of IHCC cells.
LncRNA CRNDE promotes IHCC metastasis
by regulating EMT
Metastasis is the final stage of caner-caused death,
the detailed mechanisms of tumor metastasis have been investigated
for several decades but remain obscure at present. The present
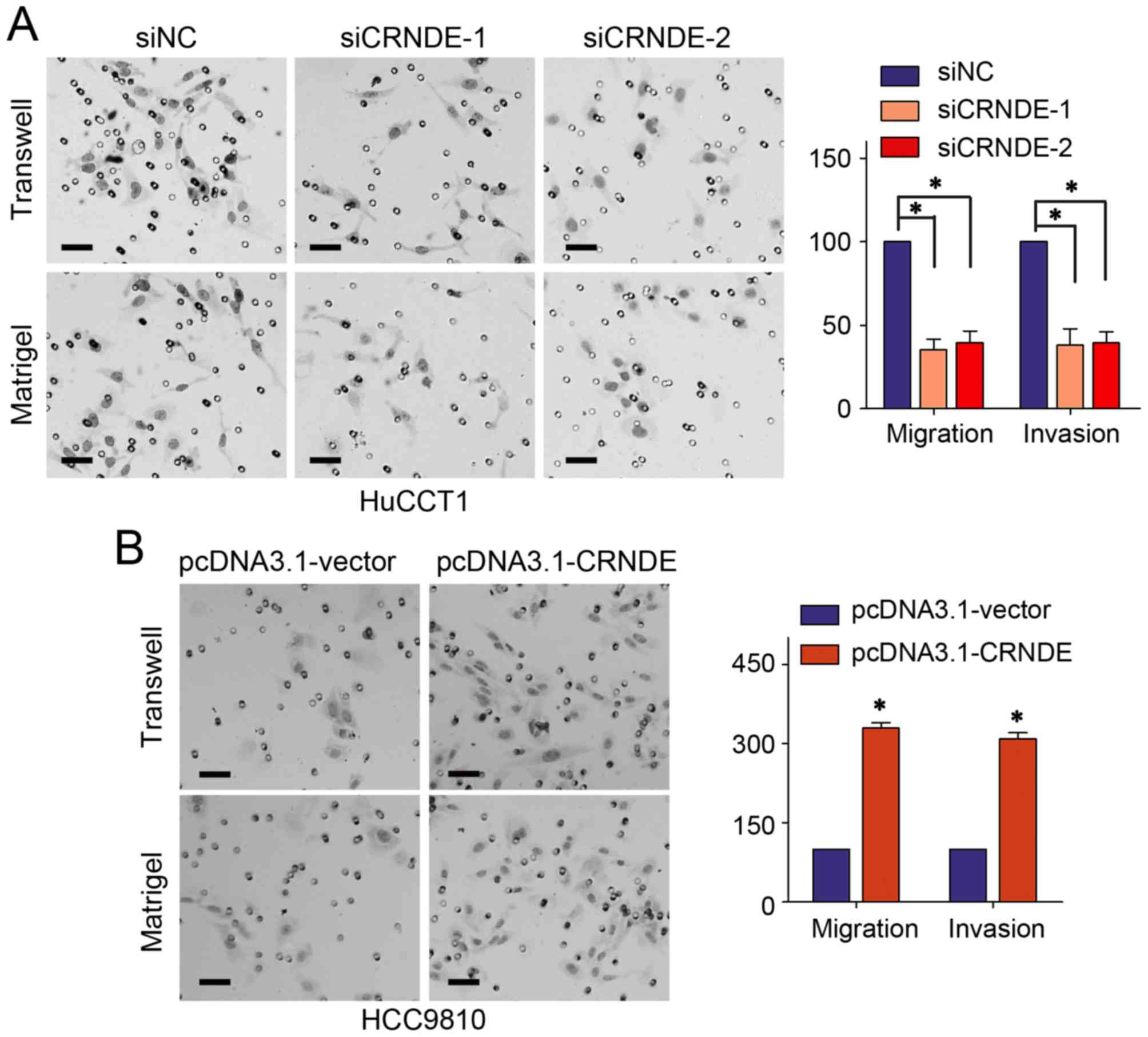
study confirmed CRNDE interference in HuCCT1 cells could repress
the migration and invasion abilities on Transwell assay and
Matrigel assay, respectively (Fig.
4A). While the motility of HCCC9810 cells was obviously
increased with CRNDE up-regulation (Fig.
4B).
