Introduction
Bladder cancer (BC) has the second most frequent
incidence of cancers of the genitourinary tract worldwide (1). In 2012 alone, 430,000 incident patients
with BC and 165,000 BC-associated mortalities were identified
(1). Although advances in treatment
have been made in previous decades, the recurrence rate of BC is
between 15 and 90% within 5 years, and the 5-year-survival-rate is
~75% (2–5). In total, ~15% of patients with papillary
BC develop muscle-invasive and metastatic cancer (3). In these patients, long-term medical care
is required, with inevitably high personnel and socioeconomic cost
(6). Therefore, it is important to
identify novel clues to diagnose and treat BC.
With the identification of long non-coding RNAs
(lncRNAs), it has been demonstrated that certain lncRNAs may serve
important roles in BC (5,7–10). LncRNAs
are a category of non-coding RNAs, containing >200 nucleotides
with limited translation potential. Unlike other non-coding RNAs,
including microRNAs, the underlying molecular mechanisms of
different lncRNAs involved in human diseases remain largely
unknown. To date, lncRNAs have been demonstrated to participate in
multiple intracellular and extracellular activities, including gene
transcription, mRNA splicing and tumorigenesis (11–15). For
example, the dysregulation of lncRNAs has been demonstrated in
different types of cancer including hepatocellular
carcinoma-upregulated lncRNA in hepatocellular carcinoma (16), metastasis-associated lung
adenocarcinoma transcript 1 in lung cancer (17) and urothelial cancer-associated 1 and
ZEB2 natural antisense transcript in BC (18,19).
Unfortunately, despite the increasing implications
of lncRNAs in bladder tumorigenesis, no lncRNA has been widely
recognized as a specific biomarker for the progression or prognosis
of BC. This fact, to a certain extent, encourages focus on novel
lncRNAs in BC. Hepatocyte nuclear factor 1A (HNF1A)-antisense RNA
(AS)1 is a newly identified lncRNA. Since its initial
identification, HNF1A-AS1 has been demonstrated to serve a critical
role in human tumorigenesis, including esophageal adenocarcinoma
(20) and lung adenocarcinoma
(21) The expression of HNF1A-AS1 was
upregulated in esophageal and lung adenocarcinoma cells (21). Functionally, HNF1A-AS1 promotes cell
viability and metastasis in these cell types (20,21),
indicating that HNF1A-AS1 may promote cancer progression. However,
whether HNF1A-AS1 exerts a robust cancer-promotion effect in BC
cells has not been extensively studied.
In the present study, the expression of HNF1A-AS1 in
patients with BC and cultured cells was examined using the
quantitative polymerase chain reaction (qPCR). The specific role of
HNF1A-AS1 in BC cell viability and metastasis was investigated
using cell viability, colony formation, cell migration and invasion
assays. Hopefully, these results may lead to the identification of
novel methods to diagnose and treat patients with BC in clinical
settings.
Materials and methods
Human samples
Between January 2011 and December 2014, a total of
30 patients with BC who were admitted to Tianjin Medical University
General Hospital (Tianjin, China) and underwent a surgical
resection were included. Cancerous tissues and their adjacent
non-cancerous tissues were obtained from each patient, and used for
the subsequent analysis. All patients provided full written
informed consent to participate in the present study, and the study
protocol was approved by the Ethics Committee of Tianjin Medical
University.
Cell lines
A total of four human BC cell lines, including T24,
J82, UMUC3 and 5637, and a normal bladder cell line SV-HUC-1 were
purchased from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China). Cells were cultured in the Dulbecco's modified
Eagle's medium (DMEM; Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) supplied with 10% fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.) without antibiotics. Cells were
maintained at 37°C in an incubator with a humidified atmosphere
containing 5% CO2. Medium was refreshed every other day,
except where otherwise stated.
RNA isolation and qPCR
To extract the total RNA from human tissues and
cultured cells, TRIzol reagent (Thermo Fisher Scientific, Inc.) was
used according to the manufacturer's protocol. A 1 µg amount of
total RNA from each sample was then reverse-transcribed into cDNA
(37°C for 15 min and 85°C for 5 sec) using a PrimeScript RT Master
Mix Perfect Real Time kit (Takara Biotechnology Co., Ltd., Dalian,
China). Subsequently, qPCR was performed using the SYBR Premix Ex
Taq kit (Takara Biotechnology Co., Ltd.) with the following
primers: HNF1A-AS1 forward, 5′-TCAAGAAATGGTGGCTAT-3′ and reverse,
5′-GCTCTGAGACTGGCTGAA-3′; GAP DH forward, 5′-GTGGACATCCGCAAAGAC-3′
and reverse, 5′-AAAGGGTGTAACGCAACTA-3′. The sequence of
Thermocycling was as follows: 95°C for 2 min followed by 40 cycles
of 95°C for 15 sec and 60°C 30 sec. The PCR reaction was normalized
to the reference gene: GAPDH. The relative level of each gene was
calculated and normalized using the 2−ΔΔCq formula
(22) Each experiment was repeated in
triplicate.
