Introduction
Tumor tissues are highly heterogeneous. Several
types of cell interact with each other, generating a diverse array
of drug sensitivities (1).
Reconstruction of this heterogeneity in a preclinical study may
provide a method to predict the efficacy of novel anticancer drugs
in clinical study (2).
Patient-derived tumor xenografts (PDXs) are established by
maintaining the tumor tissues derived from patients with cancer in
the flank of immuno-deficient mice (3,4). Compared
with conventional cancer cell lines, PDXs have been reported to be
more relevant to the original tumor tissues of patients with cancer
in terms of drug sensitivity, heterogeneity and genetic status
(5–9).
Accordingly, PDXs may be used for the evaluation of new anticancer
drugs (10).
Lung cancer is categorized into small cell lung
cancer (SCLC) and non-small cell lung cancer (NSCLC). The most
common types of NSCLC are squamous cell carcinoma, adenocarcinoma
and large cell carcinoma (11). These
types of NSCLC are further classified into several stages according
to their disease status (12).
Several anticancer drugs have been developed for each type of lung
cancer. Their molecular targets include epidermal growth factor
receptor (EGFR) tyrosine kinase [erlotinib (13), gefitinib (14) and afatinib (15)], rearranged anaplastic lymphoma kinase
[crizotinib (16), ceritinib
(17) and alectinib (18)], folate-dependent enzymes [pemetrexed
(19)], vascular endothelial growth
factor [bevacizumab (20)], C-X-C
chemokine receptor type 4 [LY2510924 (21)] and programmed cell death protein 1
[nivolumab (22)]. Although these
drugs demonstrated therapeutic effects in patients with several
types of lung cancer, novel drugs targeting underlying molecular
mechanisms are required in order to progress therapeutic approaches
to lung cancer.
Kinetochore-associated protein 2 (KNTC2), also known
as highly expressed in cancer 1, serves an important function in
the segregation of chromosomes during M phase (23). KNTC2 is upregulated in tumor tissues
of cancer patients and has been identified as a potential target
for cancer therapy (24). Antitumor
activities of KNTC2 short interfering (si)RNA have previously been
demonstrated in vivo in orthotopic hepatocellular carcinoma
tumor mouse models (25). However, it
remains unclear whether KNTC2 siRNA exhibits antitumor activities
against lung cancer PDXs.
To clarify the involvement of KNTC2 in the growth of
lung cancer PDXs, KNTC2 siRNA was encapsulated into a lipid
nanoparticle (KNTC2-LNP) and the growth inhibiting activities of
KNTC2-LNP were evaluated in a three-dimensional (3D)-culture system
and subcutaneous tumor models of lung cancer PDXs; LC-60 (resistant
to erlotinib) and LC-45 (sensitive to erlotinib).
Materials and methods
Lung cancer PDXs
Lung cancer PDXs, LC-45 (adenocarcinoma) and LC-60
(small or large cell carcinoma) were purchased from the Central
Institute for Experimental Animals (Kanagawa, Japan). PDXs were
maintained in the flank of 20 (LC-45) or 15 (LC-60) BALB/c nude
mice (20–25 g; 7–8 weeks old; female; Charles River Laboratories
International, Inc., Kanagawa, Japan). When PDXs grew to ~1,000
mm3, PDXs were excised and cryopreserved in small pieces
(40–70 µm) using Cell Banker 2 (Nippon Zenyaku Kogyo, Co., Ltd.,
Fukushima, Japan). Humane endpoint was determined by 20% weight
loss and the maximum tumor size was 2,000 mm3. Mice were
maintained in specific pathogen-free conditions with free access to
food and water, under a constant temperature of 22±2°C and a 12/12
h light/dark cycle. All experiments were approved by the
Institutional Animal Care and Use Committee in Takeda
Pharmaceutical Company Limited (Fujisawa, Japan; approval number
11387).
Chemical modification of siRNAs
KNTC2 siRNA and Luc siRNA (26) (negative control) were synthesized by
GeneDesign (Osaka, Japan) and chemically modified with
2′-O-methyl ribonucleotide to prevent immune responses, as
described previously (27).
Phosphorothioate (PS) modifications were not used in KNTC2 siRNA
because they generate optical isomers. The sequences and chemical
modifications are presented in Table
I.
 | Table I.List of chemically modified
siRNAs. |
Table I.
List of chemically modified
siRNAs.
| siRNA | Strand | Sequence |
|---|
| Luc siRNA | Sense |
5′-r(CUUACGCUGAGUACUUCGA)-dTsdT-3′ |
|
| Antisense |
5′-r(UCGAAGUACUCAGCGUAAG)-dTsdT-3′ |
| KNTC2 siRNA | Sense |
5′-r(UAGUCAACUUGGUAUAUUU)-dTdT-3′ |
|
| Antisense |
5′-r(AAAUAUACCAAGUUGACUA)-dTdT-3′ |
Encapsulation of siRNAs into a lipid
nanoparticle
KNTC2 siRNA or Luc siRNA (200 µg/ml, total 8 mg) was
encapsulated into a lipid nanoparticle (LNP) using a microfluidic
device, Asia (model no. 210; Syrris, Royston, UK). Each LNP was
composed of 3-((5-(dimethylamino)
pentanoyl)oxy)-2,2-bis(((3-pentyloctanoyl)oxy)methyl)propyl
3-pentyloctanoate (28),
dipalmitoylphosphatidylcholine (NOF Corporation, Tokyo, Japan),
cholesterol (Avanti Polar Lipids, Inc., Alabaster, Alabama) and
GS-020 (NOF Corporation) at the molar ratio of 60, 10.6, 28 and
1.4, respectively. Particle size and polydispersity index (PdI) of
LNPs were measured by dynamic light scattering using a Zetasizer
Nano ZS (Malvern Instruments Ltd., Malvern, UK). The ratio of siRNA
entrapment was calculated using Ribogreen (Thermo Fisher
Scientific, Inc., Waltham, MA) and Triton X-100 as described
previously (29). Briefly, LNPs were
dissolved in 1% Triton X-100 to release the siRNAs. The
concentrations of siRNA prior to and following the dissolution were
calculated by applying Ribogreen (29) at a final concentration of 0.25% and
measuring the fluorescence of Ribogreen using a spectrofluorometer,
Envision (Excitation, 485 nm and Emission, 535 nm) and Envision
software (version 1.13.3009.1409, PerkinElmer, Inc., Waltham, MA,
USA).
