Introduction
As a cerebrovascular disorder, intracranial aneurysm
(IA; also designated brain or cerebral aneurysm) is a ballooning or
localized dilation of the blood vessel induced by weakness of the
artery wall (1). IA cases are divided
according to size into small (diameter, <15 mm) and large
aneurysms [including large (15–25 mm), giant (25–50 mm) and
super-giant (>50 mm) aneurysms] (2). Based on the shape, IA may be classified
into saccular aneurysms, fusiform aneurysms and microaneurysms
(2). IA may not only be a result of
genetic conditions, but also lifestyle factors, including smoking,
hypertension, obesity and excess alcohol consumption (3,4).
Individuals who are between 30 and 60 years old experience the
highest incidence of IA, and women experience an increased
incidence compared with men with a ratio of 3:2 (1,5).
The genes implicated in the pathogenesis of the
rupture of IA have been evaluated previously. Through reverse
transcription-quantitative polymerase chain reaction, Guo et
al (6) identified that the mRNA
levels of caspase-3 in IA and abdominal aortic aneurysm are
8.94-fold and 6.73-fold compared with that of normal vessels, which
improves the understanding of apoptosis in ruptured intracranial
aneurysm. Nuclear factor-κB (NF-κB) functions as an
essential regulator during the initiation of IA development via
mediation of several inflammatory genes associated with macrophage
activation and recruitment, thus, NF-κB may function
as a therapeutic target for IA (7).
Increased tumor necrosis factor α and Fas-associated death domain
protein may have deleterious effects on cerebral arteries through
facilitation of inflammation and apoptosis in immune and vascular
cells, consequently weakening vessel walls (8,9). Decreased
tissue inhibitor of matrix metalloproteinases and increased matrix
metalloproteinases in the late stage of IA formation may be the
reason for extracellular matrix degradation resulting in the
progression, and rupture of IA (10).
As a main chemoattractant for monocytes and macrophages, NF-κB
activation-induced monocyte chemoattractant protein-1
(MCP-1) acts in IA formation and may serve as a potential
target for therapy inhibiting IA progression (11). Nevertheless, the molecular mechanisms
of rupture of IA have not been fully revealed.
In 2011, Kurki et al (12) explored the gene expression differences
between ruptured and unruptured saccular intracranial aneurysm
(sIA) wall samples, screening 686 upregulated and 740 downregulated
genes in the ruptured samples, and finding that hypoxia-inducible
factor-1A, ETS transcription factors, toll-like receptor signaling
and NF-κB are associated with the rupture of sIA
walls in humans. In 2010, Pera et al (13) investigated the gene expression
profiles of ruptured and unruptured IA, as well as control
intracranial arteries, and identified a total of 159 differentially
expressed genes (DEGs) and several critical biological processes,
including cell adhesion, muscle system, and the immune system, and
inflammatory response. However, comprehensive bioinformatic
analyses have not been performed to further screen the critical
genes associated with the rupture of IA. Using the data gathered by
Kurki et al (12) and Pera
et al (13), the DEGs between
ruptured, and unruptured IA samples were fully screened. Utilizing
protein-protein interaction (PPI) network construction, pathway
enrichment analysis, construction and efficiency evaluation of
support vector machine (SVM) classifier, and principal component
analysis (PCA), the key genes implicated in the rupture of IA were
further identified.
Materials and methods
Microarray data
Microarray data of GSE13353 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13353)
and GSE15629 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15629)
were downloaded from the Gene Expression Omnibus database.
GSE13353, which was deposited by Kurki et al (12) and sequenced on the platform of GPL570
(HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array,
included 11 ruptured IA wall samples and 8 unruptured IA wall
samples. Kurki et al (12)
isolated ruptured and unruptured IA wall samples from the necks of
Finnish individuals undergoing microsurgical clipping as previously
described (14–17). The IA wall samples were frozen in
liquid nitrogen and then kept in the Helsinki Neurosurgery sIA
Tissue Bank (12). GSE15629 was
deposited by Pera et al (13)
and sequenced on the platform of GPL6244 (HuGene-1_0-st) Affymetrix
Human Gene 1.0 ST Array [transcript (gene) version], from which 8
ruptured IA wall samples and 6 unruptured IA wall samples were
selected for this study. Pera et al (13) also resected full-thickness vessel wall
samples from patients who underwent microsurgical clipping. Then,
the samples were stored in RNAlater (Qiagen China Co., Ltd.,
Shanghai, China) at 80°C.
