Introduction
Human colorectal cancer (CRC) is one of the most
common types of cancer globally and one of the leading causes of
cancer-associated mortality in developed countries (1). A large proportion of patients with CRC
have already developed tumor metastasis, the primary cause of
mortality in patients with CRC, at the time of diagnosis (2). Metastasis is a multistep process that
remains poorly understood (3).
Therefore, it is necessary to reveal the pathological mechanisms
underpinning this process and to identify the critical pathways for
metastasis in CRC.
The sterol regulatory element-binding protein 1
(SREBP1) is a basic helix-loop-helix leucine zipper transcription
factor involved in regulating lipid homeostasis. High SREBP1
activity is an important feature of cancer metabolic reprogramming.
Previous studies have suggested that SREBP1 is an attractive target
for numerous types of cancer, including breast, ovarian and
pancreatic cancer (4,5). The expression of SREBP1 is elevated in
several types of cancer, and the inhibition of SREBP1 may inhibit
cancer cell growth, migration and invasion (6,7). However,
the role served by SREBP1 in CRC remains to be elucidated.
The present study demonstrated that SREBP1 is highly
expressed in CRC in order to promote the migration and invasion in
CRC cells. Furthermore, it was revealed that SREBP1 interacts with
c-Myc to promote snail family transcriptional repressor 1 (SNAIL)
expression, which is a key transcriptional repressor of cadherin-1
(CDH1) in epithelial-mesenchymal transition. Taken together, the
results of the present study demonstrated a critical role for
SREBP1 in the migration and invasion of CRC cells, providing a
potential target for the treatment of CRC.
Materials and methods
Cell culture and transfection
Fresh CRC and matched adjacent non-cancerous tissues
were collected from 44 patients (age range, 43–65 years; mean age,
52 year; 19 females and 25 males) who underwent surgery between
January 2013 and June 2016 at the Department of General Surgery,
Zhujiang Hospital of Southern Medical University (Guangzhou,
China). No patients received preoperative radiotherapy,
chemotherapy or had a history of any other treatment. CRC cell
lines SW480, HT29, HCT116, SW620, LS180 and normal colon epithelial
cells NCM460 were obtained from Foleibao Biotechnology Development
Co. (Shanghai, China). All cells were cultured in Dulbecco's
modified Eagle's medium (DMEM; HyClone; GE Healthcare Life
Sciences, Logan, UT, USA) supplemented with 10% fetal bovine serum
(FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and
were incubated at 37°C in a humidified incubator in an atmosphere
of 5% CO2. Transfection of SREBP1 (cloned into pcDNA3.1
vector; Addgene, Inc., Cambridge, MA, USA), small interfering RNA
(siRNA) of siSREBP1 and siSNAIL (IGE Biotechnology Co., Guangzhou,
China; http://www.igebio.com) were performed
using Lipofectamine 2000 reagent (Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. Empty pcDNA3.1 vectors
and scramble siRNA, a functional non-targeting siRNA provided by
the same company (IGE Biotechnology Co, Guangzhou, China,
http://www.igebio.com), were used as the negative
controls. The SREBP1 siRNA sequence was as follows:
5′-GCUCCUCACUUGAAGGCUUTT-3′. SNAIL siRNA sequence was as follows:
5′-CCAUGAGGAGUACUGCCAATT-3′. Briefly, in a 6-well plate, 5 µg
SREBP1 plasmid, 200 nm siSREBP1 and 200 nm siSNAIL were used for
transfection. A total of 48 h after transfection, the cells were
collected and prepared for analysis.
