Introduction
Gastric cancer is one of the major causes of
mortality worldwide (1). Currently,
the standard treatment regimen for gastric cancer includes surgery,
chemotherapy and radiotherapy (2). In
patients at advanced stages, chemotherapy has been demonstrated to
improve survival, by preventing tumor invasion or downsizing
distant metastatic lesions (3). One
of the leading causes of chemotherapy failure in gastric cancer is
tumor resistance. Patients who are less responsive to chemotherapy
have a poorer prognosis (4). Thus, it
is important to identify the molecular mechanisms underlying the
drug resistance of gastric cancer cells.
MicroRNAs (miRNAs/miRs) are a family of endogenous
non-coding RNA molecules that can post-transcriptionally regulate
gene expression and have been shown to perform crucial roles in
diverse biological processes, including apoptosis, proliferation,
stress response and metabolism (5).
miRNAs have been revealed to be associated with cell
chemosensitivity or chemotherapy resistance in a variety of cancer
cell types, including ovarian and breast cancer cells (6–8). However,
there is limited available data on the potential role of miRNAs in
the chemotherapy resistance of gastric cancer.
The present study reported that miR-17-5p was
upregulated in the multidrug-resistant human gastric cancer
SGC7901/cisplatin (DDP) cell line, compared with the parental
SGC7901 cell line. It was demonstrated that the downregulation of
miR-17-5p was able to inhibit drug resistance and increase
DDP-induced apoptosis. In addition, it was indicated that p21 was
upregulated in miR-17-5p-transfected cells compared with cells
transfected with control miRNA inhibitor. These results
demonstrated that miR-17-5p may perform a role in the development
of drug resistance in human gastric cell lines partially by
targeting the anti-apoptotic p21 protein.
Materials and methods
Cell culture
The human gastric adenocarcinoma SGC7901 cell line
and its multidrug-resistant variant SGC7901/DDP cells (Nanjing
KeyGen Biotech Co., Ltd., Nanjing, China) were cultured in
RPMI-1640 medium supplemented with 10% fetal calf serum (FCS;
Gibco; Thermo Fisher Scientific, Waltham, MA, USA) in a humidified
atmosphere containing 5% CO2 at 37°C. The cells were
passaged every 3–4 days. To maintain the multidrug resistance
phenotype, DDP (final concentration, 1 µg/ml; Qilu Pharmaceutical
Co., Ltd., Jinan, China) was added to the culture media used for
SGC7901/DDP cells.
RNA extraction
Total RNA from SGC7901 and SGC7901/DDP cells was
isolated using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and the miRNeasy Mini kit (Qiagen GmbH, Hilden,
Germany) according to the manufacturer's protocols.
miRNA microarray analysis
The isolated RNA from the two cell lines was labeled
using the miRCURY™ Hy3/Hy5 Power Labeling kit (Exiqon A/S, Vedbaek,
Denmark), according to the manufacturer's protocol. Subsequently, 1
µg of each sample was labeled with Hy3 fluorescent tag at the
3′-end using T4 RNA ligase. The Hy3-labeled samples were hybridized
on a miRCURY LNA microRNA array (version 18.0; Exiqon A/S)
according to the array manual. Microarray images were acquired
using the GenePix 4000B microarray scanner (Molecular Devices, LLC,
Sunnyvale, CA, USA) and processed and analyzed with GenePix Pro
software (version 6.0; Molecular Devices, LLC). The genomic
location of miRNA was obtained from CoGemiR Comparative Genomics
miRNA database (http://cogemir.tigem.it/). Prediction of miRNA
putative targets was investigated from the PicTar (http://pictar.mdc-berlin.de), Miranda (http://www.microrna.org/microrna/home.do) and
TargetScan algorithms (http://www.targetscan.org). PicTar and Miranda
algorithms were employed to investigate the potential mediator
downstream of miR-17-5p that may be involved in regulating
chemoresistance (9).
Quantitative polymerase chain reaction
(qPCR)
miRNAs were prepared as aforementioned. The
sequences of the primers were as follows: miR-17-5p, forward
5′-CGGCGGCAAAGTGCTTACAG-3′, and reverse 5′-GTGCAGGGTCCGAGGT-3′; the
internal control miR-16, forward, 5′-ATCGCCTAGCAGCACGTAA-3′, and
reverse 5′-AGCAGGGTCCGAGGTATTC-3′. qPCR (95°C for 3 min, 95°C for
12 sec, and 62°C for 40 sec for 40 cycles) was performed on the CFX
Connect Real-Time PCR system. The fold-change in miRNA expression
of SGC7901/DDP cells was calculated using the 2−ΔΔCq
method (10). All experiments were
performed in triplicate and repeated three times.
miRNA inhibitor transfection
SGC7901/DDP cells were transfected 24 h after being
seeded on 6-well plates (4×105 cells/well). A total of 2
µg miR-17-5p inhibitor or 2 µg miRNA inhibitor control (Ambion;
Thermo Fisher Scientific, Inc.) in 200 µl Opti-MEM I medium (Gibco;
Thermo Fisher Scientific, Inc.) were mixed with 2 µl Lipofectamine
2000® transfection reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) dissolved in 200 µl of the same medium and
allowed to stand at room temperature for 20 min. The resulting 400
µl transfection solutions were then added to each well. The cells
were cultured in a humidified atmosphere containing 5%
CO2 at 37°C, and after 6 h the cell medium was replaced
with fresh medium. After 24 h, transfected cells were seeded onto
96-well plates (8×103 viable cells/well) for subsequent
experiments. The expression of miR-17-5p in transfected cells was
detected by qPCR as aforementioned.