Evaluation of knockdown activities in
subcutaneous tumor models of PDXs
Small pieces (~100 mm3) of PDXs were
inoculated in the flank of BALB/c nude mice using a trocar needle
(KN-391; Natsume Seisakusho, Co., Ltd., Tokyo, Japan). Tumor sizes
were measured with calipers and defined as major axis × minor
axis2/2, as previously described (9). When the tumor sizes reached between 100
and 400 mm3, 9 mice were selected from a total of 20
mice and divided into three groups (PBS, KNTC2 and Luc) using EXSUS
2014 software (CAC Exicare Corporation, Tokyo, Japan). KNTC2 siRNA
encapsulated into LNP (KNTC2-LNP) was intravenously administered at
5 mg/kg (n=3). Luc siRNA-LNP was used as the negative control. PBS
was administered to the vehicle control group. Knockdown activity
was measured 3 days after the single administration. Total RNA was
extracted from tumor tissue using TRIzol® (Life
Technologies; Thermo Fisher Scientific, Inc.) and reverse
transcribed (thermocycling conditions: 25°C for 10 min; 42°C for 1
h; and 85°C for 5 min) using SuperScript VILO cDNA Synthesis kit
(Thermo Fisher Scientific, Inc.). The copy numbers of human
KNTC2, human β-actin (ACTB), mouse Kntc2 and
mouse Actb mRNA were individually measured by quantitative
polymerase chain reaction using a Real-Time PCR System (Thermo
Fisher Scientific, Inc.). Species-specific qPCR primers and probes
(Thermo Fisher Scientific, Inc.) are listed in Table II. Copy numbers of KNTC2 or
Kntc2 mRNA were individually normalized to ACTB or
Actb mRNA.
 | Table II.List of species specific quantitative
polymerase chain reaction primers and probes. |
Table II.
List of species specific quantitative
polymerase chain reaction primers and probes.
| Gene | Sequence
(5′-3′) |
|---|
| Human
KNTC2 |
|
|
Forward |
GAGGTACATAAACTTGAGCCCTGTATT |
|
Reverse |
TGCTGAGAATTCCAAAGGTTATGA |
|
Probe |
TGGCACCAGCCTCGGGATTAAACTTAA |
| Human
ACTB |
|
|
Forward |
CCTGGCACCCAGCACAAT |
|
Reverse | GCCGATCCACACGGAGTA
CT |
|
Probe |
ATCAAGATCATTGCTCCTCCTGAGCGC |
| Mouse
Kntc2 |
|
|
Forward |
GAATAAAAAGAGGCATCTGGAGGATAC |
|
Reverse |
CCTCCTTCAGCATCCTCACAGT |
|
Probe |
CAACTGAACACCATGAAAACGGAAAGCAA |
| Mouse
Actb |
|
|
Forward | CACTATTGGCAACGAGC
GG) |
|
Reverse |
TCCATACCCAAGAAGGAAGGC |
|
Probe |
TCCGATGCCCTGAGGCTCTTTTCC |
Evaluation of growth inhibitory
activities in subcutaneous tumor models of PDXs
A total of 10 or 15 mice were selected from 20 or 45
mice (total number of mice, 65), respectively, and divided into two
(control and erlotinib) or three (control, KNTC2 and Luc) groups
using EXSUS 2014 software. Erlotinib (Carbosynth, Ltd., Compton,
UK) was orally administered at 100 mg/kg once a day for 11 days
(LC-45) or 5 days (LC-60). In addition, 0.5% methylcellulose was
used as the control (n=5). KNTC2-LNP was intravenously administered
at 5 mg/kg at three time points, with three days between each
administration. Luc siRNA-LNP was used as the negative control. PBS
was administered to the vehicle control group. Tumor sizes were
measured as aforementioned. Growth inhibitory rate (%) was
calculated using the formula: (1-tumor growth of treated
group/tumor growth of untreated group) ×100% (9).
Cryopreservation of PDXs for 3D
culture systems
PDXs were excised from BALB/c nude mice and cut into
small pieces (~100 mm3). These pieces were digested in
Dulbecco's modified eagle's medium (DMEM) high glucose (Thermo
Fisher Scientific, Inc.) containing 75 U/ml collagenase type XI
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), 125 µg/ml dispase
type II (Thermo Fisher Scientific, Inc.), 2.5% (v/v) fetal bovine
serum (Thermo Fisher Scientific, Inc.) and 100 U/ml
penicillin/streptomycin at 37°C. Digestion was terminated prior to
complete dispersion of the PDXs. PDXs of intermediate size (between
40 and 100 µm) were collected using a cell strainer and
cryopreserved in a Cell Banker 2 (Nippon Zenyaku Kogyo, Co., Ltd.)
at −160°C.
3D-culture of cryopreserved PDXs
Cryopreserved PDXs were suspended in Advanced
DMEM/F12 (ratio, 1:1; Thermo Fisher Scientific, Inc.) containing 10
mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 2 mM
GlutaMAX-1 (Thermo Fisher Scientific, Inc.), N2 supplement (Thermo
Fisher Scientific, Inc.), B27 supplement (Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin/streptomycin, 1 mM N-acetylcysteine, 500
nM A-83-01 and 1 µM SB202190 as previously described (30). Subsequently, PDXs were seeded in
U-bottom 96-well plate (Sumitomo Bakelite Co., Ltd., Tokyo, Japan)
and cultured in a 5% CO2-humidified chamber at 37°C.
Growth of PDX was monitored using Cellavista and Cellavista Control
and Evaluation Software version 2.1.0.876 (Synentec GmbH, Elmshorn,
Germany). Data was omitted when the shape of PDX was not recognized
by Cellavista.
Evaluation of knockdown activities in
3D-culture system of PDXs
PDXs were cultured for four days as aforementioned.
KNTC2-LNP was added at concentrations of 10, 100 nM and 1 µM (n=6).
PDXs were subsequently cultured for an additional three days and
mixed together due to each tumor volume being too small to obtain a
sufficient amount of total RNA for evaluating knockdown activities.
Total RNA was extracted using RNeasy kit (Qiagen GmbH, Hilden,
Germany) and reverse-transcribed using SuperScript VILO cDNA
Synthesis kit, according to manufacturer's protocols. Knockdown
activities were calculated as aforementioned.
Evaluation of growth inhibitory
activities in a 3D-culture system of PDXs
PDXs were cultured for three (LC-60) or four (LC-45)
days as aforementioned (n=6). KNTC2-LNP was added at concentrations
ranging from 10, 100 nM and 1 µM. Erlotinib (Carbosynth) was added
at concentrations of 10, 100 nM, 1 and 10 µM using a
sample-dispensing machine (HP D300 Digital Dispenser, Tecan Japan
Co., Ltd., Kawasaki, Japan). Growth inhibitory rate was calculated
using the formula as aforementioned.