Reverse transcription-quantitative
polymerase reaction (RT-qPCR)
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and was
reverse-transcribed into cDNA at 42°C for 1 h using Premix Ex Taq
(Takara Biotechnology Co., Ltd., Dalian, China). cDNA was analyzed
using the CFX96 Real-Time system (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The PCR thermocycling conditions were as
follows: Initial denaturation at 95°C for 3 min, followed by 30
cycles at 95°C for 15 sec, 60°C for 30 sec and 72°C for 1 min. Gene
expression was calculated using the 2−∆∆Cq method for
relative quantification (8). GAPDH
was used as endogenous reference gene. Primers used were as
follows: GAPDH forward, GGAGCGAGATCCCTCCAAAA and reverse,
GGCTGTTGTCATACTTCTCATGG; SREBP1 forward, ACAGTGACTTCCCTGGCCTAT and
reverse, GCATGGACGGGTACATCTTCAA; c-Myc forward, GGCTCCTGGCAAAAGGTCA
and reverse, CTGCGTAGTTGTGCTGATGT; CDH2 forward,
TCAGGCGTCTGTAGAGGCTT and reverse, ATGCACATCCTTCGATAAGACTG;
fibronectin-1 (FN1) forward, CGGTGGCTGTCAGTCAAAG and reverse,
AAACCTCGGCTTCCTCCATAA; vimentin (VIM) forward, GACGCCATCAACACCGAGTT
and reverse, CTTTGTCGTTGGTTAGCTGGT; SNAIL forward,
TCGGAAGCCTAACTACAGCGA and reverse, AGATGAGCATTGGCAGCGAG; and
Twist-related protein 1 (TWIST1) forward, GTCCGCAGTCTTACGAGGAG and
reverse, GCTTGAGGGTCTGAATCTTGCT. All procedures were performed in
accordance with the ethical standards of the Research Committee of
Zhujiang Hospital of Southern Medical University and with the 1964
Declaration of Helsinki and its later amendments. Written informed
consent was obtained from all individual participants included in
the study.
Western blot analysis
Proteins from the nucleus and cytoplasm were
separated using the Nuclear and Cytoplasmic Protein Extraction kit
(Beyotime Institute of Biotechnology, Haimen, China). The protein
concentration was then determined using the BCA Protein Assay kit
(Beyotime Institute of Biotechnology). Briefly, the absorbance was
measured at 562 nm and protein concentration was determined
relative to bovine serum albumin. The protein concentration was
calculated from the standard curve: Protein
concentrationsample=2.624×(OD562sample-OD562blank)
(µg/µl). Then, ~20 µg of protein per lane was separated by 12%
SDS-PAGE and proteins were transferred onto polyvinylidene
difluoride membranes (EMD Millipore, Billerica, MA, USA).
Subsequent to blocking the membranes with 5% milk at room
temperature for 1 h, membranes were subsequently incubated with
primary antibodies against the following: SREBP1 (1:1,000; cat no.
ab57999; Abcam, Cambridge, UK), β-actin (1:5,000; cat no. 4967;
Cell Signaling Technology, Inc., Danvers, MA, USA), histone H3
(1:5,000; cat no. 9715; Cell Signaling Technology, Inc.). SNAIL
(1:500; cat no. C15D3; Cell Signaling Technology, Inc.) at 4°C
overnight. β-actin served as the control for total protein. Histone
H3 served as the control for nuclear proteins. Secondary antibody
horseradish peroxidase-conjugated mouse/rabbit IgG (1:5,000
dilution; cat no. 12971 and 12972; Beyotime Institute of
Biotechnology), were then incubated with the membranes at room
temperature for 1 h. The membranes were subsequently exposed to
X-ray film (Beyotime Institute of Biotechnology).
Cell migration and invasion assay
For the wound healing assay, 5×104 CRC
cells were cultured on 6-well plates for 24 h prior to a 1-mm wide
scratch being applied to the bottom of the plate. The recovery of
the scratch wound was monitored 24 h later using a light microscope
(magnification, ×10). The cell-free wound area was measured using
ImageJ software (1.41v, National Institutes of Health, Bethesda,
MD, USA) and the migration rate was determined by quantifying the
wound closure area. For the invasion assay, 10 ml Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA) was dissolved in 50 ml
serum-free DMEM (HyClone; GE Healthcare Life Sciences) and applied
to the upper chamber of 8-mm pore-size polycarbonate membrane
filters (Corning Incorporated, Corning, NY, USA), and placed in a
37°C incubator for 5 h prior to seeding the cells. CRC cells were
seeded with serum-free DMEM in the upper chamber at
1×105 cells/well, and the bottom chamber of the
apparatus contained DMEM with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.). Transwell inserts were then incubated for 48 h
at 37°C. Following incubation, the invaded cells that had invaded
through the pores of the filter and into the lower chamber were
fixed with 70% methanol for 10 min at room temperature and stained
with 1% toluidine blue (Beyotime Institute of Biotechnology) for 15
min at room temperature. Cell numbers were counted in ten randomly
selected microscopic fields per membrane using a light microscope
(magnification, ×100).