In vitro drug sensitivity assay
Following cellular adhesion for 24 h, freshly
prepared anticancer drugs, including vincristine (VCR; Zhejiang
Hisun Chemical Co., Ltd., Taizhou, China), Adriamycin (ADR;
Zhejiang Hisun Chemical Co., Ltd.), 5-fluorouracil (5-Fu; Tianjin
Jinyao Amino Acid Co., Ltd., Tianjin, China) and DDP in sequential
diluted concentrations were added to each well and incubated at
37°C. The concentrations of drugs were as follow: 0, 1.25, 2.5, 5,
7.5, 10, 15 and 20 µg/ml for VCR; 0, 0.125, 0.25, 0.5, 0.75, 1, 1.5
and 2 µg/ml for ADR; 0, 0.25, 0.5, 1.0, 2.5, 5 and 7.5, 10 µg/ml
for 5-Fu; 0, 0.625, 1.25, 2.5, 5, 7.5, 10 and 15 µg/ml for DDP. At
48 h after the addition of drugs, cell viability was assessed using
an MTT assay. PBS was used to dissolve the purple formazan. The
absorbance of each well at 450 nm was detected on a
spectrophotometer. The concentration at which each drug induced the
half-maximal inhibitory concentration (IC50) was
estimated by relative survival curves. Each assay was performed in
triplicate and repeated independently three times.
Luciferase activity assay
pGL3-p21-3′UTR was constructed as follows. The 3′
untranslated region (3′UTR) of human p21 cDNA containing the
putative target site for miR-17-5p was amplified by PCR (94°C for 4
min, 94°C for 30 sec, 58°C for 30 sec, and 70°C for 1 min for, 40
cycles) with DNA polymerase (cat no., M2101; Promega Corporation,
Madison, WI, USA), and using the primers: Forward,
5′-ATAGCTAGCCACAGGAAGCCTGCAGTCCTGG-3′ and reverse,
5′-CCTGCCCTCGAGAGGTTTACAGTCTAGG-3′. The 3′UTR was then inserted at
the Xba I site immediately downstream of the luciferase gene
in the pGL3-control vector (Promega Corporation). The sequence of
the positive control (miRNA mimic control) is
5′-GGUUCGUACGUACACUGUUCA-3′. At 24 h prior to transfection, the
cells were seeded on 12-well plates at a density of
1×105 cells/well and co-transfected with the luciferase
reporter constructs and miR-17-5p plasmid or positive control
(miRNA mimic control) sequences with Lipofectamine®
2000. After 24 h, luciferase activity was analyzed using the
Dual-Luciferase Reporter Assay system (Promega Corporation).
Relative luciferase activity was normalized to Renilla luciferase
activity. Untransfected cells were used as negative controls.
Western blot analysis
SGC7901/DDP cells were cultured in a humidified
atmosphere containing 5% CO2 at 37°C and plated in
6-well plates (3×105 cells/well) and homogenized with
lysis buffer (Beyotime Institute of Biotechnology, Haimen, China)
72 h following the transfection of the cells with miR-17-5p
inhibitor and miRNA inhibitor control. Total protein (20 µg per
lane) was separated by 12% SDS-PAGE and transferred to
polyvinylidene fluoride (PVDF) membranes. Subsequent to blocking
with 5% nonfat dried milk in TBS and Tween 20 (TBST) at 25°C for 90
min, the membrane was incubated at 4°C overnight with primary
monoclonal antibody against p21 (dilution, 1:200; cat no.,
sc-271610; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and
β-actin (dilution, 1:5,000; cat no., A1978; Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany). Subsequent to wash in TBST for 40 min,
secondary goat anti-rabbit IgG horseradish peroxidase (dilution,
1:2,000; cat no., ab97200; Abcam, Cambridge, UK) was incubated with
the PVDF membranes for 1 h at 25°C. After washing in TBST for 40
min, the protein bands were visualized using an enhanced
chemiluminescence kit (cat no., orb90504; Biorbyt Ltd., Cambridge,
UK).
Apoptosis and cell cycle assay
At 24 h following transfection, SGC7901/DDP cells
were treated with DDP at a final concentration of 1 µg/ml. The
floating and attached cells were collected and divided into two
groups. One group of cells was washed twice in PBS and re-suspended
in 100 µl Hanks' balanced salt solution (HBSS; cat no., 14025092;
Gibco; Thermo Fisher Scientific, Inc.) with 10% FCS. Next, 5 µl
Annexin V-fluorescein isothiocyanate (FITC; Beyotime Institute of
Biotechnology) and propidium iodide (PI; 0.1 mg/ml; Sigma-Aldrich;
Merck KGaA) were added and stained at 25°C for 20 min. The cells
were then re-suspended in 500 µl HBSS, and flow cytometry was used
to detect apoptosis of cells by determining the relative proportion
of Annexin V-FITC-positive/PI-negative cells within 30 min. The
other group of cells was washed with PBS and fixed with 70%
ice-cold ethanol for 24 h at 4°C. The fixed cells were washed twice
with PBS and incubated with 0.5 ml PI at 25°C for 30 min. The cell
cycle profile was determined via flow cytometry (FlowJo version
10.0.7; FlowJo LLC, Ashland, OR, USA).