Statistical analysis
Data was statistically analyzed using EXSUS 2014
software (CAC Exicare Corporation). Significance among groups was
analyzed using Bartlett's test followed by Dunnett's test (in
vivo studies) or Williams' test (in vitro studies).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Sensitivities of lung cancer PDXs to
erlotinib
To select different types of lung cancer PDXs for
the evaluation of KNTC2-LNP, the sensitivities of several PDXs to
erlotinib (an approved drug for patients with lung cancer) were
investigated. Among those PDXs, LC-45 exhibited relatively high
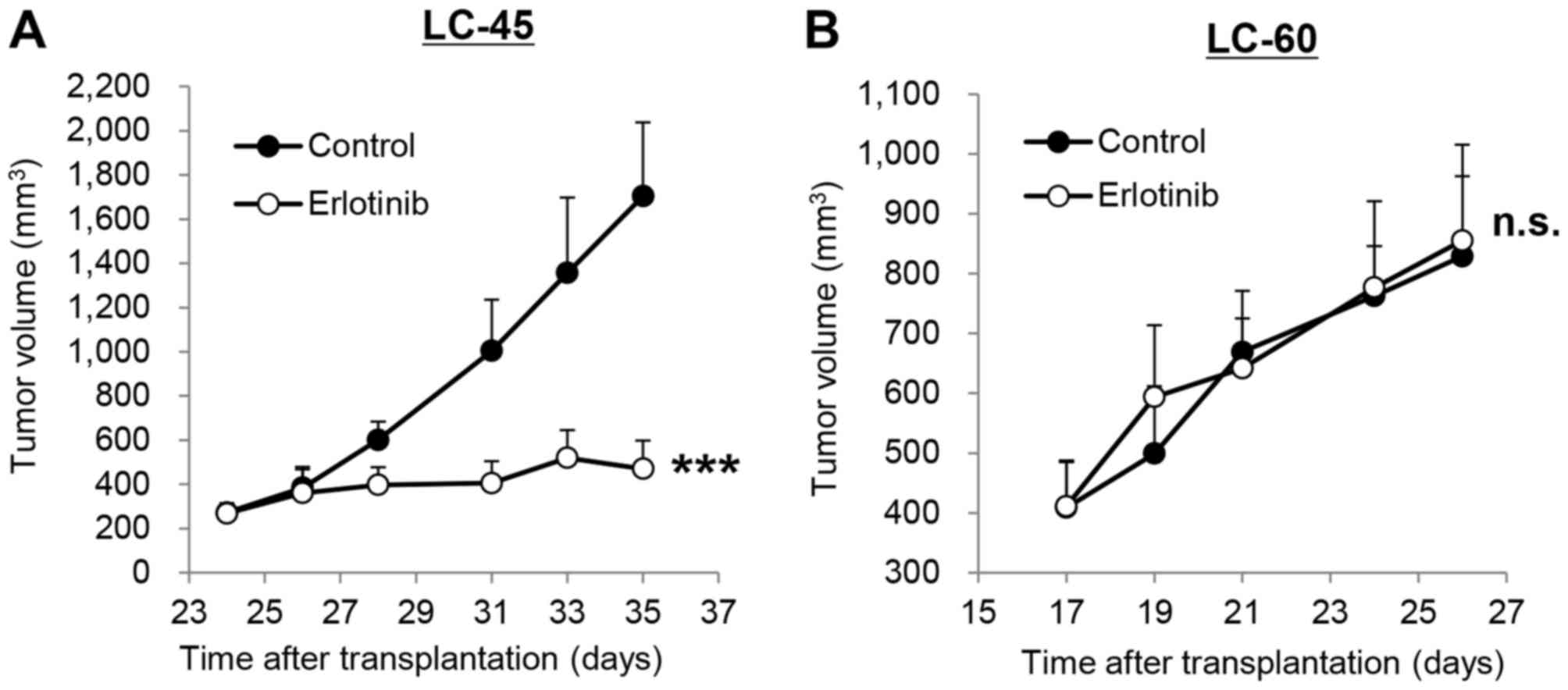
sensitivity to erlotinib in a subcutaneous tumor model (Fig. 1A). Repeated oral administration of
erlotinib (100 mg/kg, once a day) significantly (P<0.001)
inhibited the tumor growth of LC-45 by 86%. In contrast, the same
dosage of erlotinib did not inhibit the tumor growth of LC-60
(Fig. 1B).
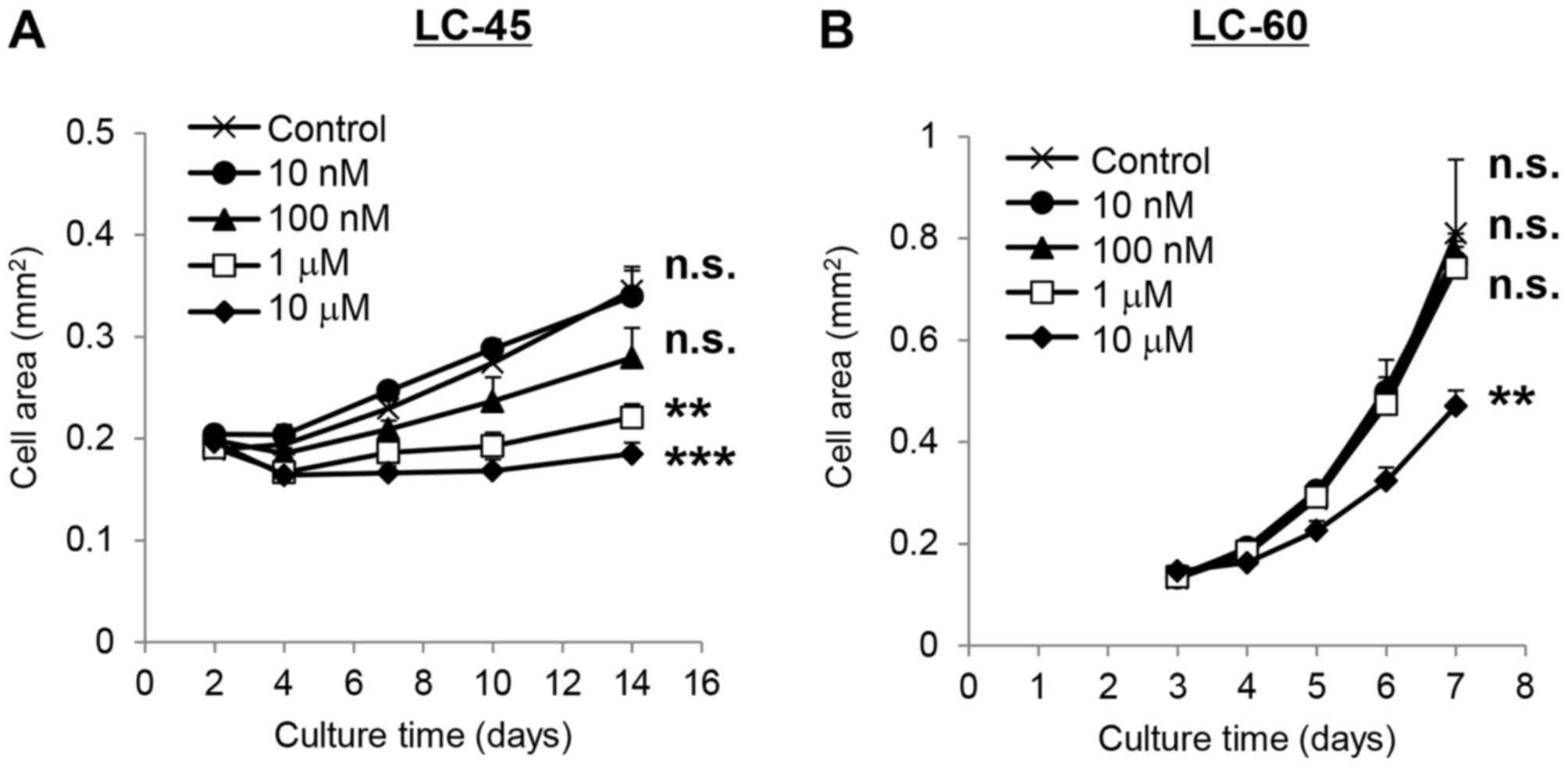
Sensitivities of LC-45 and LC-60 to erlotinib were
also investigated in 3D-culture systems. Erlotinib significantly
(P<0.001) inhibited the growth of LC-45 by 81% at the
concentration of 1 µM (Fig. 2A). The
growth of LC-60 was not inhibited by erlotinib at the same
concentration indicating that LC-60 was less sensitive to erlotinib
compared with LC-45 (Fig. 2B).