Data preprocessing and DEGs
screening
Using the oligo package (18) in R, background correction and
normalization were performed on the raw data. Using the limma
package (http://www.R-project.org) (19) in R, the genes differentially expressed
between ruptured and unruptured IA samples were selected, with
P<0.05 and |fold change | >1.5 as the thresholds. Conversely,
based on the MetaDE package (https://cran.r-project.org/web/packages/MetaDE/index.html)
(20) in R, heterogeneity tests and
differential expression analysis for each gene were conducted
successively. The genes with τ2=0 and Qpval >0.05
were homogeneous and unbiased, from which the genes with P<0.05
were further selected as DEGs.
Construction of PPI network
The interaction information of human proteins was
downloaded from the Biological General Repository for Interaction
Datasets (http://thebiogrid.org/) (21), Human Protein Reference Database
(http://www.hprd.org/) (22) and Database of Interacting Proteins
(http://dip.doe-mbi.ucla.edu/) (23) databases. Subsequently, the identified
DEGs were mapped to the interaction network of human proteins and
the PPI network for the DEGs was visualized using Cytoscape
software (version 3.1.0, http://www.cytoscape.org) (24). In the PPI network, nodes and edges
separately stand for proteins, and interactions of proteins.
Additionally, the number of edges involving one node was the
connectivity degree of the node. Furthermore, the candidate genes
in the PPI network were further screened using the betweenness
centrality (BC) method based on the following formula:
CB(v)=∑t≠v≠u∈Vσst(v)σst
σst represents the number of the shortest path
between s and t. σst(ν) stands for the number of paths past node v
among the shortest paths between s and t. The value of
CB (v) ranges from 0 to 1, a larger value
indicates an increased importance.
Additionally, hierarchical clustering analysis
(25) was performed for the candidate
genes and clustering results were visualized using Heatmap software
(version 1.0, http://www.hiv.lanl.gov/content/sequence/HEATMAP/heatmap.html)
(26).
Pathway enrichment analysis
Pathway enrichment analysis was performed for the
DEGs involved in the PPI network using the fisher algorithm
(27) based on the following
formula:
p=1–∑i=0x–1(Mi)(N–MK–i)(NK)
Among the formula, M, N and K stand for the number
of genes enriched in pathways, the total number of genes in whole
genome and the number of DEGs, respectively. p represents
the probability that no less than × DEGs are enriched in
pathways.
Construction and efficiency evaluation
of SVM classifier
As effective classifiers, SVMs may be utilized for
two-class classification of microarray data and acquire high
classification accuracy (28). The
DEGs involved in the PPI network were sorted in descending order
based on their BC values. Starting from the last 10 genes and
taking 5 genes as an interval, the DEGs were selected as
characterization factors (genes used for constructing SVM
classifier) in descending order based on BC values. With the
dataset of GSE13353 as the training dataset, the tune.svm function
of the e1071 package (http://cran.r-project.org/web/packages/e1071/)
(29) in R was used to construct an
optimal SVM classifier. Then, the dataset of GSE15629 was used as
the validation dataset for detecting the SVM classifier.
Furthermore, the Candidate Cancer Gene Database (http://ccgd-starrlab.oit.umn.edu/) (30) was used to search for the
characterization of gene-associated cancer.
PCA of characterization factors
As a multivariate technique, PCA is able to
reassemble associated-indexes into a new set of comprehensive
indexes that exhibit no correlations with each other (31). To further optimize the identified
characterization genes, the genes involved in the SVM classifier
were performed using PCA.
Results
DEGs analysis
In the ruptured samples, a total of 636 DEGs
(including 279 upregulated genes and 357 downregulated genes) in
GSE13353 and 656 DEGs (including 130 upregulated genes and 526
downregulated genes) in GSE15629 were identified using the limma
package. Using the MetaDE package, a total of 1,029 DEGs (including
527 upregulated genes and 502 downregulated genes) were screened.
To include the associated genes into our research, the DEGs
identified by limma and MetaDE packages were merged and used for
the following analyses.