Statistical analysis
Each experiment was repeated at least three times.
Numerical data are presented as the mean ± standard deviation. The
differences between two groups were analyzed with the Student's
t-test, multiple groups were analyzed by one-way analysis of
variance (ANOVA) followed by Dunnett's multiple comparisons test.
All statistical analyses were performed using SPSS software
(version 11.0; SPSS, Inc., Chicago, IL, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Comparison of miRNA expression in
parental SGC7901 and SGC7901/DDP cells
A comparison of the levels of miRNA expression
between SGC7901/DDP cells and SGC7901 cells is shown in Table I. In total, 10 out of 414 human miRNAs
examined were significantly downregulated by >2-fold in
SGC7901/DDP cells compared with SGC7901 cells. By contrast, 7
miRNAs were significantly upregulated >2-fold in SGC7901/DDP
cells. These findings suggest that these miRNAs may perform an
important role in the development of drug resistance in SGC7901/DDP
cells.
 | Table I.Differentially expressed miRNAs in
SGC7901/cisplatin and SGC7901 cells. |
Table I.
Differentially expressed miRNAs in
SGC7901/cisplatin and SGC7901 cells.
| miRNAs | Up-or down-regulated
in DDP |
Fold-changea | Locib | Putative targets
associated with multidrug resistancec |
|---|
| miR-17-5p | Up | 18.04±3.28 | 13q31.3 | E2F1, p21, MCL1 |
| miR-27a-3p | Up | 20.42±3.90 | 19p13.12 | RAS |
| miR-34a | Up | 3.05±1.89 | 1p36.23 | p27 |
| miR-199b-5p | Up | 3.43±2.08 | 9q34.11 | RAS |
| miR-222-3p | Up | 2.52±1.53 | Xp11.3 | p27, p57 |
| miR-301a-3p | Up | 7.50±2.39 | 17q22 |
|
| miR-425-5p | Up | 2.77±1.74 | 3p21.31 | BCL-2 |
| let-7a | Down | −6.21±2.43 | 9q22.2;11q24.2;
22q13.3 | RAS, ABCC5 |
| miR-9-3p | Down | −3.78±2.81 |
1q22;5q14.3;15q26.1 | RAS |
| miR-25-5p | Down | −4.11±2.20 | 7q22.1 | MCL1, RAS |
| miR-147b | Down | −2.61±1.22 | 15q21.1 |
|
| miR-150-5p | Down | −5.97±2.01 | 19q13.33 | p27, RAS |
| miR-219-1-3p | Down | −7.20±3.00 | 6p22.3 |
|
| miR-340-3p | Down | −4.32±1.23 | 5q35.3 |
|
| miR-431-5p | Down | −13.22±3.39 | 14q32.31 | RAS |
| miR-553 | Down | −3.64±2.52 | 1p21.2 |
|
| miR-654 | Down | −9.10±2.59 | 14q32.31 | RAS |
miR-17-5p expression is upregulated in
SGC7901/DDP cells compared with SGC7901 cells
miRNA microarray analysis of SGC7901 and SGC7901/DDP
cells demonstrated that miR-17-5p was significantly upregulated in
SGC7901/DDP cells compared with the parental SCG7901 cells
(18.04±3.28-fold; P<0.01). qPCR for miR-17-5p confirmed that
miR-17-5p was upregulated by 3.20±0.66-fold (P<0.01) in
SGC7901/DDP cells compared with SGC7901 cells, where the expression
was set as 1 (Table II).
 | Table II.Validation of microRNA-17-5p
upregulation in SGC7901/DDP cells by quantitative polymerase chain
reaction analysis. |
Table II.
Validation of microRNA-17-5p
upregulation in SGC7901/DDP cells by quantitative polymerase chain
reaction analysis.