Short hairpin (sh)RNA infection
The specific shRNA against HNF1A-AS1 (shHNF1A-AS1)
and the scrambled negative control shRNA (shNC) were chemically
designed and synthesized by GenePharma Co., Ltd. (Shanghai, China).
Lipofectamine® 2000 was purchased from Invitrogen;
Thermo Fisher Scientific, Inc., and used according to the
manufacturer's protocol. The transfection efficiency of the
specific shRNA against HNF1A-AS1 in T24 and 5637 cell lines was
confirmed by qPCR, as aforementioned.
Cell viability assay
Cell viability was determined using an MTT assay.
Briefly, 1×104 T24 and 5637 cells were seeded in 96-well
plates and transfected with HNF1A-AS1 shRNA (shHNF1A-AS1 group) or
scrambled shRNA (shNC group) for 48 h. Cells that were transfected
with no shRNA were synchronously co-cultured as a control group.
Following treatment, 10 µl MTT (5 µg/ml) was mixed with the medium
in each well and cells were incubated for an additional 3 h at 37°C
in the dark. The formazan crystals that formed were dissolved in
100 µl dimethylsulfoxide and the absorbance of each well of the
96-well plate was measured at 570 nM with a plate reader (Tecan
Schweiz AG, Mannedorf, Switzerland).
Colony formation assay
A total of 10×103 T24 and 5637 cells were
seeded into 6-well plates and treated with specific shRNA against
HNF1A-AS1 (shHNF1A-AS1 group) or scrambled shRNA (shNC group).
Control cells were also synchronously cultured (control group).
Following treatment for 24 h, the cell lines were suspended and
re-seeded into 12-well plates (100 cells/well) in triplicate. The
cultured medium was replaced every other day. Following incubation
for 10 days, the colonies from three groups were fixed with
ice-cold methanol and stained with 1% crystal violet. A colony was
considered to be present when the number of assembled cells was
>50. Subsequently, the number of colonies was determined under a
light microscope at ×200 magnification (Nikon Corporation, Tokyo,
Japan). The following formula was used to calculate the rate of
colony formation: Colony formation rate=(number of colonies/number
of seeded cells) × 100%.
Transwell assay
A total of 5×103 T24 and 5637 cells were
seeded into 12-well plates and transfected with specific shRNA or
scrambled shRNA for 48 h. Subsequently, cells were harvested and
diluted to a concentration of 5×105/ml with the DMEM
without FBS. For the migration assay, a 100 µl aliquot of cells was
added into the upper chambers (Corning Incorporated, Corning, NY,
USA) and 200 µl medium with 10% FBS was added to the lower chamber.
After 8 h of incubation at 37°C, cells were washed with PBS three
times, fixed with methanol and stained with 1% crystal violet for 5
min. For the invasion assay, the Transwell chambers were coated
with Matrigel (BD Biosciences, San Jose, CA, USA) 6 h before the
experiment at 37°C in an incubator. Images of the cells on the
lower surface of the chamber were captured with a light microscope,
and counted in six random fields. All experiments were repeated at
least three times in triplicate.
Cell cycle assay
A total of 5×103 T24 and 5637 cells were
seeded into 12-well plates and transfected with control and
specific shRNAs. After 48 h of incubation, cells were harvested and
fixed with 75% ice-cold ethanol overnight at 4°C. Subsequently,
cells were washed with PBS twice and stained with 50 mg/ml
propidium iodide supplied with 50 mg/ml RNase A (DNase-free) at
37°C for an additional 30 min. The percentage of cells in each
phase in the two cell lines was calculated using flow cytometry.
The experiments were repeated three times with each test in
triplicate.