PPI network analysis and pathway
enrichment analysis
The PPI network constructed for the merged DEGs
identified 510 nodes (including 290 upregulated genes and 220
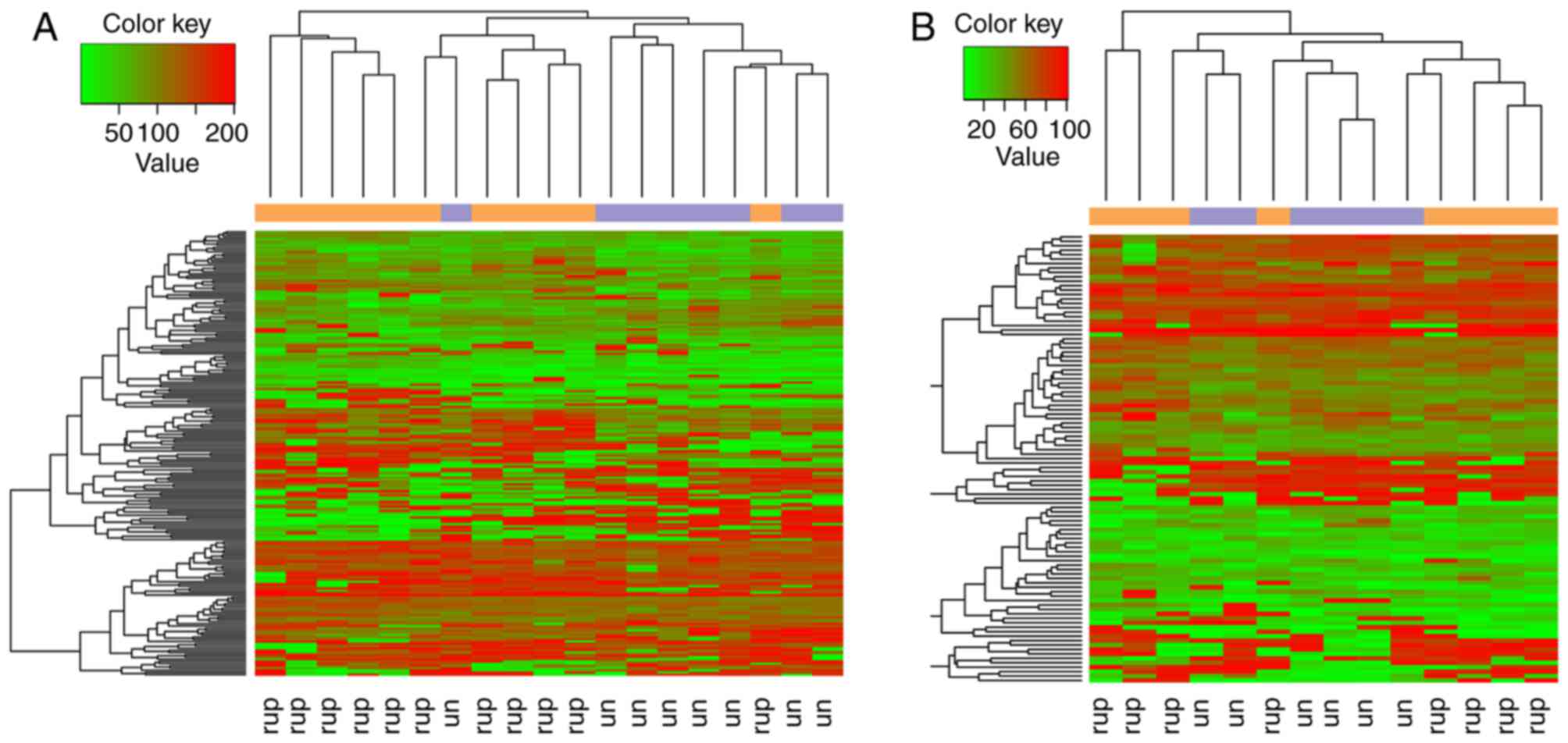
downregulated genes) and 907 interactions (Fig. 1). Combined with connectivity degrees
and BC scores, the top 100 genes [including fibronectin 1 (FN1),
amyloid β (A4) precursor protein (APP), nuclear RNA export factor 1
(NXF1) and signal transducer and activator of transcription 3
(STAT3)] in the PPI network were selected as candidate genes.
Hierarchical clustering analysis revealed that the candidate genes
were able to separate the ruptured samples from the unruptured
samples (Fig. 2).
To further understand the biological pathways
involved the genes in PPI network, pathway enrichment analysis was
conducted. A total of 7 pathways were enriched, including the
mitogen-activated protein kinase signaling pathway (P=0.004304),
pathways in cancer (P=0.007181) and Toll-like receptor signaling
pathway (P=0.040118), which involved toll-like receptor 3
(TLR3) (Table I).
 | Table I.Pathways enriched for the genes
involved in the protein-protein interaction network. |
Table I.
Pathways enriched for the genes
involved in the protein-protein interaction network.