Chromatin immunoprecipitation
(ChIP)
ChIP experiments were performed using a
SimpleChIP® Plus Sonication Chromatin IP kit (Cell
Signaling Technology, Inc.), according to the manufacturer's
protocol. Normal rabbit IgG antibody was used as a control.
Antibodies used were as follows: Normal rabbit IgG (1:500 dilution;
cat. no. 2729; Cell Signaling Technology, Inc.) and anti-c-Myc
(1:500 dilution; cat. no. 13978; Cell Signaling Technology, Inc.)
at 4°C overnight. Subsequent qPCR was performed using the CFX96
Real-Time system (Bio-Rad Laboratories, Inc.). Data was calculated
using the 2−∆∆Cq method for relative quantification
(8). The PCR thermocycling conditions
were as follows: Initial denaturation at 95°C for 3 min, followed
by 30 cycles at 95°C for 15 sec, 60°C for 30 sec and 72°C for 1
min. Primers for the SNAIL promoter region were as follows:
Forward, 3′-CTTGACTCAGTGTCCCTCC-5′ and reverse,
3′-GCCAGAACTAATCGCATC-5′.
Co-immunoprecipitation (Co-IP)
Transfected CRC cells were lysed in cell lysis
buffer [50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% NP40,
protease inhibitors] for 30 min. Lysates were incubated with
Proteins A and G (Invitrogen; Thermo Fisher Scientific, Inc.) at
4°C overnight, which were coupled with c-Myc antibodies overnight
at 4°C. Following immunoprecipitation, Proteins A and G was washed
with lysis buffer five times to remove any un-precipitated proteins
and were boiled in SDS buffer for 5 min to elute the proteins. The
eluent was analyzed for precipitated SREBP1 protein using western
blot analysis as aforementioned. A normal IgG antibody was used as
a control. Antibodies against the following were used: SREBP1
(1:500 dilution, cat. no. 28481; Abcam), c-Myc (1:100 dilution,
cat. no. 13978; Cell Signaling Technology, Inc.) and normal rabbit
IgG (1:1,000 dilution, cat. no. 2729; Cell Signaling Technology,
Inc.).