Statistical analysis
All data are presented as the mean ± standard
deviation. Each experiment was repeated at least three times in
triplicate. A two-tailed Student's t-test was used to compare means
of two groups, whereas one-way analysis of variance was used for
comparisons among multiple groups (≥3 groups), followed by a least
significant difference post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
HNF1A-AS1 is upregulated in clinical
BC tissues and cultured BC cells
Initially, the expression of lncRNA HNF1A-AS1 was
detected in patients with BC and cultured BC cell lines. As no
antibody is currently available for HNF1A-AS1, only the transcript
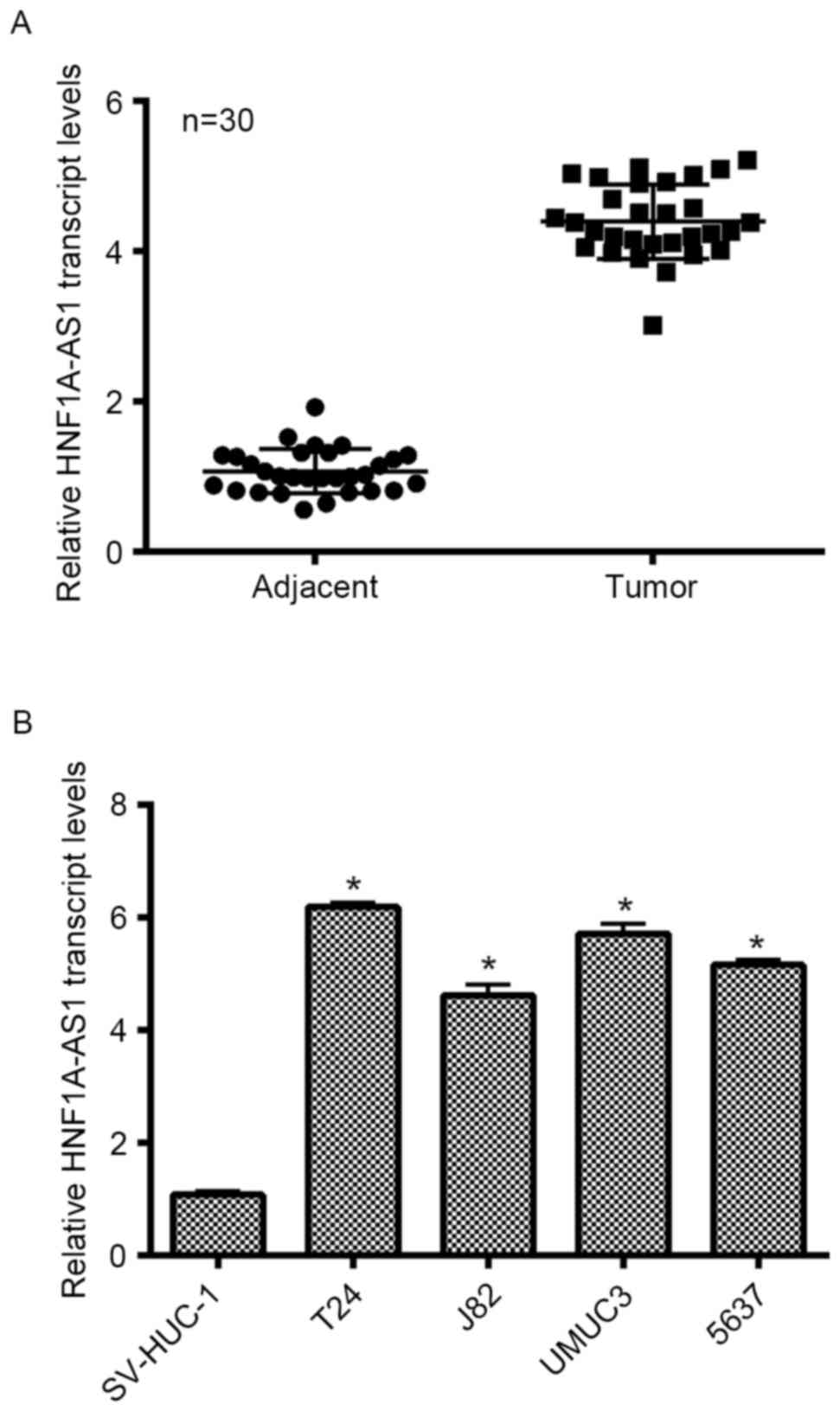
level was detected in the present study. As presented in Fig. 1A, the transcript levels of HNF1A-AS1
in 30 clinical BC tissues extracted from patients were
significantly increased (>4-fold) compared with the adjacent
non-cancerous tissues. SV-HUC-1 cells are normal human urothelial
cells that are used as control cells. This cell line was cultured
as an internal control in the present study. Simultaneously, four
other BC cell lines (T24, J82, UMUC3 and 5637) were synchronously
cultured. The relative expression of HNF1A-AS1 in different BC cell
lines was examined, and it was demonstrated that the transcript
levels of HNF1A-AS1 in the entire BC cell lines were notably
upregulated compared with that in SV-HUC-1 cells. Remarkably, the
transcript level of HNF1A-AS1 was increased 6.2-fold in T24 cells
and 5.8-fold in UMUC3 cells compared with the control cells.
Furthermore, the transcript level of HNF1A-AS1 was increased
4.6-fold in J82 cells and 5.1-fold in 5637 cells. T24 and 5637
cells were selected as representative BC cells for subsequent
analyses. These results revealed that HNF1A-AS is significantly
overexpressed in clinical BC tissues and BC cells cultured in
vitro.