Encapsulation of siRNAs into LNP
The particle sizes of KNTC2 siRNA-LNP and Luc
siRNA-LNP were 75 and 73 nm, respectively. PdIs were 0.012 and
0.022. The entrapment efficiencies were 98.2 and 97.6%,
demonstrating that each siRNA was successfully encapsulated into
LNP.
Knockdown and growth inhibitory
activities of KNTC2-LNP in 3D-culture systems of LC-60
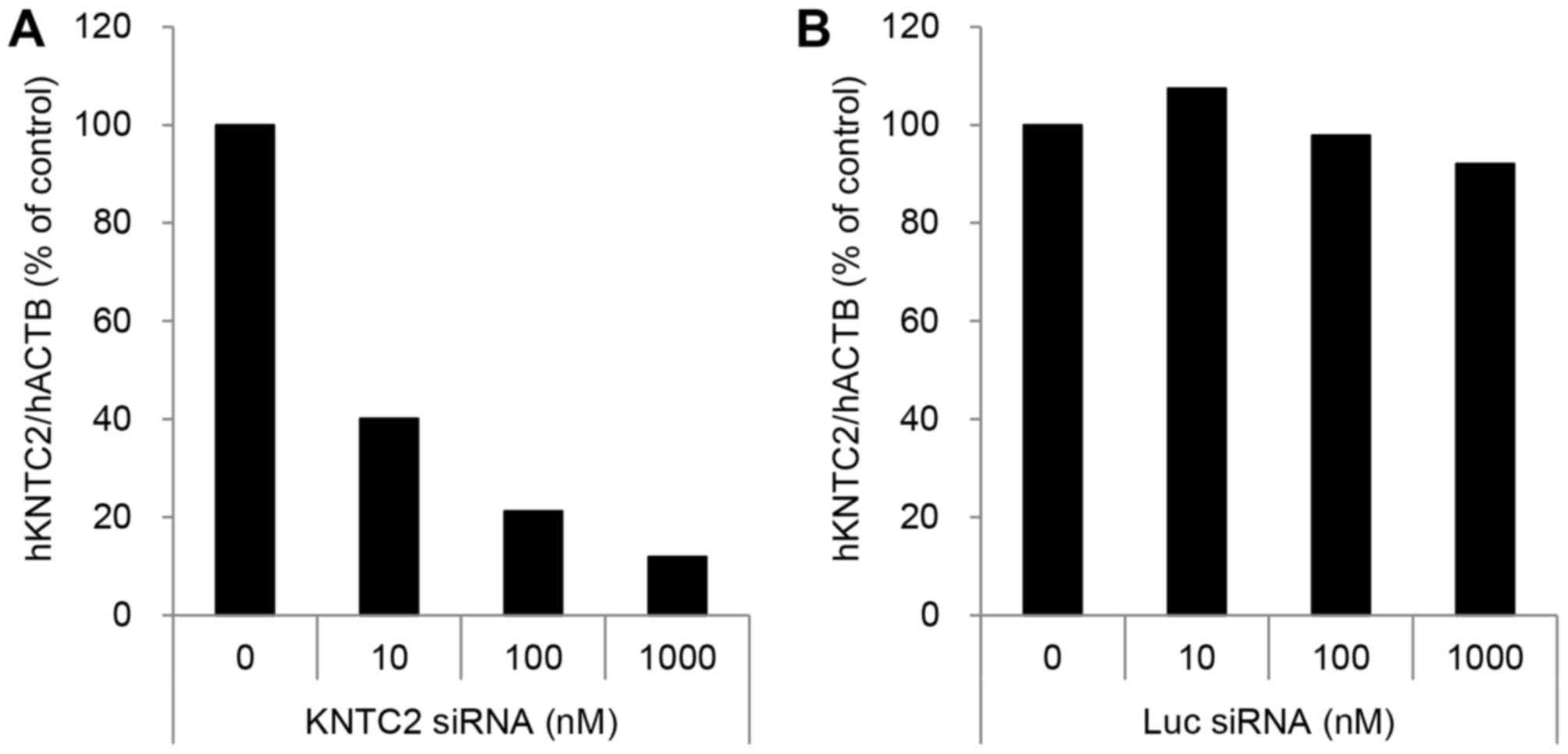
The knockdown activities of KNTC2-LNP were 60% at 10
nM, 79% at 100 nM and 88% at 1 µM in 3D-culture systems of LC-60
(Fig. 3A). The knockdown activities
of Luc siRNA-LNP (negative control) were markedly decreased
compared with KNTC2-LNP, indicating the specificity of KNTC2 siRNA
(Fig. 3B).