| Description | Gene number | P-value | Gene symbol |
|---|
| hsa04010: MAPK
signaling pathway | 18 | 0.004304 | FGFR2, ZAK,
BRAF, MAP2K3, RELA, MKNK2, MAPK10, STK4, MAX, MAPK1, MAPK12, DUSP1,
MAP3K8, SOS2, MAPK8IP2, MAPK8, PAK1, STMN1 |
| hsa05200: Pathways
in cancer | 20 | 0.007181 | FGFR2, E2F3,
RET, BRAF, RELA, STAT5B, ARNT2, EGLN1, MAPK10, STK4, BIRC2, STAT3,
MAPK1, MAX, SOS2, RALB, JAK1, CSF3R, MAPK8, FN1 |
| hsa04621: NOD-like
receptor signaling pathway | 7 | 0.013053 | MAPK1, NOD2,
MAPK12, RELA, MAPK8, MAPK10, BIRC2 |
| hsa04012: ErbB
signaling pathway | 8 | 0.019615 | MAPK1, BRAF,
ERBB3, STAT5B, SOS2, MAPK8, MAPK10, PAK1 |
| hsa00270: Cysteine
and methionine metabolism | 5 | 0.021434 | ADI1, LDHB,
LDHA, SRM, TRDMT1 |
| hsa04620: Toll-like
receptor signaling pathway | 8 | 0.040118 | MAPK1, MAPK12,
RELA, MAP2K3, MAP3K8, TLR3, MAPK8, MAPK10 |
| hsa04722:
Neurotrophin signaling pathway | 9 | 0.042021 | MAPK1, MAPK12,
BRAF, RELA, SOS2, NGFRAP1, MAPK8, MAPK10, ARHGDIA |
Construction and efficiency evaluation
of SVM classifier
With the dataset of GSE13353 as the training
dataset, optimization of the SVM classifier was performed. When the
number of characterization genes reached 15, the SVM classifier
could completely and accurately distinguish all samples (Fig. 3A). Thus, the 15 genes (including FN1)
were considered as the genes required for constructing the SVM
classifier (parameters: γ, 0.5; cost, 4; cross, 10). To confirm
that the SVM classifier had repeatability and portability, the
validation dataset of GSE15629 was used to detect the SVM
classifier. In the dataset of GSE15629, the SVM classifier was able
to fully differentiate the ruptured samples from the unruptured
samples (Fig. 3B). Therefore, the
expression pattern characteristics of the 15 characterization genes
in ruptured and unruptured samples were notable. Furthermore, the
characterization of gene-associated cancer was assessed, revealing
that NXF1 was associated with nervous system cancer (Table II).
 | Table II.The 15 characterization genes
involved in the support vector machine classifier and the types of
cancer associated with them. |
Table II.
The 15 characterization genes
involved in the support vector machine classifier and the types of
cancer associated with them.
| Gene | BC_score | Degree | P-value | logFC | Pubmed ID | Cancer type |
|---|
| APP | 0.9309 | 113 | 0.022006 | −1.40543 | 22057237 | Colorectal
cancer |
| BMI1 | 0.5413 | 19 | 0.000794 | −1.21654 |
|
|
| CCDC8 | 0.5700 | 26 | 0.003424 | −1.04234 |
|
|
| CFL1 | 0.5344 | 14 | 0.049459 | 0.817437 |
|
|
| FBL | 0.5457 | 17 | 0.032831 | −0.77802 |
|
|
| FBXW7 | 0.5380 | 19 | 0.024609 | −1.45185 | 22370638 | Blood cancer |