Knockdown of HNF1A-AS1 is successfully
achieved by specific shRNA transfection
To elucidate the detailed role of HNF1A-AS1 in human
BC, specific shRNA against HNF1A-AS1 (shHNF1A-AS1) was designed and
synthesized. Transfection of scrambled shRNA (shNC) did not
markedly change the transcript level of HNF1A-AS1 in the T24 and
5637 cell lines. However, when the cells were treated with the
specific shRNA, the transcript level of HNF1A-AS1 was significantly
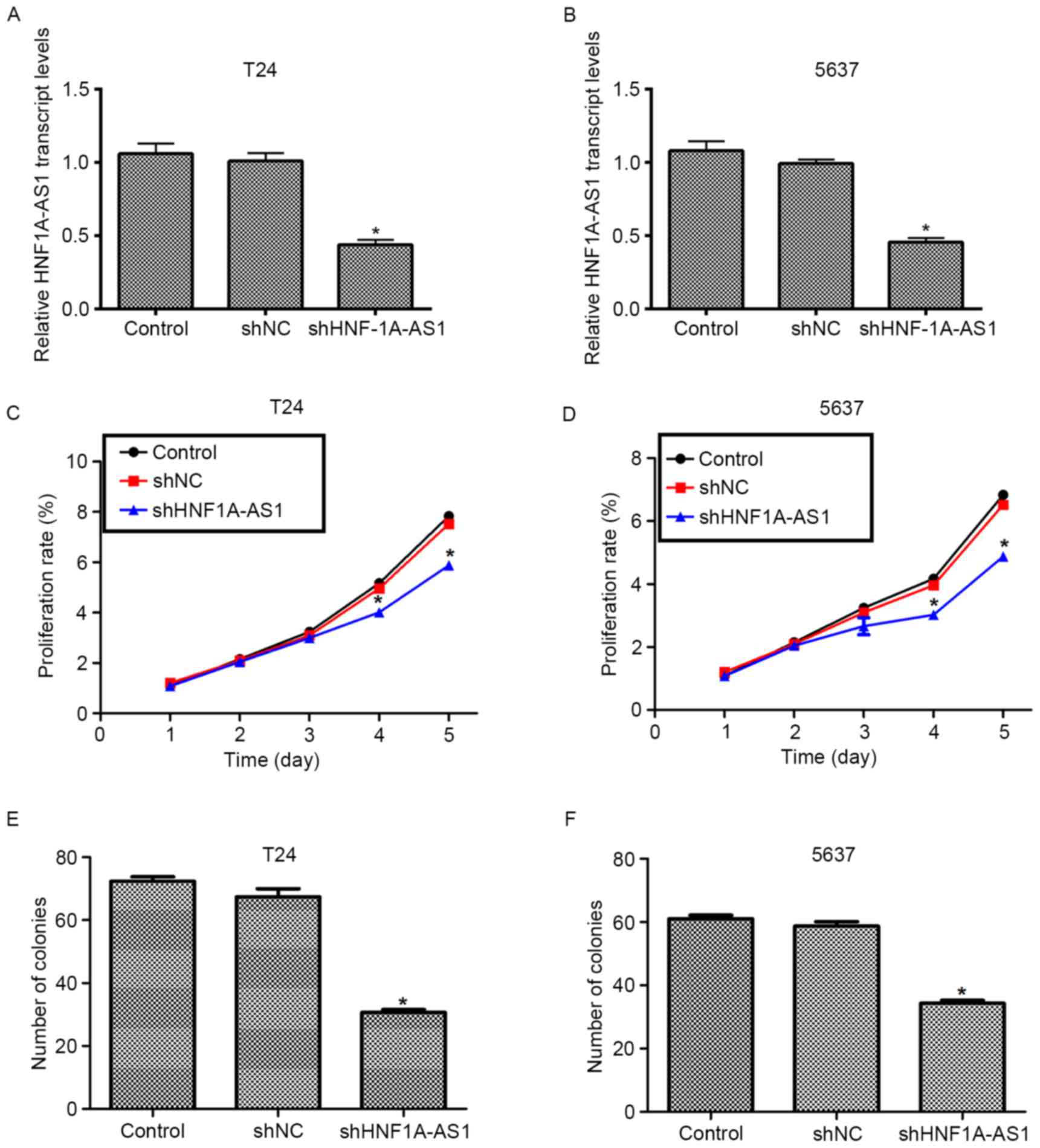
decreased (~50%) in the two cell lines (Fig. 2A and B). These results indicated the
high efficiency of specific shRNA to deplete HNF1A-AS1.