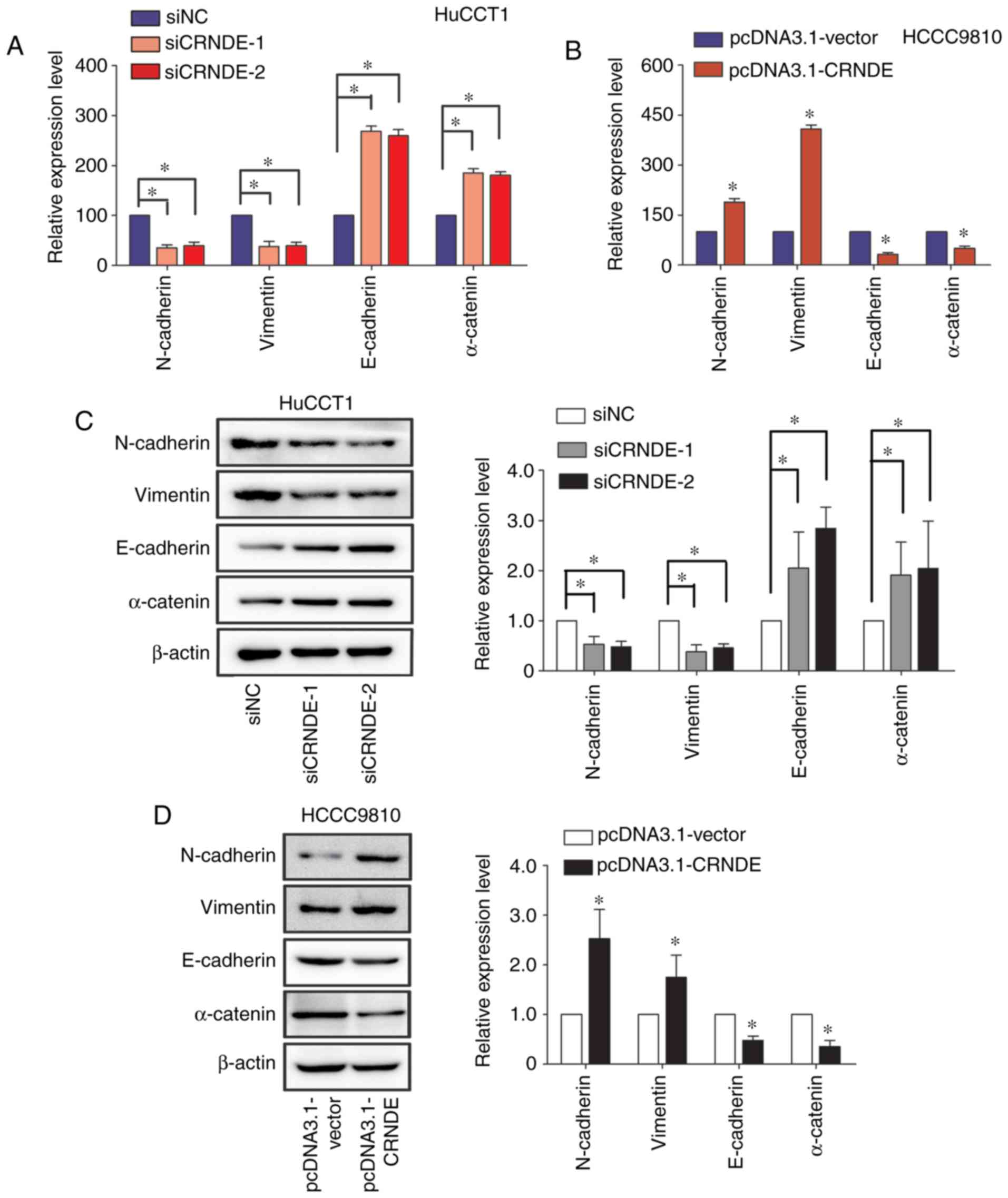
Epithelial-mesenchymal transition (EMT) is an
essential process in the initiation and progression of cancer
metastasis (12,13). This study investigated the role of
CRNDE on IHCC EMT by evaluating the expression of epithelial
markers (E-cadherin and α-catenin) and mesenchymal markers
(N-cadherin and Vimentin) (14). As
expected, the mRNAs levels of mesenchymal markers were notably
reduced with CRNDE knock-down in HuCCT1 cells, while the mRNAs
levels of epithelial markers were greatly improved with CRNDE
silencing (Fig. 5A). Consistently,
CRNDE overexpression in HCCC9810 cells increased the mRNAs levels
of mesenchymal markers, accompanied with down-regulation of
epithelial markers (Fig. 5B). The
results of the western blot assay further confirmed the findings of
the RT-qPCR assay (Fig. 5C and D).