| Groups | ΔCq |
2−ΔΔCq | P-value |
|---|
| SGC7901 | 15.229±0.373 | 1 |
|
| SGC7901/DDP | 13.579±0.327 | 3.200±0.66 | <0.01 |
Downregulation of miR-17-5p reverses
drug resistance of gastric cancer cells
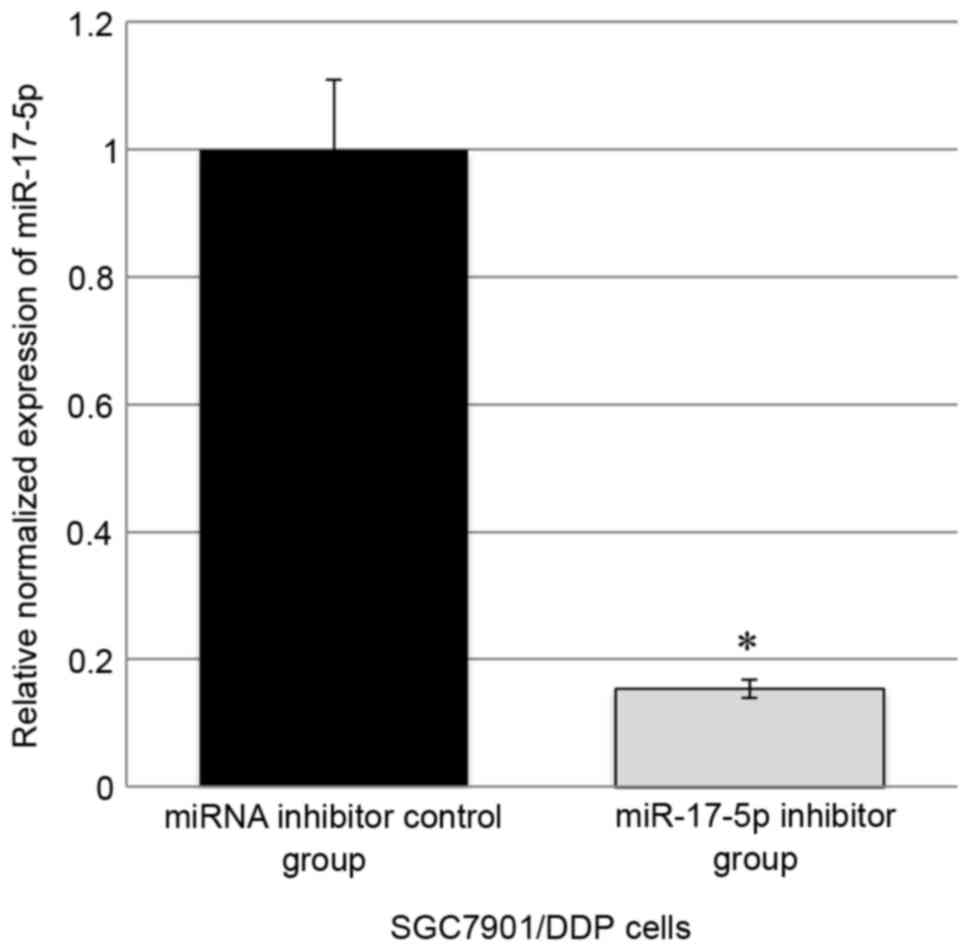
In SGC7901/DDP cells, transfection of the miR-17-5p
inhibitor was able to significantly inhibit the expression of
miR-17-5p by ~85% compared with the control (Fig. 1). MTT assay revealed that miR-17-5p
inhibitor-transfected cells exhibited markedly decreased resistance
to DDP, ADR, VCR and 5-Fu compared with the cells transfected with
miRNA inhibitor control, indicating that the downregulation of
miR-17-5p was able to reverse drug resistance of SGC7901/DDP cells
(Table III).
 | Table III.IC50 values of drugs for
gastric cancer cells. |
Table III.
IC50 values of drugs for
gastric cancer cells.
|
| IC50,
µg/ml |
|---|
|
|
|
|---|
| Groups | DDP | Adriamycin | Vincristine | 5-fluorouracil |
|---|
| SGC7901/DDP +
miR-17-5p inhibitor |
3.964±0.508a |
0.413±0.039a |
8.673±0.433a |
3.664±0.458a |
| SGC7901/DDP +
inhibitor control | 7.082±0.605 | 0.705±0.118 | 12.335±0.556 | 5.898±0.817 |
| Untreated SGC7901
cells |
2.165±0.218a |
0.220±0.016a |
7.057±0.179a |
3.582±0.139a |
p21 may be a target gene of
miR-17-5p
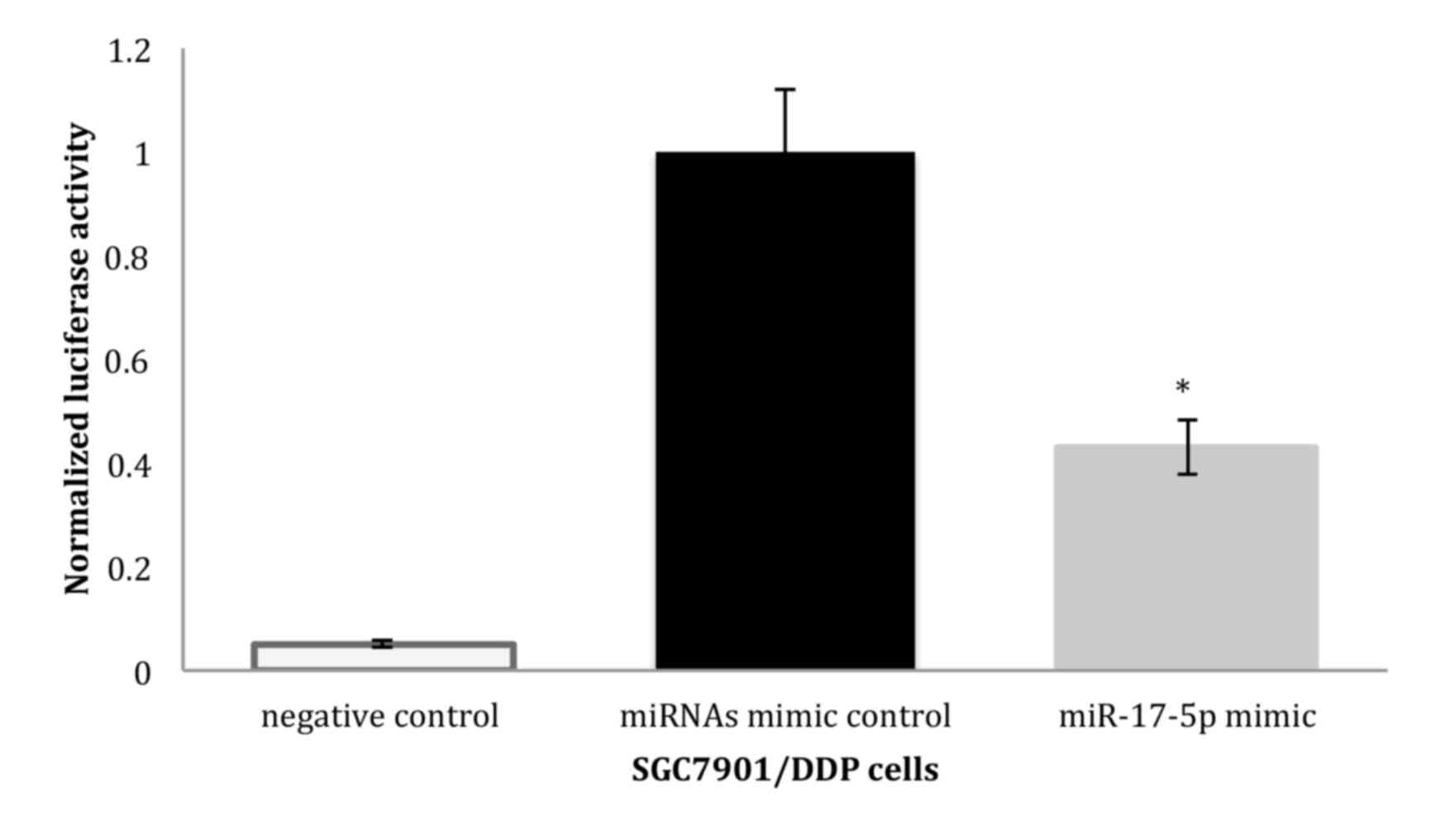
PicTar and Miranda algorithms predicted that p21 was
a potential target for miR-17-5p. To investigate whether p21 is the