Knockdown of HNF1A-AS1 inhibits cell
viability and colony formation in T24 and 5637 cells
The role of HNF1A-AS in human BC was investigated
using a cell viability assay. No significant differences in cell
viability among the three groups in the first 3 days for the T24
and 5637 cells were identified (Fig. 2C
and D). However, the viability of the T24 cells was suppressed
by 27% on the fourth day and by 30% on the fifth day (Fig. 2C). For the 5637 cells, the viability
was decreased by 27% on the fourth day and the inhibition of 5637
and 35% on the fifth day (Fig. 2D).
The colony formation assay additionally demonstrated that the
knockdown of HNF1A-AS by specific shRNA remarkably decreased the
numbers of colonies for the two BC cell lines (Fig. 2E and F). Quantification of the number
of colonies for T24 cells revealed that ~70 colonies were formed in
control shRNA-treated cells, whereas only 34 colonies were observed
in shHNF1A-AS1-transfected cells. Similarly, in 5637 cells, the
number of colonies formed on agar plates was also decreased by 42%
in specific shRNA-treated cells compared with the control cells.
These results indicate that HNF1A-AS1 promotes cell viability and
colony formation in the BC cell lines.
Knockdown of HNF1A-AS1 arrests cell
cycle in G0/G1 phase in T24 and 5637
cells
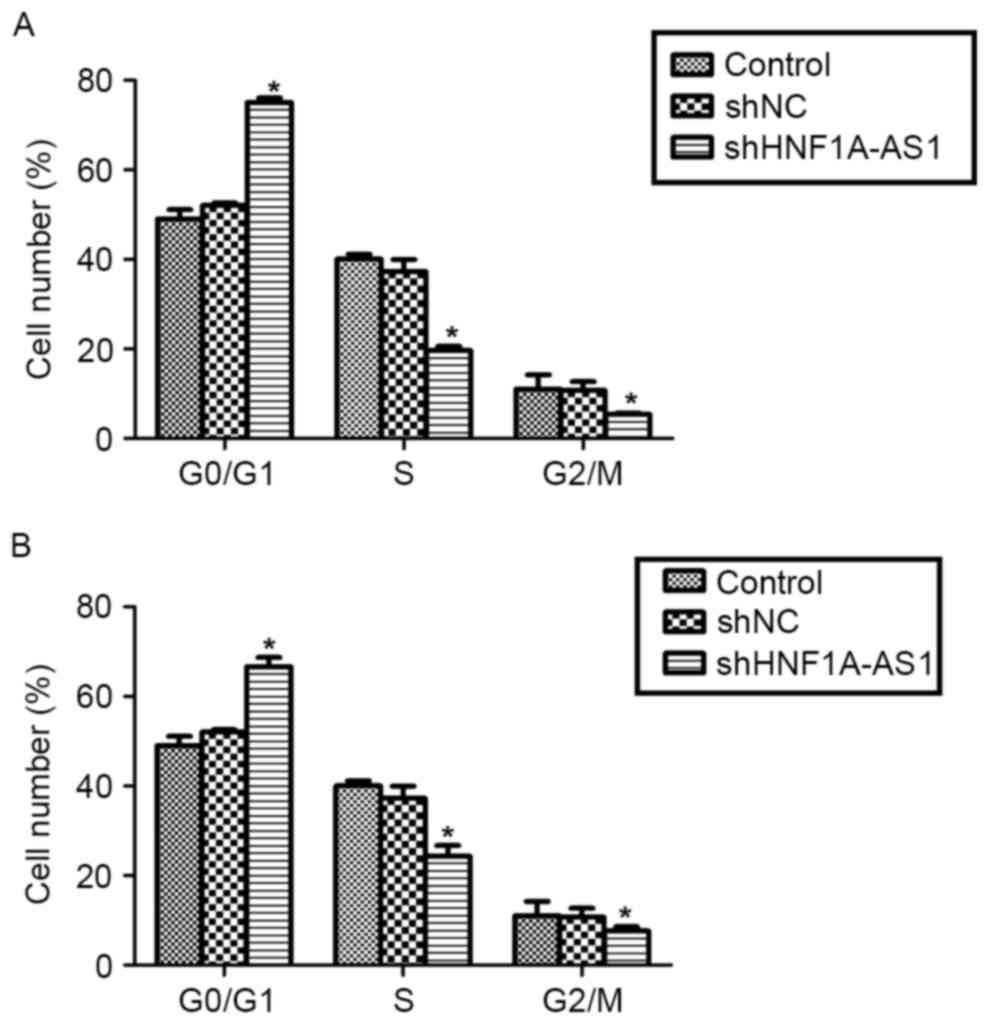
To explain the inhibitive effects by shHNF1A-AS1 on
viability, the cell cycle distribution in T24 and 5637 cells was
analyzed using flow cytometry. The knockdown of HNF1A-AS1 in T24
cells increased the proportion of cells in
G0/G1 phase by 60%, whereas it decreased the
proportion of cells in S phase by 50% and that in G2/M
phase by 48% (Fig. 3A). Similarly,
the knockdo wn of HNF1A-AS1 in 5637 cells increased the proportion
of cells in G0/G1 phase by 36%, whereas it
decreased the proportion of cells in S phase and G2/M
phase by 40 and 31%, respectively (Fig.