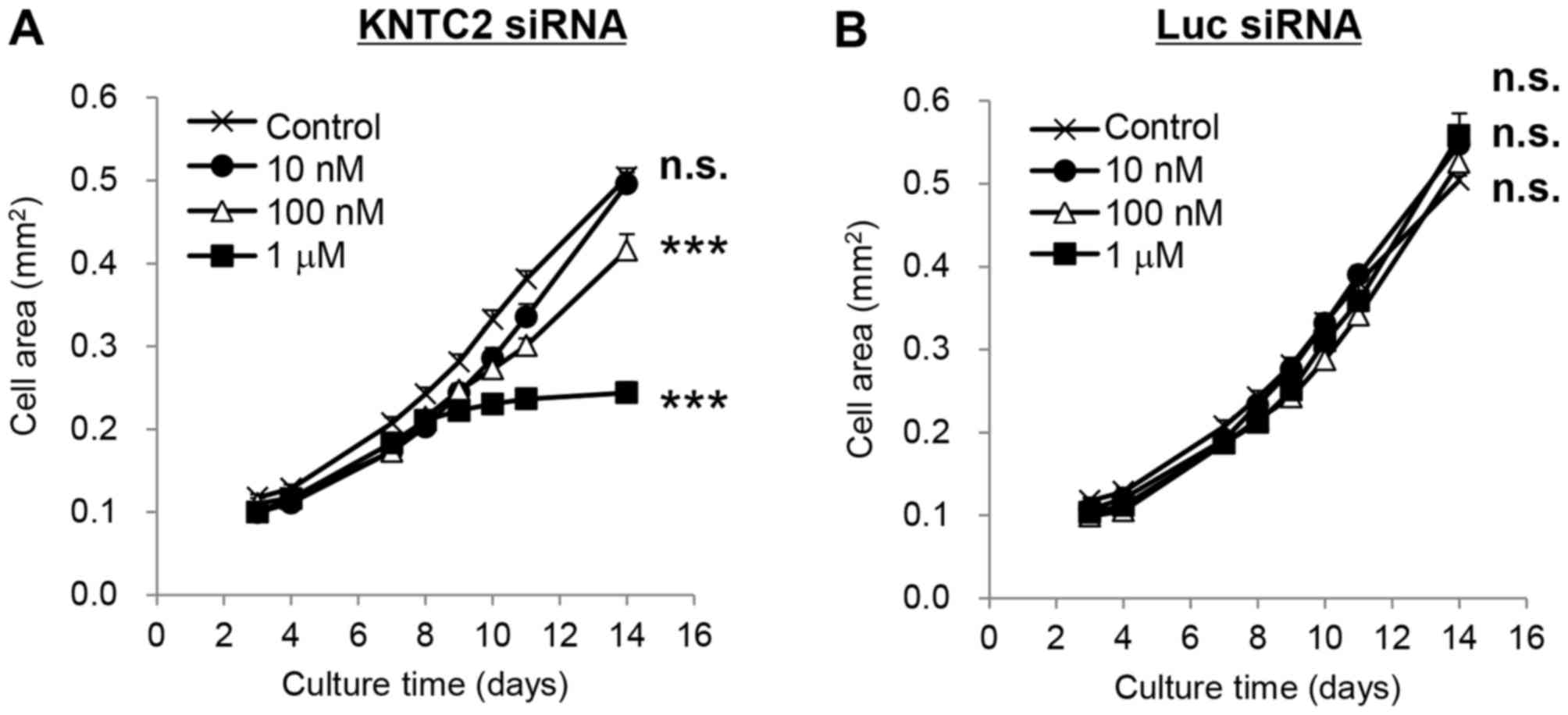
Tumor growth of LC-60 was significantly inhibited by
KNTC2-LNP at 100 nM and 1 µM (P<0.001; Fig. 4A). The growth inhibition rates were 21
and 63%, respectively. Luc siRNA-LNP (negative control) did not
significantly inhibit the growth of LC-60 at the same
concentrations, indicating that growth inhibition was specifically
caused by the suppression of human KNTC2 mRNA expression
(Fig. 4B).
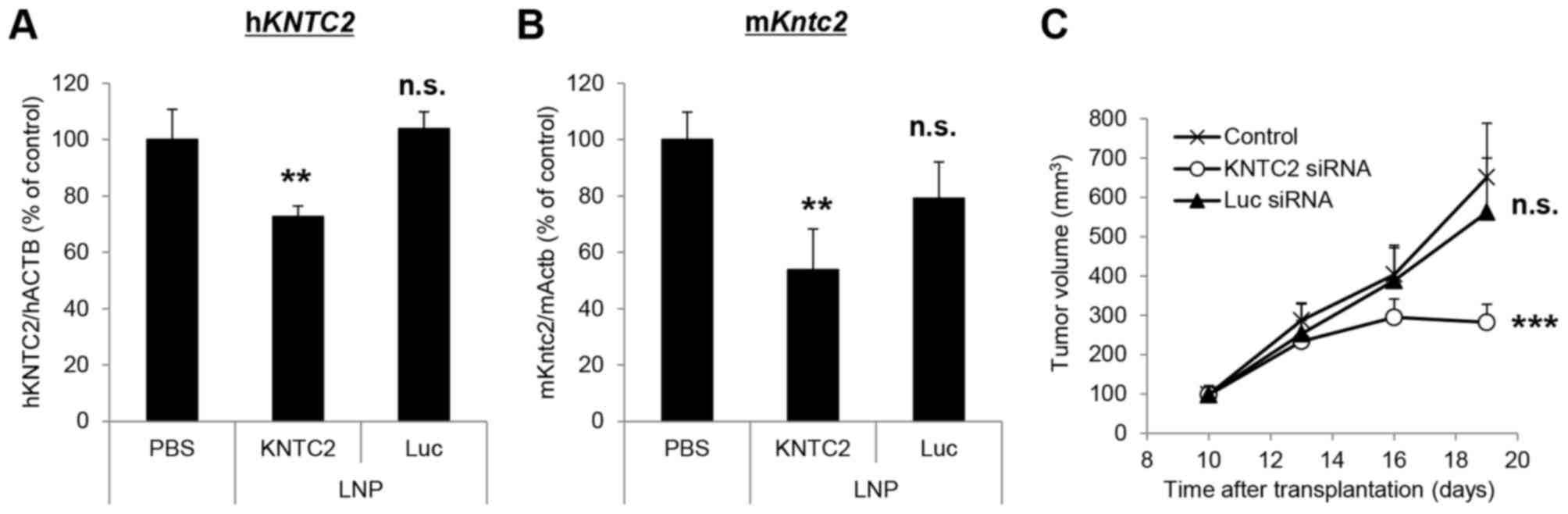
Knockdown and growth inhibitory
activities of KNTC2-LNP in subcutaneous tumor models of lung cancer
PDXs
Knockdown and growth inhibitory activities of
KNTC2-LNP were further investigated in the subcutaneous tumor model
of LC-60. Single intravenous administration of KNTC2-LNP (5 mg/kg)
significantly suppressed the expression levels of human
KNTC2 and mouse Kntc2 mRNA in LC-60 by 27 and 46%,
respectively (P<0.01; Fig. 5A and
B). Luc siRNA-LNP (negative control) did not exhibit knockdown
activities at the same dosage. Repeated intravenous administration
of KNTC2-LNP (5 mg/kg, twice a week) significantly inhibited the
growth of LC-60 by 67% (P<0.001; Fig.