target gene of the miR-17-5p, a luciferase reporter vector with the
putative p21 3′UTR target site for the miR-17-5p downstream of the
luciferase gene (pGL3-p21-3′UTR) was constructed (please see the
method and Materials). Luciferase reporter constructs, together
with the miR-17-5p plasmid or the control miRNAs, were transfected
into SGC7901/DDP cells. In SGC7901/DDP cells, a significant
decrease in relative luciferase activity was noted when
pGL3-p21-3′UTR was co-transfected with the miR-17-5p plasmid but
not with the control miRNAs, indicating that p21 may be the target
gene of miR-17-5p (Fig. 2).
Downregulation of miR-17-5p increases
p21 expression in SGC7901/DDP cells
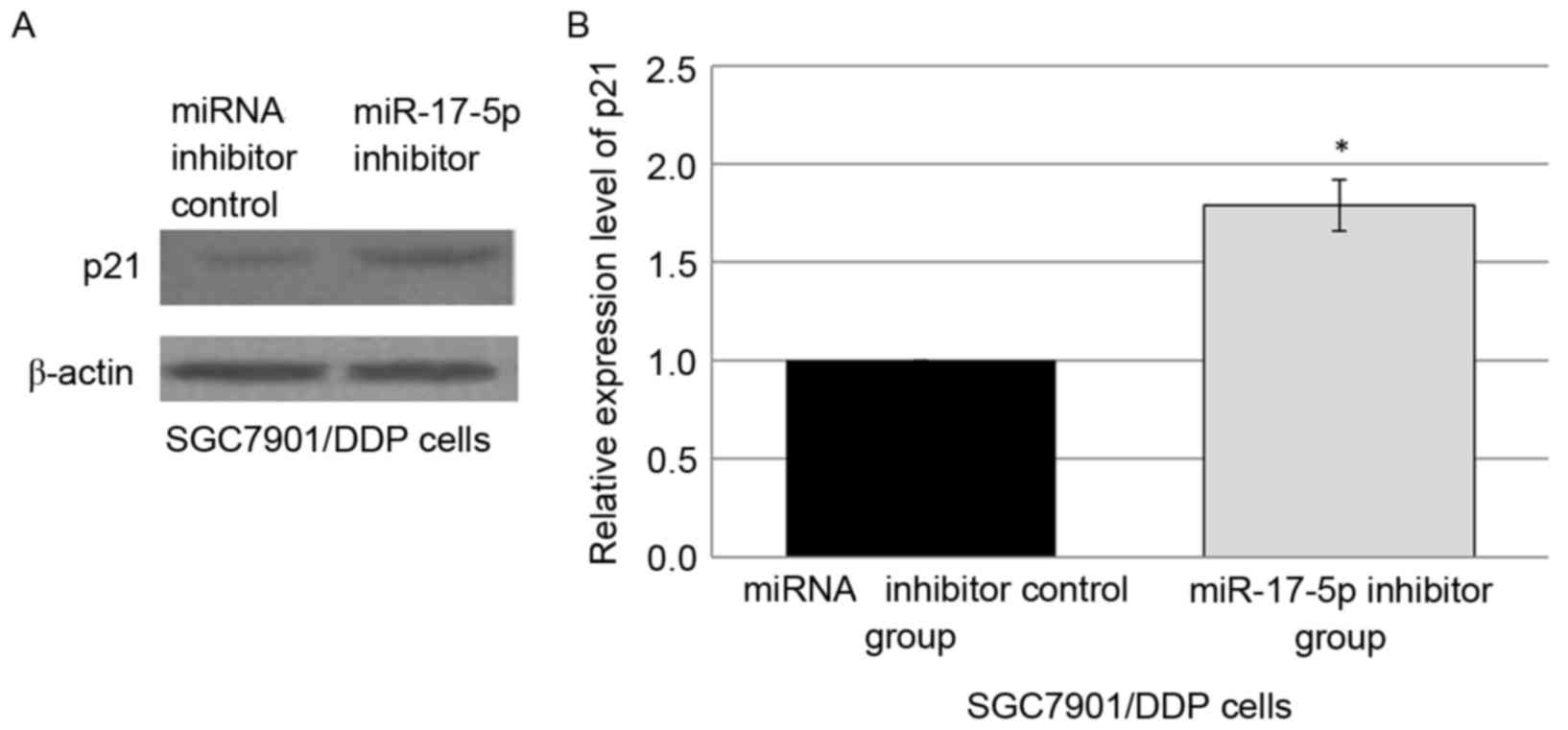
Since it was indicated that p21 may be the target of
miR-17-5p and that p21 was associated with apoptosis, it was
hypothesized that miR-17-5p may perform a role in the development
of drug resistance, at least in part by modulation of apoptosis via
targeting p21. Western blot analysis was employed to analyze p21
levels in the SGC7901/DDP cells transfected with either control
miRNA inhibitor or miR-17-5p inhibitor. The downregulation of
miR-17-5p expression in the miR-17-5p inhibitor-transfected
SGC7901/DDP cells was observed in concurrence with the
overexpression of p21 protein, compared with miRNA inhibitor
control group (Fig. 3A and B).