3B). These results indicate that the knockdown of HNF1A-AS1
arrested BC cell cycle progression by notably accumulating cells in
G0/G1 phase in T24 and 5637 cells.
Knockdown of HNF1A-AS1 inhibits BC
cell migration and invasion in vitro
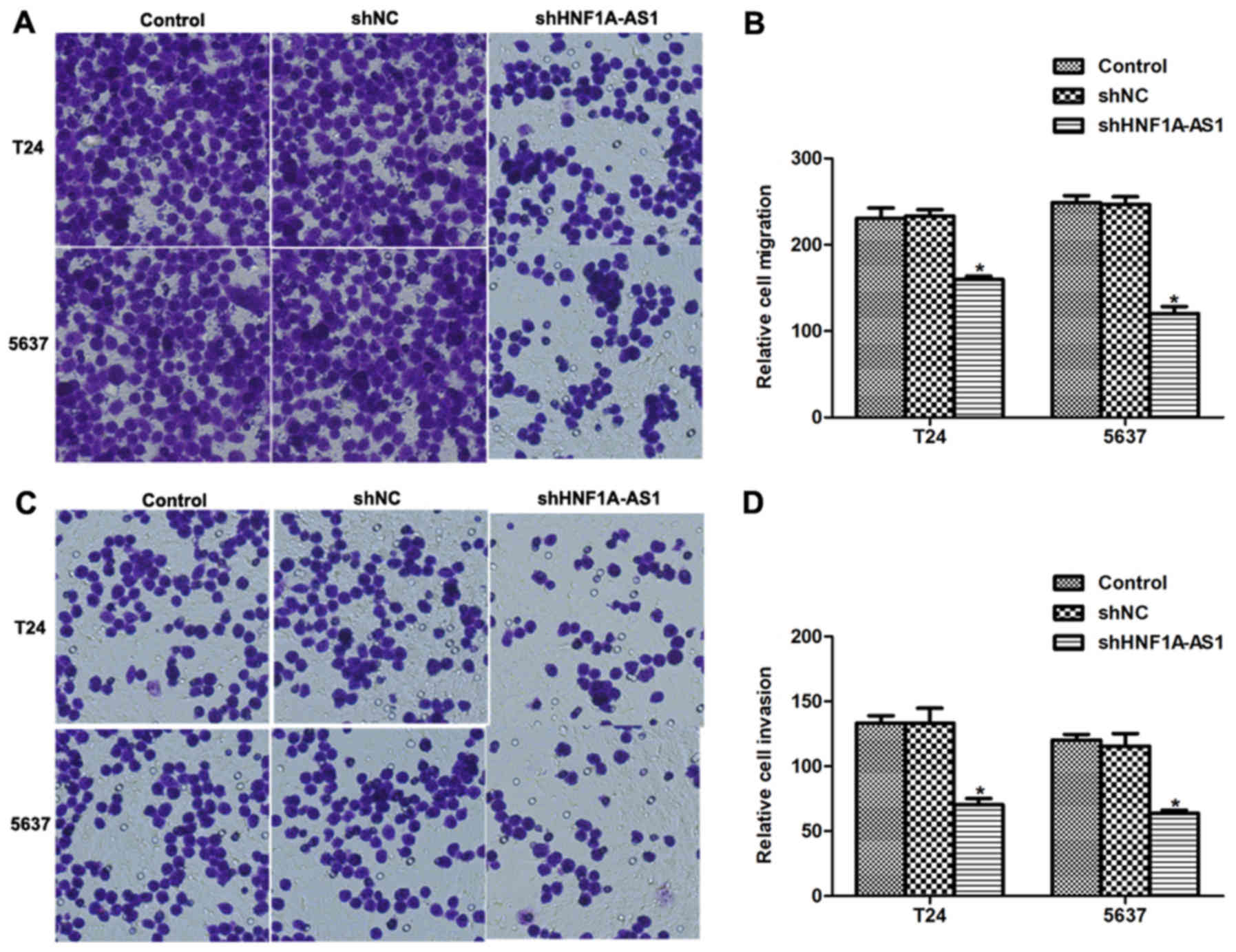
The effects of HNF1A-AS1 knockdown on cell migration
and invasion in vitro were investigated. Numerous T24 and
5637 cells migrated through the membrane (Fig. 4A and B); however, notable disparities
between the control and HNF1A-AS1 shRNA-treated groups were
observed. For the migration assays, the number of cells migrating
to the lower surface of the chamber was decreased by 31% for T24
cells and 44% for 5637 cells following knockdown of HNF1A-AS1
(Fig. 4B). For the invasion assays,
the differences between specific shRNA-treated and control groups
were 45% for T24 cells and 48% for 5637 cells (Fig. 4C and D). These results suggest that
HNF1A-AS1 may promote T24 and 5637 cell metastases in
vitro.