5C). Luc siRNA-LNP did not inhibit the growth of LC-60
indicating that the growth inhibition was specifically caused by
the suppression of human KNTC2 and mouse Kntc2 mRNA
expression levels.
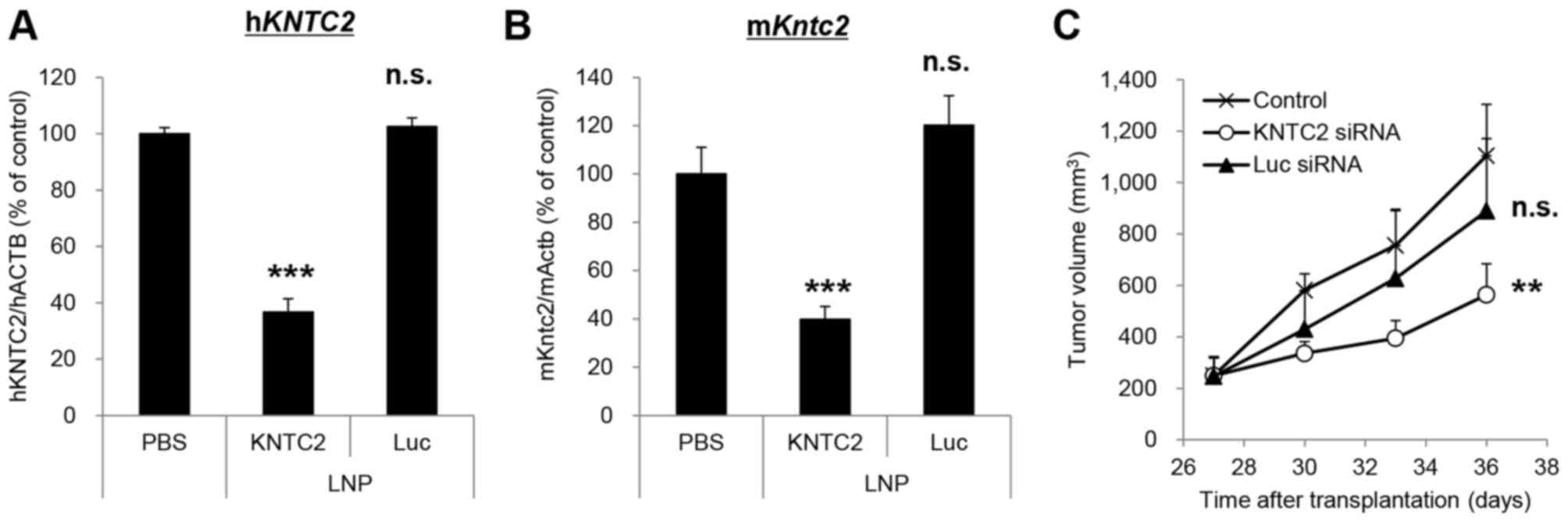
Knockdown and growth inhibitory activities of
KNTC2-LNP were also investigated using another lung cancer PDX,
LC-45. Single intravenous administration of KNTC2-LNP (5 mg/kg)
significantly suppressed the expression levels of human
KNTC2 and mouse Kntc2 mRNA in LC-45 by 63 and 60%,
respectively (P<0.001; Fig. 6A and
B). Repeated intravenous administration of KNTC2-LNP (5 mg/kg,
twice a week) significantly inhibited the growth of LC-45 by 63%
(P<0.01; Fig. 6C) suggesting that
KNTC2-LNP exhibits antitumor activity against various types of lung
cancer PDXs.
Discussion
To clarify the involvement of KNTC2 in the growth of
lung cancer PDXs, the growth inhibitory activities of KNTC2-LNP
were evaluated in 3D-culture systems and subcutaneous tumor models
of lung cancer PDXs, LC-45 and LC-60.
Lung cancer PDXs implanted into the flank of
immuno-deficient mice were estimated to be composed of lung cancer
patient-derived human cells and host mouse-derived stromal cells
(31). Stromal cells have been
reported to affect the proliferation of cancer cells in tumor
tissues (32). Therefore, in the
present study, KNTC2 siRNA that suppressed not only human
KNTC2 mRNA but also mouse Kntc2 mRNA was selected in
order to clarify the involvement of KNTC2 in human-derived cancer
cells and mouse-derived stromal cells. The knockdown activities of
KNTC2 siRNA were sufficient to inhibit the in vivo growth of
LC-60 and LC-45. Negative control Luc siRNA did not inhibit the
growth of these PDXs, indicating that their growths were dependent
on KNTC2.
Notably, KNTC2 siRNA inhibited the growth of LC-60
that was resistant to an approved drug, erlotinib. According to
previous studies, KNTC2 siRNA was estimated to impair the
chromosome segregation of LC-60 leading to cell cycle arrest and
apoptosis (33,34). This molecular mechanism is distinct
from that of erlotinib, which inhibits the auto-phosphorylation of
EGFR tyrosine kinase (35).
Inhibitors of EGFR tyrosine kinase were reported to be ineffective
in patients with lung cancer with mutations in KRAS
(36) or EGFR (37). LC-60 exhibited a mutated KRAS
gene (G12V). Therefore, KNTC2 may be a therapeutic target for
patients with lung cancer that is resistant to erlotinib.
Several types of 3D-culture systems have been
developed for PDXs to increase the efficiency of drug screening
(38–40). Efficiency of evaluating KNTC2 siRNA
was increased by establishing 3D-culture systems of LC-45 and
LC-60. Their sensitivities to erlotinib were similar to that of
subcutaneous tumors. These results were consistent with previous
reports demonstrating the similarities of 3D-culture systems to
subcutaneous tumor models using other lung cancer PDXs (41,42).
The maximum knockdown activity of KNTC2-LNP in the
3D-culture system was sufficient to inhibit the growth of LC-60 and
predict the result of in vivo study. In contrast, growth
inhibitory activity of KNTC2-LNP was not detected in the 3D-culture
system of LC-45 (data not shown). The reason for these results was
speculated to be that the maximum knockdown activity of KNTC2-LNP
was insufficient to inhibit the growth of LC-45. Applicability of
the 3D-culture system to the evaluation of KNTC2-LNP was considered
to be different between PDXs.
In conclusion, the involvement of KNTC2 in the
growth of lung cancer PDXs LC-45 (sensitive to erlotinib) and LC-60
(resistant to erlotinib) was clarified. The results markedly
indicated that KNTC2 is a promising target for the treatment of
lung cancer.