Downregulation of miR-17-5p increases
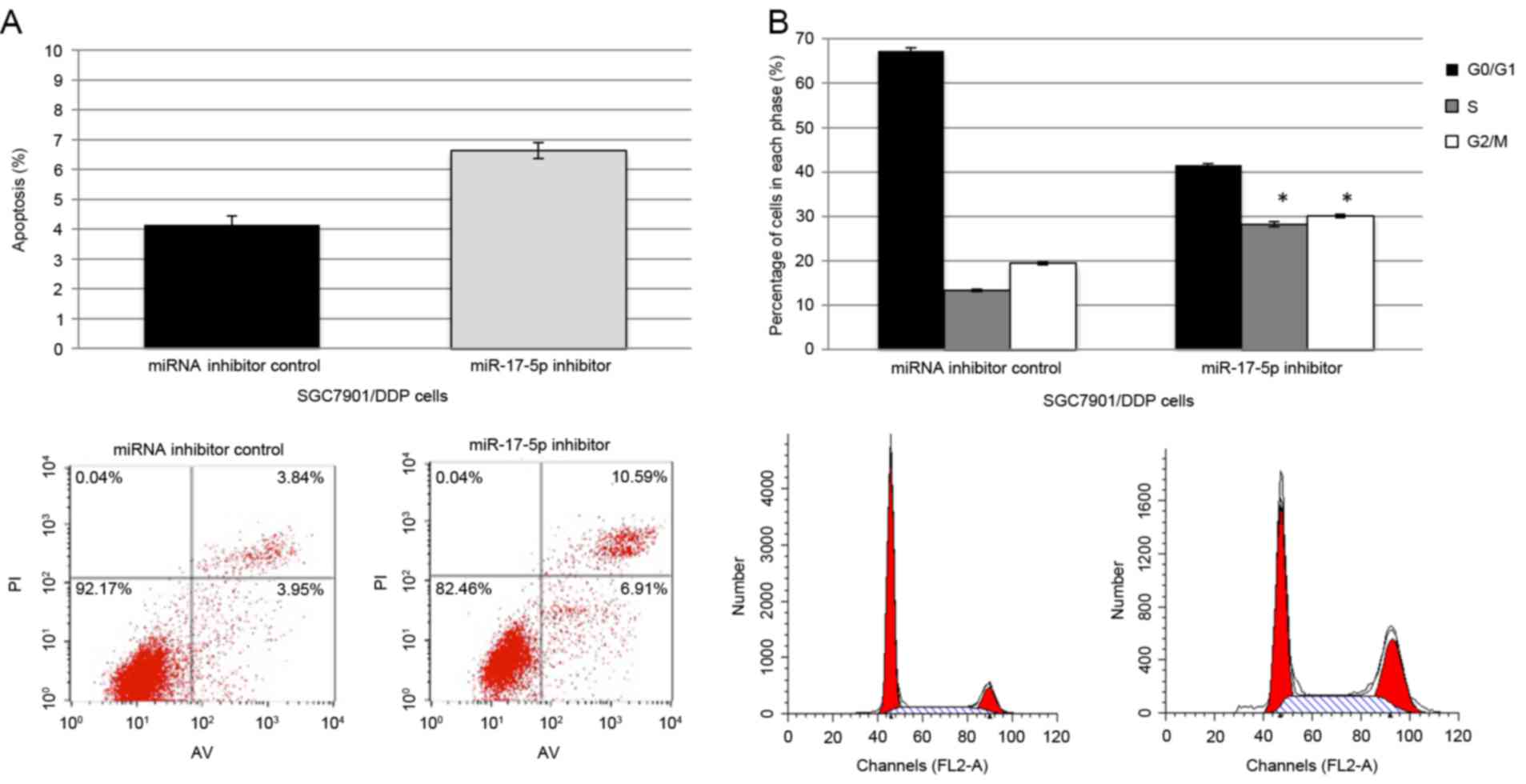
the sensitivity of SGC7901/DDP cells to DDP-induced apoptosis
DDP-induced apoptosis and cell cycle arrest
following transfection with the miR-17-5p inhibitor and the control
miRNAs inhibitor were assessed by flow cytometry in SGC7901/DDP
cells. Following transfection with miR-17-5p inhibitor, a marked
increase in apoptosis was observed in the DDP-treated cells,
compared with the cells transfected with miRNA inhibitor control
(Fig. 4A). Simultaneously, an
increase in the percentage of cells that are arrested at G1/S in
the cells that were transfected with the miR-17-5p inhibitor was
observed, as indicated by an increase in the percentage of cells at
S and G2/M and a decrease at G0/G1 (Fig.