Statistical analysis
Data are presented as the mean ± SD. Statistical
comparisons were performed using unpaired two-tailed Student's
t-tests or one-way analysis of variance followed by a Newman-Keuls
test. P≤0.05 was considered statistically significant.
Results
SREBP1 is highly expressed in
colorectal cancer tissues and cell lines
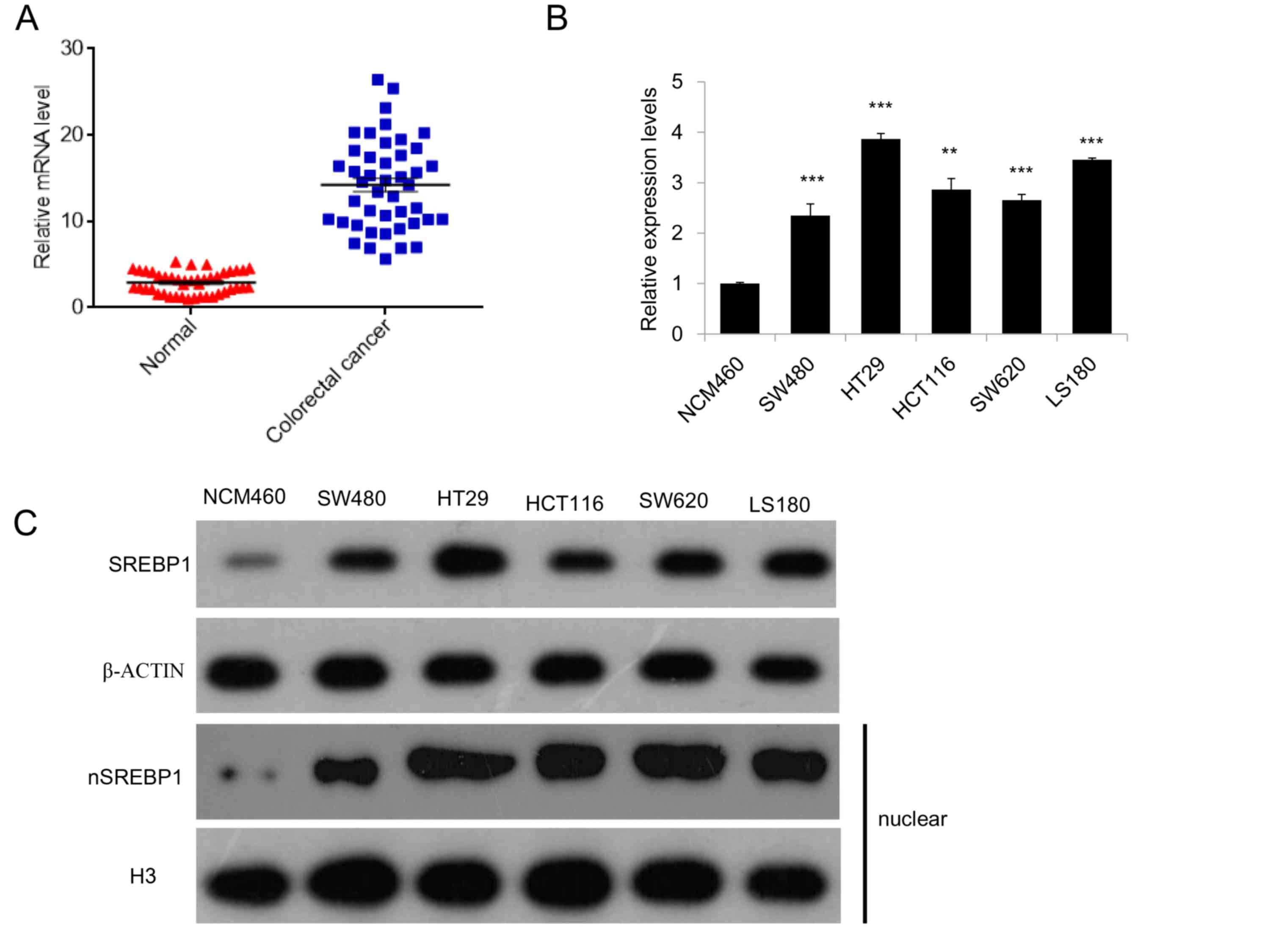
To evaluate the potential roles for SREBP1 in CRC,
the expression of SREBP1 in 44 paired CRC samples and adjacent
normal tissues were analyzed using RT-qPCR. The results of the
present study demonstrated that SREBP1 is highly expressed in CRC
samples, compared with expression in healthy controls (Fig. 1A). The expression of SREBP1 was also
measured in several CRC cell lines, including SW480, HT29, HCT116,
SW620 and LS180, and a higher expression of SREBP1 was observed in
these CRC cell lines compared with normal colon epithelial cells
(NCM460; Fig. 1B). Additionally,
western blot analysis of total and nuclear SREBP1 further confirmed
that SREBP1 is highly expressed and activated in these CRC cell
lines (Fig. 1C), suggesting a
potential role for SREBP1 in colorectal cancer.
SREBP1 regulates the proliferation,
migration and invasion in CRC cells
SREBP1 is a well-known key regulator of lipid
metabolism. Recently, growing evidence has indicated that SREBP1 is
involved in cancer development and progression through targeting
lipid metabolism and cancer cell migration. To determinate the
effects of SREBP1 on the growth of CRC cells, the proliferation
rate of CRC SW480 cells upon SREBP1 overexpression or SREBP1
knockdown was measured. It was revealed that SREBP1 overexpression
increased the proliferation rate of CRC cells, while SREBP1
knockdown using siRNA decreased the proliferation ability of CRC
cells (Fig. 2A). These results
suggested that SREBP1 may regulate CRC cell growth.