| FN1 | 0.6292 | 42 | 0.01076 | 1.299781 |
|
|
| HNRNPU | 0.5447 | 24 | 0.049798 | 0.906204 | 27006499 | Gastric cancer |
| NDEL1 | 0.5339 | 10 | 0.036456 | 0.840461 | 24316982 | Liver cancer |
| NXF1 | 0.7250 | 57 | 0.004859 | 1.123627 | 23685747 | Nervous system
cancer |
| RELA | 0.5382 | 18 | 0.037826 | 0.860905 |
|
|
| STAT3 | 0.5487 | 15 | 0.00697 | 1.266099 | 22699621 | Pancreatic
cancer |
| TAGLN2 | 0.5337 | 12 | 0.000581 | 1.101816 |
|
|
| TCF4 | 0.5357 | 12 | 0.045941 | −0.95848 | 23045694 | Nervous system
cancer |
| UBQLN1 | 0.5464 | 16 | 0.016495 | 0.942896 | 24316982 | Liver cancer |
PCA of characterization factors
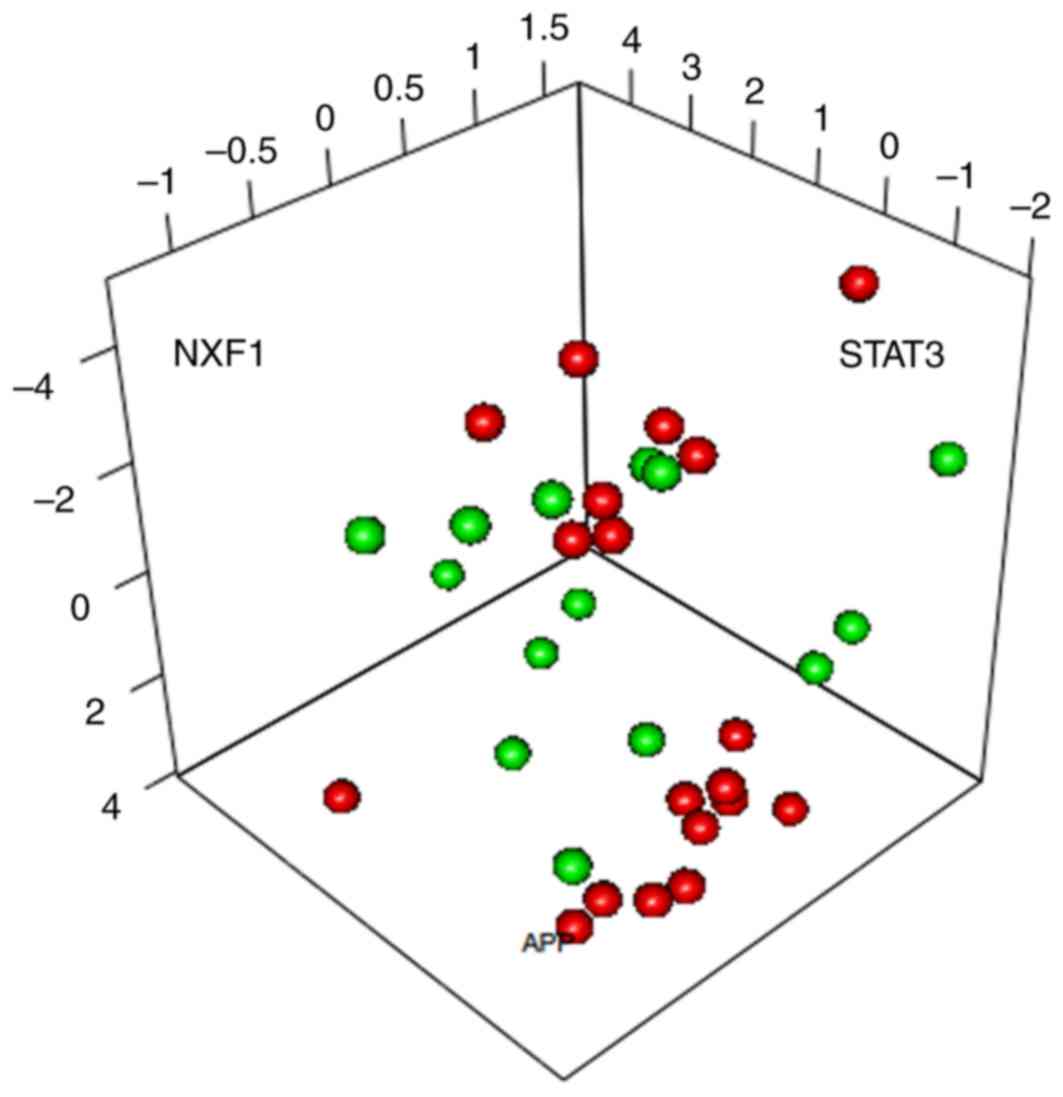
To further optimize the identified characterization
genes, PCA was conducted on the 15 characterization genes involved
in the SVM classifier. APP, NXF1 and STAT3 were the 3 principal
components able to separate the samples (Fig. 4).
Discussion
In the present study, the DEGs between ruptured and
unruptured samples were identified using limma and MetaDE packages.
In the ruptured samples, a total of 636 DEGs in GSE13353 and 656
DEGs in GSE15629 were identified by the limma package. Using the
MetaDE package, a total of 1,029 DEGs were screened. These DEGs
were merged and a PPI network analysis was performed. Combined with
connectivity degrees and BC scores, the top 100 genes (including
FN1, APP, NXF1 and STAT3) in the PPI network were identified as
candidate genes.
Pathway enrichment analysis of the genes in the PPI
network revealed that TLR3 was enriched in the TLR signaling
pathway. TLRs have been reported to function in vascular
inflammatory diseases including aneurysm and atherosclerosis
(32). TLR4, which is
expressed in IA walls of humans and rats, may be implicated in IA
formation via NF-κB activation in endothelial cells (33). FN1 was one of the 15 genes used
for constructing the SVM classifier. Alternatively spliced extra
domain A (EDA) of fibronectin is essential for tissue repair, and
decreased expression of EDA may promote susceptibility to aneurysm
of patients with a bicuspid aortic valve (34). Wang and Astrof (35) demonstrated that the local synthesis of
FN1 serves essential functions in spatial regulation of Notch
signaling and cardiovascular development. As a multi-domain
extracellular matrix glycoprotein, fibronectin functions in blood
vessel morphogenesis during pathological angiogenesis and embryonic
development, and is expressed during pathological angiogenesis in
multiple diseases, including late stage artherosclerosis, lung
cancer and in abnormal ocular conditions (36–39). Thus,
TLR3 and FN1 may serve functions in the rupture of
IA.