Discussion
BC is one of the most common types of cancer, with
>12 million incident diagnoses and >1.3 million mortalities
each year (23). BC may be
categorized into non-muscle-invasive BC (80%) and muscle-invasive
BC (20%) on the basis of the scale of tumor infiltration into the
musculature of the bladder of the patient (24). Non-muscle-invasive BC exhibits a high
tendency for recurrence, which leads to its high prevalence in the
USA (24). In addition, between 10
and 30% of non-muscle-invasive BC progresses to the muscle-invasive
phenotype, which results in a markedly decreased 5-year survival
rate (25). Therefore, the early
detection of BC is a priority for improving the outcomes of
patients with BC.
In the present study, the role of the previously
seldom studied but widely expressed lncRNA HNF1A-AS1 was explored.
First, it was identified that HNFA-AS1 was upregulated in clinical
BC tissues and cultured BC cells. The knockdown of HNF1A-AS1
resulted in a decreased cell viability rate in vitro.
Concurrently, it was observed that the cell cycle was arrested in
G0/G1 phase, indicating that the inhibitory
effects on viability by the knockdown of HNF1A-AS1 may be achieved
through interrupting cell cycle progression. Additionally, the
knockdown of HNF1A-AS1 inhibited cell migratory and invasive
abilities in the two cell lines used in the present study. To the
best of our knowledge, the present study is the first to identify
the role of HNF1A-AS1 in BC. Taken together, these results suggest
that HNF1A-AS1 serves critical roles in BC cell viability and
migration. The dysregulation of HNF1A-AS1 in BC may contribute to
the development and progression of BC.
HNF1A-AS1 was indicated to be associated with the
progression of lung adenocarcinoma, esophageal adenocarcinoma
(20,21) and, most recently, in gastric cancer
(26). Functionally, HNF1A-AS1 is
consistently recognized as a regulator of cell viability and
metastasis in the aforementioned types of cancer. However, no
detailed mechanisms underlying its biological activity have been
revealed. Previously, Yang et al (20) have demonstrated that HNF1A-AS1 was
involved in the regulation of lncRNA H19, another widely studied
cancer-associated lncRNA. Concurrently, it was indicated that
HNF1A-AS1 knockdown preferentially affected genes that are linked
to assembly of chromatin and the nucleosome, a mechanism essential
to cell cycle progression (15). In
that study, the well-known cancer-associated lncRNA H19 was the
gene most markedly inhibited by HNF1A-AS1 knockdown. These results
suggest that lncRNA H19 may elicit its effects through HNF1A-AS1.
H19 may be a critical downstream effector that transduces signals
from HNF1A-AS1, and therefore may contribute to HNF1A-AS1-mediated
cell viability and metastasis, particularly in the regulation of
cell cycle progression. Therefore, it would be interesting to
investigate the association between HNF1A-AS1 and H19 in BC in
additional studies.
In conclusion, the present study suggests that
HNF1A-AS1 was overexpressed in BC. The knockdown of HNF1A-AS1
inhibited cell viability and metastasis in BC cells. These results
may lead to the identification of novel methods to diagnose and
treat BC in clinical settings.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sylvester RJ, van der Meijden AP,
Oosterlinck W, Witjes JA, Bouffioux C, Denis L, Newling DW and
Kurth K: Predicting recurrence and progression in individual
patients with stage Ta T1 bladder cancer using EORTC risk tables: A
combined analysis of 2596 patients from seven EORTC trials. Eur
Urol. 49:466–477. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kurth KH, Denis L, Bouffioux C, Sylvester
R, Debruyne FM, Pavone-Macaluso M and Oosterlinck W: Factors
affecting recurrence and progression in superficial bladder
tumours. Eur J Cancer. 31A:1–1846. 1995.
|
|
4
|
Fernandez-Gomez J, Madero R, Solsona E,
Unda M, Martinez-Piñeiro L, Gonzalez M, Portillo J, Ojea A, Pertusa
C, Rodriguez-Molina J, et al: Predicting nonmuscle invasive bladder
cancer recurrence and progression in patients treated with bacillus
Calmette-Guerin: The CUETO scoring model. J Urol. 182:2195–2203.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhu H, Li X, Song Y, Zhang P, Xiao Y and
Xing Y: Long non-coding RNA ANRIL is up-regulated in bladder cancer
and regulates bladder cancer cell proliferation and apoptosis
through the intrinsic pathway. Biochem Biophys Res Commun.