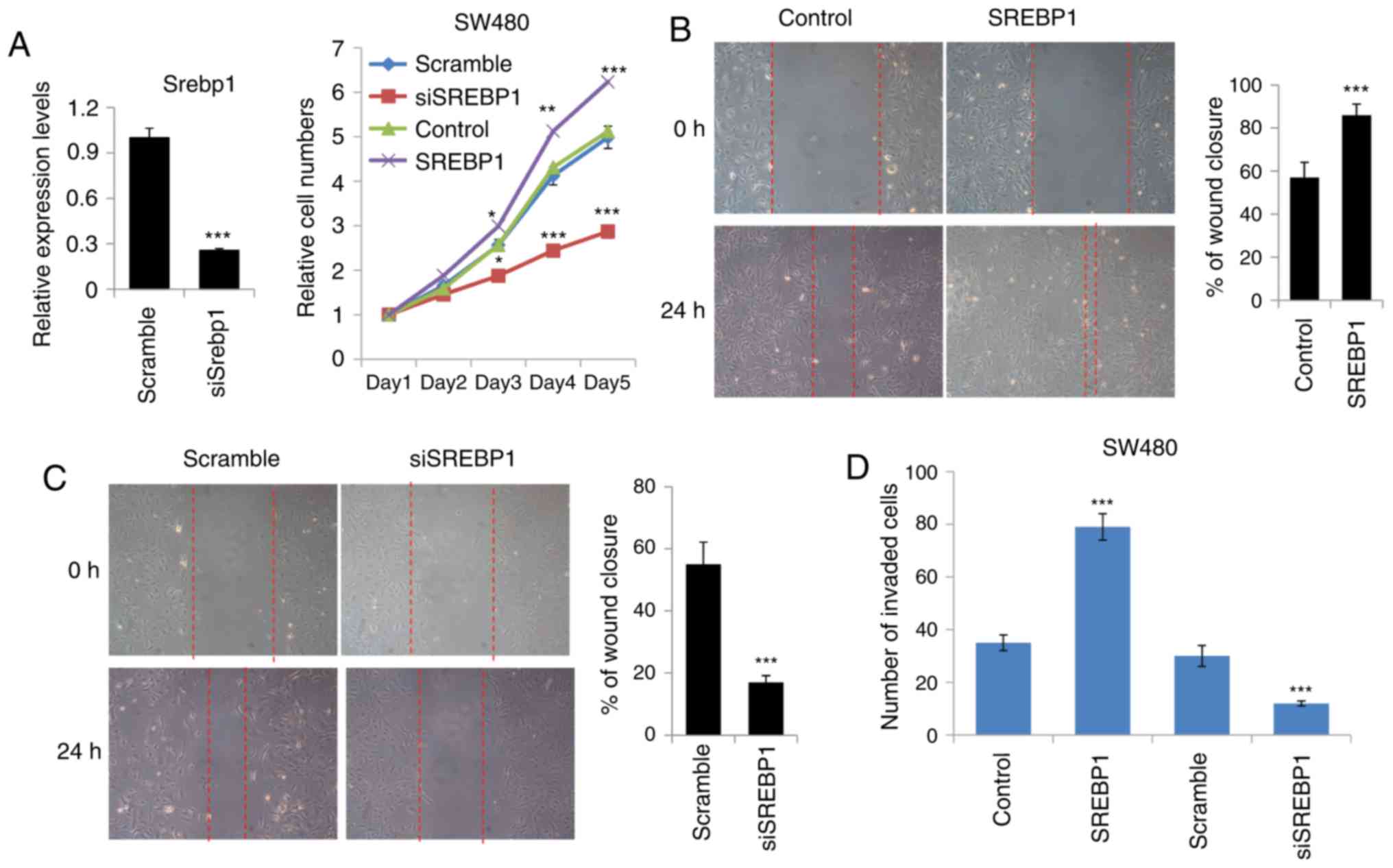 | Figure 2.SREBP1 regulates the proliferation,
migration and invasion of CRC cells. (A) Cell proliferation assays
of CRC cells transfected with SREBP1 or SREBP1 siRNA. Right,
knockdown efficiency of siSREBP1 and left, cell proliferation
assays. Data are presented as the mean ± standard deviation of
three independent experiments. *P<0.05, **P<0.01 and
***P<0.001 vs. scramble or control. (B) Wound healing assays of
CRC cells transfected with SREBP1. Right, representative image of
wound healing assays and left, analysis of the migration rate. The
migration rate was determined by quantifying the wound closure area
after 24 h of migration. Data are presented as the mean ± standard
deviation of three independent experiments. ***P<0.001 vs.