Acknowledgements
The authors wish to thank Dr Keiji Yamamoto, Dr
Kazuhiro Ogi, Dr Michiyasu Takeyama, Dr Hirokazu Matsumoto and Dr
Toshiyuki Nomura for encouraging the accomplishment of this work.
The authors also thank Dr Tadahiro Nambu, Mr Yuji Baba, Dr Kazuhide
Nakamura and Dr Toshiya Tamura for the helpful discussion to
establish 3D culture systems of PDXs, Dr Rumiko Ochiai for helpful
discussion on the formulation of lipid nanoparticles and Ms Syu
Morita and Ms Kaori Konno for technical assistance. KNTC2 siRNAs
were designed and selected by Dr Nobuyuki Miyajima, Mr Yoshiki
Katou, Ms Kuniko Kikuchi and Dr Hiroshi Uejima. All of the
aforementioned individuals were affiliated to Takeda Pharmaceutical
Company, Ltd., Kanagawa, Japan at the time when the present study
was undertaken.
References
|
1
|
Tentler JJ, Tan AC, Weekes CD, Jimeno A,
Leong S, Pitts TM, Arcaroli JJ, Messersmith WA and Eckhardt SG:
Patient-derived tumour xenografts as models for oncology drug
development. Nat Rev Clin Oncol. 9:338–350. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pompili L, Porru M, Caruso C, Biroccio A
and Leonetti C: Patient-derived xenografts: A relevant preclinical
model for drug development. J Exp Clin Cancer Res. 35:1892016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fichtner I, Rolff J, Soong R, Hoffmann J,
Hammer S, Sommer A, Becker M and Merk J: Establishment of
patient-derived non-small cell lung cancer xenografts as models for
the identification of predictive biomarkers. Clin Cancer Res.
14:6456–6468. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang D, Pham NA, Tong J, Sakashita S, Allo
G, Kim L, Yanagawa N, Raghavan V, Wei Y, To C, et al: Molecular
heterogeneity of non-small cell lung carcinoma patient-derived
xenografts closely reflect their primary tumors. Int J Cancer.
140:662–673. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Perez-Soler R, Kemp B, Wu QP, Mao L, Gomez
J, Zeleniuch-Jacquotte A, Yee H, Lee JS, Jagirdar J and Ling YH:
Response and determinants of sensitivity to paclitaxel in human
non-small cell lung cancer tumors heterotransplanted in nude mice.
Clin Cancer Res. 6:4932–4938. 2000.PubMed/NCBI
|
|
6
|
Daniel VC, Marchionni L, Hierman JS,
Rhodes JT, Devereux WL, Rudin CM, Yung R, Parmigiani G, Dorsch M,
Peacock CD and Watkins DN: A primary xenograft model of small-cell
lung cancer reveals irreversible changes in gene expression imposed
by culture in vitro. Cancer Res. 69:3364–3373. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang XC, Zhang J, Li M, Huang XS, Yang
XN, Zhong WZ, Xie L, Zhang L, Zhou M, Gavine P, et al:
Establishment of patient-derived non-small cell lung cancer
xenograft models with genetic aberrations within EGFR, KRAS and
FGFR1: Useful tools for preclinical studies of targeted therapies.
J Transl Med. 11:1682013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ilie M, Nunes M, Blot L, Hofman V,
Long-Mira E, Butori C, Selva E, Merino-Trigo A, Vénissac N, Mouroux
J, et al: Setting up a wide panel of patient-derived tumor
xenografts of non-small cell lung cancer by improving the
preanalytical steps. Cancer Med. 4:201–211. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Owonikoko TK, Zhang G, Kim HS, Stinson RM,
Bechara R, Zhang C, Chen Z, Saba NF, Pakkala S, Pillai R, et al:
Patient-derived xenografts faithfully replicated clinical outcome
in a phase II co-clinical trial of arsenic trioxide in relapsed
small cell lung cancer. J Transl Med. 14:1112016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nukatsuka M, Saito H, Nakagawa F,
Tsujimoto H, Sakamoto K, Tsukioka S, Uchida J, Kiniwa M, Kobunai T
and Takechi T: Combination therapy using oral S-1 and targeted
agents against human tumor xenografts in nude mice. Exp Ther Med.
3:755–762. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hu Y, Wang L, Gu J, Qu K and Wang Y:
Identification of microRNA differentially expressed in three
subtypes of non-small cell lung cancer and in silico functional
analysis. Oncotarget. 8:74554–74566. 2017.PubMed/NCBI
|
|
12
|
Nicholson AG, Chansky K, Crowley J,
Beyruti R, Kubota K, Turrisi A, Eberhardt WE, van Meerbeeck J and
Rami-Porta R; Staging and Prognostic Factors Committee, ; Advisory
Boards, and Participating Institutions, ; Staging and Prognostic
Factors Committee Advisory Boards and Participating Institutions, :
The international association for the study of lung cancer lung
cancer staging project: Proposals for the revision of the clinical
and pathologic staging of small cell lung cancer in the forthcoming
Eighth edition of the TNM classification for lung cancer. J Thorac
Oncol. 11:300–311. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Johnson JR, Cohen M, Sridhara R, Chen YF,
Williams GM, Duan J, Gobburu J, Booth B, Benson K, Leighton J, et
al: Approval summary for erlotinib for treatment of patients with
locally advanced or metastatic non-small cell lung cancer after
failure of at least one prior chemotherapy regimen. Clin Cancer
Res. 11:6414–6421. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sequist LV, Martins RG, Spigel D, Grunberg
SM, Spira A, Jänne PA, Joshi VA, McCollum D, Evans TL, Muzikansky
A, et al: First-line gefitinib in patients with advanced
non-small-cell lung cancer harboring somatic EGFR mutations. J Clin
Oncol. 26:2442–2449. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dungo RT and Keating GM: Afatinib: First
global approval. Drugs. 73:1503–1515. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Malik SM, Maher VE, Bijwaard KE, Becker
RL, Zhang L, Tang SW, Song P, Liu Q, Marathe A, Gehrke B, et al:
U.S. food and drug administration approval: Crizotinib for
treatment of advanced or metastatic non-small cell lung cancer that
is anaplastic lymphoma kinase positive. Clin Cancer Res.
20:2029–2034. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kim DW, Mehra R, Tan DS, Felip E, Chow LQ,
Camidge DR, Vansteenkiste J, Sharma S, De Pas T, Riely GJ, et al:
Activity and safety of ceritinib in patients with ALK-rearranged
non-small-cell lung cancer (ASCEND-1): Updated results from the
multicentre, open-label, phase 1 trial. Lancet Oncol. 17:452–463.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Song Z, Wang M and Zhang A: Alectinib: A
novel second generation anaplastic lymphoma kinase (ALK) inhibitor
for overcoming clinically-acquired resistance. Acta Pharm Sin B.