467:223–228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dancik GM, Owens CR, Iczkowski KA and
Theodorescu D: A cell of origin gene signature indicates human
bladder cancer has distinct cellular progenitors. Stem Cells.
32:974–982. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Peter S, Borkowska E, Drayton RM, Rakhit
CP, Noon A, Chen W and Catto JW: Identification of differentially
expressed long noncoding RNAs in bladder cancer. Clin Cancer Res.
20:5311–5321. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Q, Su M, Lu G and Wang J: The
complexity of bladder cancer: Long noncoding RNAs are on the stage.
Mol Cancer. 12:1012013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun X, Du P, Yuan W, Du Z, Yu M, Yu X and
Hu T: Long non-coding RNA HOTAIR regulates cyclin J via inhibition
of microRNA-205 expression in bladder cancer. Cell Death Dis.
6:e19072015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhuang C, Li J, Liu Y, Chen M, Yuan J, Fu
X, Zhan Y, Liu L, Lin J, Zhou Q, et al: Tetracycline-inducible
shRNA targeting long non-coding RNA PVT1 inhibits cell growth and
induces apoptosis in bladder cancer cells. Oncotarget.
6:41194–41203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Martin L and Chang HY: Uncovering the role
of genomic ‘dark matter’ in human disease. J Clin Invest.
122:1589–1595. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Y, He L, Du Y, Zhu P, Huang G, Luo J,
Yan X, Ye B, Li C, Xia P, et al: The long noncoding RNA lncTCF7
promotes self-renewal of human liver cancer stem cells through
activation of Wnt signaling. Cell Stem Cell. 16:413–425. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Han X, Yang F, Cao H and Liang Z: Malat1
regulates serum response factor through miR-133 as a competing
endogenous RNA in myogenesis. FASEB J. 29:3054–3064. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li P, Ruan X, Yang L, Kiesewetter K, Zhao
Y, Luo H, Chen Y, Gucek M, Zhu J and Cao H: A liver-enriched long
non-coding RNA, lncLSTR, regulates systemic lipid metabolism in
mice. Cell Metab. 21:455–467. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Han P, Li W, Lin CH, Yang J, Shang C,
Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al: A long noncoding
RNA protects the heart from pathological hypertrophy. Nature.
514:102–106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M and Zatloukal K: Characterization of HULC, a novel
gene with striking up-regulation in hepatocellular carcinoma, as
noncoding RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu W, Zhang S, Li X, Xue M, Cao S and Chen
W: Ets-2 regulates cell apoptosis via the Akt pathway, through the
regulation of urothelial cancer associated 1, a long non-coding
RNA, in bladder cancer cells. PLoS One. 8:e739202013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhuang J, Lu Q, Shen B, Huang X, Shen L,
Zheng X, Huang R, Yan J and Guo H: TGFβ1 secreted by
cancer-associated fibroblasts induces epithelial-mesenchymal
transition of bladder cancer cells through lncRNA-ZEB2NAT. Sci Rep.
5:119242015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang X, Song JH, Cheng Y, Wu W, Bhagat T,
Yu Y, Abraham JM, Ibrahim S, Ravich W, Roland BC, et al: Long
non-coding RNA HNF1A-AS1 regulates proliferation and migration in
oesophageal adenocarcinoma cells. Gut. 63:881–890. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wu Y, Liu H, Shi X, Yao Y, Yang W and Song
Y: The long non-coding RNA HNF1A-AS1 regulates proliferation and
metastasis in lung adenocarcinoma. Oncotarget. 6:9160–9172.
2015.PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ploeg M, Aben KK and Kiemeney LA: The
present and future burden of urinary bladder cancer in the world.
World J Urol. 27:289–293. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Millán-Rodríguez F, Chéchile-Toniolo G,
Salvador-Bayarri J, Palou J, Algaba F and Vicente-Rodríguez J:
Primary superficial bladder cancer risk groups according to
progression, mortality and recurrence. J Urol. 164:680–684. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rübben H, Lutzeyer W, Fischer N, Deutz F,
Lagrange W and Giani G: Natural history and treatment of low and
high risk superficial bladder tumors. J Urol. 139:283–285. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dang Y, Lan F, Ouyang X, Wang K, Lin Y, Yu
Y, Wang L, Wang Y and Huang Q: Expression and clinical significance
of long non-coding RNA HNF1A-AS1 in human gastric cancer. World J
Surg Oncol. 13:3022015. View Article : Google Scholar : PubMed/NCBI
|