control. (C) Wound healing assays of CRC cells transfected with
SREBP1 siRNA. Right, representative image of wound healing assays
and left, analysis of the migration rate. The migration rate was
determined by quantifying the wound closure area after 24 h of
migration. Data are presented as the mean ± standard deviation of
three independent experiments. ***P<0.001 vs. scramble. (D)
Transwell assays of CRC cells transfected with SREBP1 or SREBP1
siRNA. Cells that had spread through the pores of the filter and
into the lower chamber were fixed with 70% methanol and were
counted. Data are presented as the mean ± standard deviation of
three independent experiments. ***P<0.001 vs. scramble or
control. SREBP1, sterol regulatory element-binding protein 1; CRC,
colorectal cancer; si, small interfering RNA. |
As metastasis is a major problem in CRC and is
associated with cancer cell migration and invasion (9–11), the
present study aimed to determine whether SREBP1 may affect the
migration or invasion of CRC cells. Wound healing assays were
performed and it was revealed that SREBP1 overexpression
significantly increased the wound closure area, suggesting that
SREBP1 may accelerate the migration rate in CRC cells (Fig. 2B). However, knockdown of SREBP1 using
siRNA resulted in impairment of the migration ability in CRC cells
(Fig. 2C). Furthermore, Transwell
assays were performed in order to confirm the effects of SREBP1 on
CRC cell migration. Compared with the control vector-expressing
cells, SREBP1-overexpressing cells exhibited a 2-fold increase in
the number of cells penetrating the Transwell membrane (Fig. 2D). In line with this, SREBP1 knockdown
reduced the number of cells penetrating the membrane, indicating
important roles for SREBP1 in CRC cell invasion (Fig. 2D). Taken together, these data
demonstrated that SREBP1 may promote CRC cell growth, migration and
invasion.
SREBP1 promotes EMT and migration by
increasing SNAIL expression
Given that EMT is the key process involved in cancer
migration and that SREBP1 has been reported to serve a role in
breast cancer migration (6), it is
reasonable to hypothesize that SREBP1 promotes CRC cell migration
through EMT. In order to elucidate the mechanism by which SREBP1
promotes CRC cell migration, the expression of several mesenchymal
genes, including CDH2, FN1, VIM, SNAIL and TWIST1, was measured.
Data obtained through qPCR demonstrated that overexpression of
SREBP1 increased the expression of all these mesenchymal genes,
while knockdown of SREBP1 using siRNA inhibited these genes
(Fig. 3A and B). Furthermore, the
change in SNAIL expression was most marked upon SREBP1