5:34–37. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hazarika M, White RM, Johnson JR and
Pazdur R: FDA drug approval summaries: Pemetrexed (Alimta).
Oncologist. 9:482–488. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cohen MH, Gootenberg J, Keegan P and
Pazdur R: FDA drug approval summary: Bevacizumab (Avastin) plus
Carboplatin and Paclitaxel as first-line treatment of
advanced/metastatic recurrent nonsquamous non-small cell lung
cancer. Oncologist. 12:713–718. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Salgia R, Stille JR, Weaver RW, McCleod M,
Hamid O, Polzer J, Roberson S, Flynt A and Spigel DR: A randomized
phase II study of LY2510924 and carboplatin/etoposide versus
carboplatin/etoposide in extensive-disease small cell lung cancer.
Lung Cancer. 105:7–13. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Brahmer J, Reckamp KL, Baas P, Crinò L,
Eberhardt WE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE,
Holgado E, et al: Nivolumab versus docetaxel in advanced
Squamous-cell Non-small-cell lung cancer. N Engl J Med.
373:123–135. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Y, Riley DJ, Chen PL and Lee WH: HEC,
a novel nuclear protein rich in leucine heptad repeats specifically
involved in mitosis. Mol Cell Biol. 17:6049–6056. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ferretti C, Totta P, Fiore M, Mattiuzzo M,
Schillaci T, Ricordy R, Di Leonardo A and Degrassi F: Expression of
the kinetochore protein Hec1 during the cell cycle in normal and
cancer cells and its regulation by the pRb pathway. Cell Cycle.
9:4174–4182. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Makita Y, Murata S, Katou Y, Kikuchi K,
Uejima H, Teratani M, Hoashi Y, Kenjo E, Matsumoto S, Nogami M, et
al: Anti-tumor activity of KNTC2 siRNA in orthotopic tumor model
mice of hepatocellular carcinoma. Biochem Biophys Res Commun.
493:800–806. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Elbashir SM, Harborth J, Lendeckel W,
Yalcin A, Weber K and Tuschl T: Duplexes of 21-nucleotide RNAs
mediate RNA interference in cultured mammalian cells. Nature.
411:494–498. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Frank-Kamenetsky M, Grefhorst A, Anderson
NN, Racie TS, Bramlage B, Akinc A, Butler D, Charisse K, Dorkin R,
Fan Y, et al: Therapeutic RNAi targeting PCSK9 acutely lowers
plasma cholesterol in rodents and LDL cholesterol in nonhuman
primates. Proc Natl Acad Sci USA. 105:pp. 11915–11920. 2008;
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hoashi Y: Cationic Lipid Patent
WO2016021683 A1. August 6–2015, issued February 11, 2016.
|
|
29
|
Heyes J, Palmer L, Bremner K and
MacLachlan I: Cationic lipid saturation influences intracellular
delivery of encapsulated nucleic acids. J Control Release.
107:276–287. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sato T, Stange DE, Ferrante M, Vries RG,
Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J,
Siersema PD and Clevers H: Long-term expansion of epithelial
organoids from human colon, adenoma, adenocarcinoma, and Barrett's
epithelium. Gastroenterology. 141:1762–1772. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schneeberger VE, Allaj V, Gardner EE,
Poirier JT and Rudin CM: Quantitation of murine stroma and
selective purification of the human tumor component of
patient-derived xenografts for genomic analysis. PLoS One.
11:e01605872016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Taromi S, Kayser G, Catusse J, von
Elverfeldt D, Reichardt W, Braun F, Weber WA, Zeiser R and Burger
M: CXCR4 antagonists suppress small cell lung cancer progression.
Oncotarget. 7:85185–85195. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu G, Qiu XL, Zhou L, Zhu J, Chamberlin R,
Lau J, Chen PL and Lee WH: Small molecule targeting the Hec1/Nek2
mitotic pathway suppresses tumor cell growth in culture and in
animal. Cancer Res. 68:8393–8399. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hu CM, Zhu J, Guo XE, Chen W, Qiu XL, Ngo
B, Chien R, Wang YV, Tsai CY, Wu G, et al: Novel small molecules
disrupting Hec1/Nek2 interaction ablate tumor progression by
triggering Nek2 degradation through a death-trap mechanism.
Oncogene. 34:1220–1230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pollack VA, Savage DM, Baker DA,
Tsaparikos KE, Sloan DE, Moyer JD, Barbacci EG, Pustilnik LR,
Smolarek TA, Davis JA, et al: Inhibition of epidermal growth factor
receptor-associated tyrosine phosphorylation in human carcinomas
with CP-358,774: Dynamics of receptor inhibition in situ and
antitumor effects in athymic mice. J Pharmacol Exp Ther.
291:739–748. 1999.PubMed/NCBI
|
|
36
|
Pao W, Wang TY, Riely GJ, Miller VA, Pan
Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG and Varmus HE: KRAS
mutations and primary resistance of lung adenocarcinomas to
gefitinib or erlotinib. PLoS Med. 2:e172005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pao W, Miller VA, Politi KA, Riely GJ,
Somwar R, Zakowski MF, Kris MG and Varmus H: Acquired resistance of
lung adenocarcinomas to gefitinib or erlotinib is associated with a
second mutation in the EGFR kinase domain. PLoS Med. 2:e732005.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ekert JE, Johnson K, Strake B, Pardinas J,
Jarantow S, Perkinson R and Colter DC: Three-dimensional lung tumor
microenvironment modulates therapeutic compound responsiveness in
vitro-implication for drug development. PLoS One. 9:e922482014.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Friedrich J, Seidel C, Ebner R and
Kunz-Schughart LA: Spheroid-based drug screen: Considerations and
practical approach. Nat Protoc. 4:309–324. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Perche F and Torchilin VP: Cancer cell
spheroids as a model to evaluate chemotherapy protocols. Cancer
Biol Ther. 13:1205–1213. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Endo H, Okami J, Okuyama H, Kumagai T,
Uchida J, Kondo J, Takehara T, Nishizawa Y, Imamura F, Higashiyama
M and Inoue M: Spheroid culture of primary lung cancer cells with
neuregulin 1/HER3 pathway activation. J Thorac Oncol. 8:131–139.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Onion D, Argent RH, Reece-Smith AM, Craze
ML, Pineda RG, Clarke PA, Ratan HL, Parsons SL, Lobo DN, Duffy JP,
et al: 3-Dimensional patient-derived lung cancer assays reveal
resistance to Standards-of-Care promoted by stromal cells but
sensitivity to histone deacetylase inhibitors. Mol Cancer Ther.
15:753–763. 2016. View Article : Google Scholar : PubMed/NCBI
|