Together, CRNDE could promote the metastasis of IHCC cells by
facilitating EMT.
Discussion
Accumulating evidence has revealed that lncRNAs are
essential participants in cancer biology. LncRNAs are involved in
almost every aspect of cellular biology by acting as
transcriptional modulators, splicing regulators,
posttranscriptional processors, enhancers, molecular decoys for
miRNAs, chromatin remodelers, or as guides for protein-protein,
protein-DNA and protein-RNA interactions (15,16).
Recent literatures have indicated that lncRNAs are often expressed
in a disease-, tissue- or developmental-specific manner, which
makes a foundation for utilizing lncRNAs as biomarkers and
therapeutic targets in different diseases. Moreover, the number of
lncRNAs has recently been greatly expanded to triple the number of
protein-coding genes (5). Therefore,
lncRNAs are likely to serve as the basis for clinical diagnosis and
treatment in oncology.
The present study discovered that lncRNA CRNDE was
up-regulated in IHCC cells and tissues, and high CRNDE expression
significantly promoted clinical progression of IHCC, including
poorer differentiation, larger tumor size, lymph node metastasis,
and advanced TNM stage. Further investigations found CRNDE
up-regulation predicted poorer OS and earlier recurrence of IHCC
patients. Moreover, CRNDE was revealed to be an independent risk
factor of poor OS and PFS in IHCC. The results of in vivo
studies verified that CRNDE could increase the proliferation
ability of IHCC cells on CCK-8 assay and colony formation assay. By
detecting the mRNA levels and protein levels of EMT markers, we
confirmed CRNDE could facilitate the EMT of IHCC cells and thus
promote the metastasis of IHCC cells, which was also confirmed by
Transwell assay and Matrigel assay. All these results suggested
that CRNDE could promote the progression of IHCC and may serve as a
promising prognostic marker and therapeutic target of IHCC
patients.
Increased CRNDE expression was first discovered by
Graham and his colleagues (7) in the
early stage of colorectal neoplasia, and was regarded as a
promising tissue and plasma biomarker in colorectal cancer early
detection. Since then, the functional roles and mechanisms of CRNDE
in cancer development and progression have been gradually
exhibited. It has been demonstrated that CRNDE was up-regulated in
various solid tumors, including hepatocellular carcinoma, renal
cancer, adrenocortical carcinoma, pancreatic cancer, prostate
cancer, ovarian cancer, and leukemias (17). CRNDE is a multifunctional lncRNA and
functions through different splice forms, which could provide
specific functional scaffolds for regulatory complexes, such as the
polycomb repressive complex 2 (PRC2) and CoREST chromatin-modifying
complexes (18). Besides, CRNDE
exhibited a very time- and tissue-specific pattern of expression.
Recent literatures have discovered some other signaling pathways
through which CRNDE exerts its functions. Zheng et al
(19) demonstrated that CRNDE played
an oncogenic role in human glioma stem cells through negatively
regulating miR-186 expression. Besides, CRNDE was discovered to
recruit the Deleted in Malignant Brain Tumors 1 (DMBT1) and
Baculoviral IAP repeat Containing 1 (c-IAP1) through acting as a
scaffold, which could accelerate gallbladder cancer development by
promoting the PI3K-AKT pathway (20).
In breast cancer, CRNDE was revealed to promote tumor growth
through acting as a molecular sponge of miR-136, and thus
hyperactivating the Wnt/β-catenin signaling pathway (11). Therefore, various signaling pathways
may underlie the functions and mechanisms of CRNDE in different
cancers. The detailed mechanisms of CRNDE exerting its functions
deserve further investigations.
In conclusion, the present study confirmed that
CRNDE was up-regulated in IHCC cells and tissues. Statistical
analyses found CRNDE expression was positively correlated with IHCC
clinical progression, poor OS and PFS. Furthermore, univariate and
multivariate COX regression analyses discovered that CRNDE
overexpression was an independent risk factor of IHCC poor OS and
PFS. The function of CRNDE was also confirmed in vitro
assays by detecting its role in affecting the proliferation and
metastasis of IHCC cells. Mechanistically, EMT was demonstrated to
be accelerated with CRNDE up-regulation, which may be the
underlying mechanism of CRNDE promoting IHCC metastasis. These
findings indicated that CRNDE may be an essential prognostic marker
and therapeutic target in IHCC.