4B).
Discussion
Chemotherapy resistance is a common occurrence and
contributes to cancer mortality, since it often leads to failure in
the inhibition of disease progression. The mechanisms of drug
resistance are complex and multifactorial, and one of the main
mechanisms is the apoptotic pathway being defective in cancer cells
(11). The development of drug
resistance in various types of cancer cells has been reported to be
associated with a reduced susceptibility to drug-induced apoptosis,
which is partially due to the overexpression of anti-apoptotic
proteins (5,12).
miRNAs have been shown to perform a role in the
initiation of cancer, tumor progression and sensitivity to
treatment (13,14). Current studies suggest that miRNAs
also perform important roles in mediating resistance to
chemotherapy (11,15). In the present study, a subset of
differentially-expressed miRNAs were identified, and in particular,
it was observed that miR-17-5p was significantly upregulated in
SGC7901/DDP cells compared with SGC7901 cells.
miR-17-5p is part of the miR-17-92 polycistronic
cluster, a family of oncogenic miRNAs (miR-17-5p, miR-17-3p,
miR-18a, miR-19a, miR-20a, miR-19b-1 and miR-92-1) commonly
deregulated in cancer, on chromosome 13 (16). It was previously reported that miR-17
is upregulated in colorectal (17–19),
gastric (19,20) and pancreatic adenocarcinomas (19). A previous study demonstrated that
miR-17-5p directly controls the expression of the type II
transforming growth factor-β (TGF-β) receptor in colorectal cancer
progression, inhibits the transcription of individual TGF-β
responsive genes and indirectly stimulates angiogenesis through
inhibition of a wide repertoire of anti-angiogenic factors
(21). It has also been reported that
miR-17-5p, which may function as a pro-proliferative factor,
increases the proliferation and growth of gastric cancer cells
in vitro and in vivo (22). Fontana et al (23) revealed that miR-17-5p is expressed at
higher levels in neuroblastoma cell lines that exhibit
overexpression of neuroblastoma-derived V-Myc Avian
myelocytomatosis (MYCN) compared with cell lines with a
lower expression level of MYCN. It was also demonstrated
that the overexpression of the miR-17-92 cluster markedly inhibits
hypoxia-induced apoptosis in colon cancer cell lines through the
regulation of p53-mediated transcriptional repression (24). In the present study, it was
demonstrated that miR-17-5p may induce resistance to chemotherapy
by regulating p21 expression in gastric cancer cells. Similar
observations have been reported in neuroblastoma cells (23) and endometrial cancer cells (25). p21 is a tumor suppressor gene, which
was identified to be involved in various biological processes,
including tumor apoptosis, cell cycle blockage, metastasis and
chemotherapy resistance (26).
However, the underlying mechanism was not completely clarified.
Considering the well-characterized role of p21 in apoptosis and
drug resistance, miR-17-5p may perform a role in the development of
drug resistance of cancer cells at least in part by modulating
apoptosis via targeting p21.
Since a single miRNA may potentially regulate a wide
range of target genes, the functions of miRNAs are cell-type
specific. Studies have indicated that the function of miR-17-5p
appears to be conflicting in different cancer cell types. Fan et
al (27) reported that miR-17-5p
serves a role in suppressing epithelial mesenchymal transition and
metastasis in breast cancer cells. However, another study
demonstrated that the downregulation of miR-17-5p contributes to
the resistance of lung cancer cells to paclitaxel (28). One potential explanation for these
contradictory functions of miR-17-5p on regulating apoptosis may be
due to the balance of expression levels between its targeted pro-
and anti-apoptotic genes in different cell types.
In summary, to the best of our knowledge, the
current study presented the first evidence that miR-17-5p may be
involved in the development of drug resistance in human gastric
cancer cell lines. miR-17-5p may modulate the resistance of gastric
cancer cell lines to certain anticancer drugs, at least in part,
through targeting p21 expression. The present study has provided a
rationale for the development of novel therapeutics to overcome
drug resistance in gastric cancer. One limitation of the present
study is that the present data are derived from cell lines that
have been removed from their in vivo context and cannot be
considered accurate surrogates for clinical tumors. Therefore,
future studies to assess the roles of miR-17-5p in vivo and
in clinical context are required.
References
|
1
|
Du Y, Xu Y, Ding L, Yao H, Yu H, Zhou T
and Si J: Down-regulation of miR-141 in gastric cancer and its
involvement in cell growth. J Gastroenterol. 44:556–561. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cunningham D, Allum WH, Stenning SP,
Thompson JN, Van de Velde CJ, Nicolson M, Scarffe JH, Lofts FJ,
Falk SJ, Iveson TJ, et al: Perioperative chemotherapy versus
surgery alone for resectable gastroesophageal cancer. N Engl J Med.