PCA identified that APP, NXF1 and
STAT3 were the three principal components that were able to
separate the samples. Duplication of the APP locus, which
leads to the increase of amyloid-β peptides, is able to induce
autosomal dominant early-onset Alzheimer's disease with cerebral
amyloid angiopathy (40). As a result
of assessing the characterization of gene-associated cancer,
NXF1 was identified to be associated with nervous system
cancer. NXF1, which belongs to a family of evolutionarily
conserved proteins, includes an NTF2-like domain, a noncanonical
RNP-type RNA binding domain, a ubiquitin-associated domain and four
leucine-rich repeats (41). In
vivo blockade of STAT3 signaling or inhibition of
interleukin-17A (IL-17A) lead to an apparent increase in
fatal rupture and aneurysm severity in mouse models, and the
prevalence of vascular abnormalities are high in patients with
STAT3 deficiency (42). Romain et
al (43) identified that T
cell-specific STAT3 signaling serves a central function in
promoting vascular aneurysm and IL-17 serves a protective
function in the process. Phosphorylated STAT3 is associated with
TLR4-dependent abdominal aortic aneurysm (AAA) formation, and
STAT3 and/or TLR4 may be utilized for therapy of AAA
(44,45). The level of phosphorylated STAT1
increases during aneurysmal degeneration, and the loss of
STAT1 is associated with aneurysm formation and an increased
rate of aortic rupture in a model of aortic dissection (46). These findings indicate that APP,
NXF1 and STAT3 may be involved in the rupture of IA.
In conclusion, the DEGs between ruptured and
unruptured samples were identified using limma, and MetaDE
packages. Additionally, TLR3, FN1, APP, NXF1 and
STAT3 may function in the rupture of IA. However, a further
validation of the roles of these genes in the rupture of IA is
required.
Acknowledgements
The present study was supported by Shanghai
Municipal Commission of Health and Family Planning (grant no.
201440319). The authors would like to thank to Fenghe (Shanghai)
Information Technology Co., Ltd for their help.
Glossary
Abbreviations
Abbreviations:
|
IA
|
intracranial aneurysm
|
|
DEGs
|
differentially expressed genes
|
|
PPI
|
protein-protein interaction
|
|
SVM
|
support vector machine
|
|
PCA
|
principal component analysis
|
|
NF-κB
|
Nuclear factor-κB
|
|
MCP-1
|
monocyte chemoattractant protein-1
|
|
sIA
|
saccular intracranial aneurysm
|
|
BC
|
betweenness centrality
|
|
TLRs
|
the toll-like receptors
|
References
|
1
|
Brisman JL, Song JK and Newell DW:
Cerebral aneurysms. N Eng J Med. 355:928–939. 2006. View Article : Google Scholar
|
|
2
|
Bhidayasiri R, Waters MF and Giza C:
Neurological differential diagnosis: A prioritized approach. John
Wiley & Sons; pp. 5602005
|
|
3
|
Goljan EF: Rapid Review Pathology. 3rd.
Mosby Inc.; Maryland Heights, MO: 2011, View Article : Google Scholar
|
|
4
|
Flemming KD: Stroke essentials for primary
care: A practical guide. Mayo Clin Proc. 85:pp. e762010; View Article : Google Scholar
|
|
5
|
Haberland C: Clinical Neuropathology. Text
and Color Atlas (1). Demos Medical. 2006.
|
|
6
|
Guo F, Li Z, Song L, Han T, Feng Q, Guo Y,
Xu J, He M and You C: Increased apoptosis and cysteinyl aspartate
specific protease-3 gene expression in human intracranial aneurysm.
J Clin Neurosci. 14:550–555. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Aoki T, Kataoka H, Shimamura M, Nakagami
H, Wakayama K, Moriwaki T, Ishibashi R, Nozaki K, Morishita R and
Hashimoto N: NF-kappaB is a key mediator of cerebral aneurysm
formation. Circulation. 116:2830–2840. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jayaraman T, Berenstein V, Li X, Mayer J,
Silane M, Shin YS, Niimi Y, Kiliç T, Gunel M and Berenstein A:
Tumor necrosis factor α is a key modulator of inflammation in
cerebral aneurysms. Neurosurgery. 57:558–564. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fontanella M, Rainero I, Gallone S, Rubino
E, Fenoglio P, Valfrè W, Garbossa D, Carlino C, Ducati A and
Pinessi L: Tumor necrosis factor-alpha gene and cerebral aneurysms.