References
|
1
|
Sirica AE, Dumur CI, Campbell DJ, Almenara
JA, Ogunwobi OO and Dewitt JL: Intrahepatic cholangiocarcinoma
progression: Prognostic factors and basic mechanisms. Clin
Gastroenterol Hepatol. 7 11 Suppl:S68–S78. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Razumilava N and Gores GJ:
Cholangiocarcinoma. Lancet. 383:2168–2179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aljiffry M, Abdulelah A, Walsh M,
Peltekian K, Alwayn I and Molinari M: Evidence-based approach to
cholangiocarcinoma: A systematic review of the current literature.
J Am Coll Surg. 208:134–147. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen LL: Linking long noncoding RNA
localization and function. Trends Biochem Sci. 41:761–772. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Evans JR, Feng FY and Chinnaiyan AM: The
bright side of dark matter: lncRNAs in cancer. J Clin Invest.
126:2775–2782. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Adams BD, Parsons C, Walker L, Zhang WC
and Slack FJ: Targeting noncoding RNAs in disease. J Clin Invest.
127:761–771. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Graham LD, Pedersen SK, Brown GS, Ho T,
Kassir Z, Moynihan AT, Vizgoft EK, Dunne R, Pimlott L, Young GP, et
al: Colorectal neoplasia differentially expressed (CRNDE), a novel
gene with elevated expression in colorectal adenomas and
adenocarcinomas. Genes Cancer. 2:829–840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao H, Song X, Kang T, Yan B, Feng L, Gao
L, Ai L, Liu X, Yu J and Li H: Long noncoding RNA CRNDE functions
as a competing endogenous RNA to promote metastasis and oxaliplatin
resistance by sponging miR-136 in colorectal cancer. Onco Targets
Ther. 10:205–216. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu B, Ye X, Du Q, Zhu B, Zhai Q and Li XX:
The long non-coding RNA CRNDE promotes colorectal carcinoma
progression by competitively binding miR-217 with TCF7L2 and
enhancing the Wnt/β-catenin signaling pathway. Cell Physiol
Biochem. 41:2489–2502. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shao K, Shi T, Yang Y, Wang X, Xu D and
Zhou P: Highly expressed lncRNA CRNDE promotes cell proliferation
through Wnt/β-catenin signaling in renal cell carcinoma. Tumour
Biol. Oct 6–2016.(Epub ahead of print). View Article : Google Scholar
|
|
11
|
Huan J, Xing L, Lin Q, Xui H and Qin X:
Long noncoding RNA CRNDE activates Wnt/β-catenin signaling pathway
through acting as a molecular sponge of microRNA-136 in human
breast cancer. Am J Transl Res. 9:1977–1989. 2017.PubMed/NCBI
|
|
12
|
Tsai JH and Yang J: Epithelial-mesenchymal
plasticity in carcinoma metastasis. Genes Dev. 27:2192–2206. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeisberg M and Neilson EG: Biomarkers for
epithelial-mesenchymal transitions. J Clin Invest. 119:1429–1437.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartonicek N, Maag JL and Dinger ME: Long
noncoding RNAs in cancer: Mechanisms of action and technological
advancements. Mol Cancer. 15:432016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chandra Gupta S and Nandan Tripathi Y:
Potential of long non-coding RNAs in cancer patients: From
biomarkers to therapeutic targets. Int J Cancer. 140:1955–1967.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ellis BC, Molloy PL and Graham LD: CRNDE:
A long non-coding RNA involved in canceR, neurobiology, and
DEvelopment. Front Genet. 3:2702012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Khalil AM, Guttman M, Huarte M, Garber M,
Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van
Oudenaarden A, et al: Many human large intergenic noncoding RNAs
associate with chromatin-modifying complexes and affect gene
expression. Proc Natl Acad Sci USA. 106:pp. 11667–11672. 2009;
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zheng J, Li XD, Wang P, Liu XB, Xue YX, Hu
Y, Li Z, Li ZQ, Wang ZH and Liu YH: CRNDE affects the malignant
biological characteristics of human glioma stem cells by negatively
regulating miR-186. Oncotarget. 6:25339–25355. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shen S, Liu H, Wang Y, Wang J, Ni X, Ai Z,
Pan H, Liu H and Shao Y: Long non-coding RNA CRNDE promotes
gallbladder carcinoma carcinogenesis and as a scaffold of DMBT1 and
C-IAP1 complexes to activating PI3K-AKT pathway. Oncotarget.
7:72833–72844. 2016. View Article : Google Scholar : PubMed/NCBI
|