355:11–20. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fang L, Li H, Wang L, Hu J, Jin T, Wang J
and Yang BB: MicroRNA-17-5p promotes chemotherapeutic drug
resistance and tumour metastasis of colorectal cancer by repressing
PTEN expression. Oncotarget. 5:2974–2987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rivera F, Vega-Villegas ME and Lόpez-Brea
MF: Chemotherapy of advanced gastric cancer. Cancer Treat Rev.
33:315–324. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhu W, Zhu D, Lu S, Wang T, Wang J, Jiang
B, Shu Y and Liu P: MiR-497 modulates multidrug resistance of human
cancer cell lines by targeting BCL2. Med Oncol. 29:384–391. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNA in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kovalchuk O, Filkowski J, Meservy J,
Ilnytskyy Y, Tryndyak VP, Chekhun VF and Pogribny IP: Involvement
of microRNA-451 in resistance of the MCF-7 breast cancer cells to
chemotherapeutic drug doxorubicin. Mol Cancer Ther. 7:2152–2159.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hu H, Li S, Cui X, Lv X, Jiao Y, Yu F, Yao
H, Song E, Chen Y, Wang M and Lin L: The overexpression of
hypomethylated miR-663 induces chemotherapy resistance in human
breast cancer cells by targeting heparin sulfate proteoglycan 2
(HSPG2). J Biol Chem. 288:10973–10985. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luo HC: MiRNAs expression profiling of
gastric carcinoma and function of significantly down-regulated
miR-9 and miR-433. Chongqing Chongqing Med Univ. 1–85. 2010.
|
|
10
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fojo T: Multiple paths to a drug
resistance phenotype: Mutations, translocations, deletions and
amplification of coding genes or promoter regions, epigenetic
changes and microRNA. Drug Resist Updat. 10:59–67. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun
S, Hong L, Liu J and Fan D: MiR-15b and miR-16 modulate multidrug
resistance by targeting BCL2 in human gastric cancer cells. Int J
Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kastl L, Brown I and Schofield AC:
MiRNAs-34a is associated with docetaxel resistance in human breast
cancer cells. Breast Cancer Res Treat. 131:445–454. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ueda T, Volinia S, Okumura H, Shimizu M,
Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: A microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Szakács G, Paterson JK, Ludwig JA,
Booth-genthe C and Gottesman MM: Targeting multidrug resistance in
cancer. Nat Rev Drug Discov. 5:219–234. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
He L, Thomson JM, Hemann MT,
Hernando-monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe
SW, Hannon GJ and Hammond SM: A microRNA polycistron as a potential
human oncogene. Nature. 435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Motoyama K, Inoue H, Takatsuno Y, Tanaka
F, Mimori K, Uetake H, Sugihara K and Mori M: Over- and
under-expressed microRNAs in human colorectal cancer. Int J Oncol.
34:1069–1075. 2009.PubMed/NCBI
|
|
18
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:pp. 2257–2261.
2006; View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li X, Zhang Y, Zhang Y, Ding J, Wu K and
Fan D: Survival prediction of gastric cancer by a seven-microRNA
signature. Gut. 59:579–585. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dews M, Fox JL, Hultine S, Sundaram P,
Wang W, Liu YY, Furth E, Enders GH, El-Deiry W, Schelter JM, et al:
The myc-miR-17~92 axis blunts TGF{beta} signaling and production of
multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res.
70:8233–8246. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu Q, Luo G, Yang Z, Zhu F, An Y, Shi Y
and Fan D: miR-17-5p promotes proliferation by targeting SOCS6 in
gastric cancer cells. FEBS Lett. 588:2055–2062. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fontana L, Fiori ME, Albini S, Cifaldi L,
Giovinazzi S, Forloni M, Boldrini R, Donfrancesco A, Federici V,
Giacomini P, et al: Antagomir-17-5p abolishes the growth of
therapy-resistant neuroblastoma through p21 and BIM. PLoS One.
3:e22362008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yan HL, Xue G, Mei Q, Wang YZ, Ding FX,
Liu MF, Lu MH, Tang Y, Yu HY and Sun SH: Repression of the
miR-17-92 cluster by p53 has an important function in
hypoxia-induced apoptosis. EMBO J. 28:2719–2732. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen Y, Lu L, Xu J, Meng W, Qing Y, Liu Y,
Zhang B and Hu H: Bortezomib induces apoptosis of endometrial
cancer cells through microRNA-17-5p by targeting p21. Cell Biol
Int. 37:1114–1121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huether A, Höpfner M, Baradari V, Schuppan
D and Scherübl H: EGFR blockade by cetuximab alone or as
combination therapy for growth control of hepatocellular cancer.
Biochem Pharmacol. 70:1568–1578. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan M, Sethuraman A, Brown M, Sun W and
Pfeffer LM: Systematic analysis of metastasis-associated genes
identifies miR-17-5p as a metastatic suppressor of basal-like
breast cancer. Breast Cancer Res Treat. 146:487–502. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chatterjee A, Chattopadhyay D and
Chakrabarti G: miR-17-5p downregulation contributes to paclitaxel
resistance of lung cancer cells through altering beclin1
expression. PLoS One. 9:e957162014. View Article : Google Scholar : PubMed/NCBI
|