Neurosurgery. 60:668–673. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Aoki T, Kataoka H, Moriwaki T, Nozaki K
and Hashimoto N: Role of TIMP-1 and TIMP-2 in the progression of
cerebral aneurysms. Stroke. 38:2337–2345. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Aoki T, Kataoka H, Ishibashi R, Nozaki K,
Egashira K and Hashimoto N: Impact of monocyte chemoattractant
protein-1 deficiency on cerebral aneurysm formation. Stroke.
40:942–951. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kurki MI, Häkkinen SK, Frösen J, Tulamo R,
von und zu Fraunberg M, Wong G, Tromp G, Niemelä M, Hernesniemi J,
Jääskeläinen JE and Ylä-Herttuala S: Upregulated signaling pathways
in ruptured human saccular intracranial aneurysm wall: An emerging
regulative role of toll-like receptor signaling and nuclear
factor-κB, hypoxia-inducible factor-1A, and ETS transcription
factors. Neurosurgery. 68:1667–1676. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pera J, Korostynski M, Krzyszkowski T,
Czopek J, Slowik A, Dziedzic T, Piechota M, Stachura K, Moskala M,
Przewlocki R and Szczudlik A: Gene expression profiles in human
ruptured and unruptured intracranial aneurysms: What is the role of
inflammation? Stroke. 41:224–231. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Frösen J, Piippo A, Paetau A, Kangasniemi
M, Niemelä M, Hernesniemi J and Jääskeläinen J: Remodeling of
saccular cerebral artery aneurysm wall is associated with rupture:
Histological analysis of 24 unruptured and 42 ruptured cases.
Stroke. 35:2287–2293. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Frösen J, Piippo A, Paetau A, Kangasniemi
M, Niemelä M, Hernesniemi J and Jääskeläinen J: Growth factor
receptor expression and remodeling of saccular cerebral artery
aneurysm walls: Implications for biological therapy preventing
rupture. Neurosurgery. 58:534–541. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tulamo R, Frösen J, Junnikkala S, Paetau
A, Pitkäniemi J, Kangasniemi M, Niemelä M, Jääskeläinen J, Jokitalo
E, Karatas A, et al: Complement activation associates with saccular
cerebral artery aneurysm wall degeneration and rupture.
Neurosurgery. 59:1069–1077. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Laaksamo E, Tulamo R, Baumann M, Dashti R,
Hernesniemi J, Juvela S, Niemelä M and Laakso A: Involvement of
mitogen-activated protein kinase signaling in growth and rupture of
human intracranial aneurysms. Stroke. 39:886–892. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rao Y, Lee Y, Jarjoura D, Ruppert AS, Liu
CG, Hsu JC and Hagan JP: A comparison of normalization techniques
for microRNA microarray data. Stat Appl Genet Mol Biol. 7:Article
222008. View Article : Google Scholar
|
|
19
|
Smyth GK: Limma: Linear models for
microarray data = Bioinformatics and computational biology
solutions using R and Bioconductor. Springer; New York, NY: pp.
397–420. 2005
|
|
20
|
Chang LC, Lin HM, Sibille E and Tseng GC:
Meta-analysis methods for combining multiple expression profiles:
Comparisons, statistical characterization and an application
guideline. BMC Bioinformatics. 14:3682013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Stark C, Breitkreutz BJ, Reguly T, Boucher
L, Breitkreutz A and Tyers M: BioGRID: A general repository for
interaction datasets. Nucleic Acids Res. 34(Database Issue):
D535–D539. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Keshava Prasad TS, Goel R, Kandasamy K,
Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R,
Shafreen B, Venugopal A, et al: Human protein reference
database-2009 update. Nucleic Acids Res. 37(Database Issue):
D767–D772. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xenarios I, Rice DW, Salwinski L, Baron
MK, Marcotte EM and Eisenberg D: DIP: The database of interacting
proteins. Nucleic Acids Res. 28:289–291. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Köhn HF and Hubert LJ: Hierarchical
cluster analysis. Wiley StatsRef: Statistics Reference Online;
2015
|
|
26
|
Wilkinson L and Friendly M: The history of
the cluster heat map. Am Stat. 63:179–184. 2012. View Article : Google Scholar
|
|
27
|
Damar İH, Altunkaş F, Çelik A, Koç F,
Karayakalı M, Karaman K, Arısoy A and Ceyhan K: Fragmented QRS
frequency in patients with cardiac syndrome X. Anatol J Cardiol.
16:616–620. 2016.PubMed/NCBI
|
|
28
|
Ma S, Lv M, Deng F, Zhang X, Zhai H and Lv
W: Predicting the ecotoxicity of ionic liquids towards Vibrio
fischeri using genetic function approximation and least squares
support vector machine. J Hazard Mater. 283:591–598. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dimitriadou E, Hornik K, Leisch F, Meyer D
and Weingessel A: Misc functions of the Department of Statistics
(e1071), TU Wien. R package. 1:5–24. 2008.
|
|
30
|
Abbott KL, Nyre ET, Abrahante J, Ho YY,
Vogel RI and Starr TK: The candidate cancer gene database: A
database of cancer driver genes from forward genetic screens in
mice. Nucleic Acids Res. 43(Database Issue): D844–D848. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Abdi H and Williams LJ: Principal
component analysis. Wiley Interdisciplinary Reviews: Comput Stat.
2:433–459. 2010. View Article : Google Scholar
|
|
32
|
Huggins C, Pearce S, Peri F, Neumann F,
Cockerill G and Pirianov G: A novel small molecule TLR4 antagonist
(IAXO-102) negatively regulates non-hematopoietic toll like
receptor 4 signalling and inhibits aortic aneurysms development.
Atherosclerosis. 242:563–570. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Aoki T, Nishimura M, Ishibashi R, Kataoka
H, Takagi Y and Hashimoto N: Toll-like receptor 4 expression during
cerebral aneurysm formation. Laboratory investigation. J Neurosurg.
113:851–858. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Paloschi V, Kurtovic S, Folkersen L, Gomez
D, Wågsäter D, Roy J, Petrini J, Eriksson MJ, Caidahl K, Hamsten A,
et al: Impaired splicing of fibronectin is associated with thoracic
aortic aneurysm formation in patients with bicuspid aortic valve.
Arterioscler Thromb Vasc Biol. 31:691–697. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang X and Astrof S: Neural crest
cell-autonomous roles of fibronectin in cardiovascular development.
Development. 143:88–100. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Astrof S and Hynes RO: Fibronectins in
vascular morphogenesis. Angiogenesis. 12:165–175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Neri D and Bicknell R: Tumour vascular
targeting. Nat Rev Cancer. 5:436–446. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Pedretti M, Rancic ZA, Herzog BA,
Soltermann A, Herzog BA, Schliemann C, Lachat M, Neri D and
Kaufmann PA: Comparative immunohistochemical staining of
atherosclerotic plaques using F16, F8 and L19: Three clinical-grade
fully human antibodies. Atherosclerosis. 208:382–389. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pedretti M, Soltermann A, Arni S, Weder W,
Neri D and Hillinger S: Comparative immunohistochemistry of L19 and
F16 in non-small cell lung cancer and mesothelioma: Two human
antibodies investigated in clinical trials in patients with cancer.
Lung Cancer. 64:28–33. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rovelet-Lecrux A, Hannequin D, Raux G, Le
Meur N, Laquerrière A, Vital A, Dumanchin C, Feuillette S, Brice A,
Vercelletto M, et al: APP locus duplication causes autosomal
dominant early-onset Alzheimer disease with cerebral amyloid
angiopathy. Nat Genet. 38:24–26. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
41
|
Herold A, Suyama M, Rodrigues JP, Braun
IC, Kutay U, Carmo-Fonseca M, Bork P and Izaurralde E: TAP (NXF1)
belongs to a multigene family of putative RNA export factors with a
conserved modular architecture. Mol Cell Biol. 20:8996–9008. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chandesris MO, Azarine A, Ong KT, Taleb S,
Boutouyrie P, Mousseaux E, Romain M, Bozec E, Laurent S, Boddaert
N, et al: Frequent and widespread vascular abnormalities in human
signal transducer and activator of transcription 3 deficiency. Circ
Cardiovasc Genet. 5:25–34. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Romain M, Taleb S, Dalloz M, Ponnuswamy P,
Esposito B, Pérez N, Wang Y, Yoshimura A, Tedgui A and Mallat Z:
Overexpression of SOCS3 in T lymphocytes leads to impaired
interleukin-17 production and severe aortic aneurysm formation in
mice-brief report. Arterioscler Thromb Vasc Biol. 33:581–584. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liao M, Xu J, Clair AJ, Ehrman B, Graham
LM and Eagleton MJ: Local and systemic alterations in signal
transducers and activators of transcription (STAT) associated with
human abdominal aortic aneurysms. J Surg Res. 176:321–328. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Qin Z, Bagley J, Sukhova G, Baur WE, Park
HJ, Beasley D, Libby P, Zhang Y and Galper JB: Angiotensin
II-induced TLR4 mediated abdominal aortic aneurysm in
apolipoprotein E knockout mice is dependent on STAT3. J Mol Cell
Cardiol. 87:160–170. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Eagleton MJ, Xu J, Liao M, Parine B,
Chisolm GM and Graham LM: Loss of STAT1 is associated with
increased aortic rupture in an experimental model of aortic
dissection and aneurysm formation. J Vasc Surg. 51:951–961. 2010.
View Article : Google Scholar : PubMed/NCBI
|