overexpression or knockdown (Fig. 3A and
B). Western blot analysis of SNAIL revealed similar results
(Fig. 3C). This led to the hypothesis
that SNAIL is responsible for the SREBP1-driven EMT in CRC cells.
In order to confirm this, wound healing and Transwell assays were
performed in CRC cells transfected with SNAIL siRNA or SREBP1 plus
SNAIL siRNA. Wound healing assays demonstrated that SREBP1 does not
promote CRC cell migration when SNAIL is inhibited by siRNA
(Fig. 3D and E), suggesting that the
function of SREBP1 is SNAIL-dependent. Transwell assays further
confirmed the results of the wound healing assays (Fig. 3D). Taken together, these results
indicated that SREBP1 promotes migration and ETM by increasing
SNAIL expression.
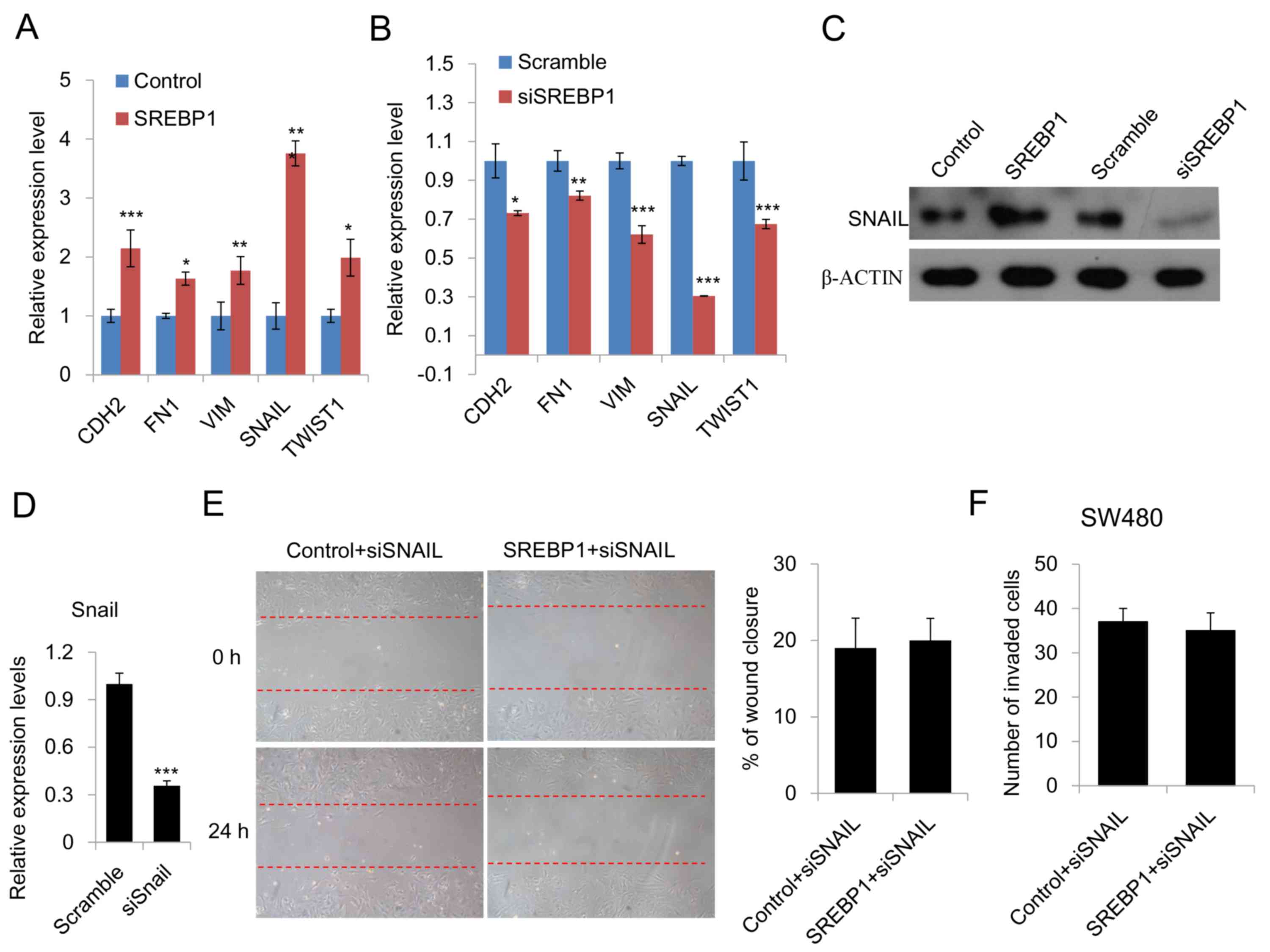 | Figure 3.SREBP1 promotes EMT by increasing
SNAIL expression. (A) qPCR analysis of the expression of
EMT-related genes in CRC cells transfected with SREBP1 or a control
vector. Data are presented as the mean ± standard deviation of
three independent experiments. *P<0.05, **P<0.01 and
***P<0.001 vs. control. (B) qPCR analysis of the expression of
EMT-related genes in CRC cells transfected with SREBP1 siRNA or
scramble siRNA. Data are presented as the mean ± standard deviation
of three independent experiments. *P<0.05, **P<0.01 and
***P<0.001 vs. scramble. (C) Western blot analysis of SNAIL
expression in CRC cells transfected with control vector, SREBP1,
scramble or siSREBP1. (D) qPCR analysis of the knockdown efficiency
of siSNAIL. Data are presented as the mean ± standard deviation of
three independent experiments. ***P<0.001 vs. scramble. (E)
Wound healing assays of CRC cells transfected with SREBP1 and SNAIL
siRNA. Right, representative image of wound healing assays and
left, analysis of the migration rate. The migration rate was
determined by quantifying the wound closure area after 24 h of
migration. Data are presented as the mean ± standard deviation of
three independent experiments. (F) Transwell assays of CRC cells
transfected with SREBP1 and SNAIL siRNA. Cells that had invaded
through the pores of the filter and into the lower chamber were
fixed with 70% methanol and counted. Data are presented as the mean
± standard deviation of three independent experiments. SREBP1,
sterol regulatory element-binding protein 1; EMT,
epithelial-mesenchymal transition; qPCR, quantitative polymerase
chain reaction; CRC, colorectal cancer; si, small interfering RNA;
CDH2, cadherin 2; FN1, fibronectin-1; VIM, vimentin; TWIST1,
Twist-related protein 1. |
SREBP1 facilitates the binding of
c-Myc to the SNAIL promoter
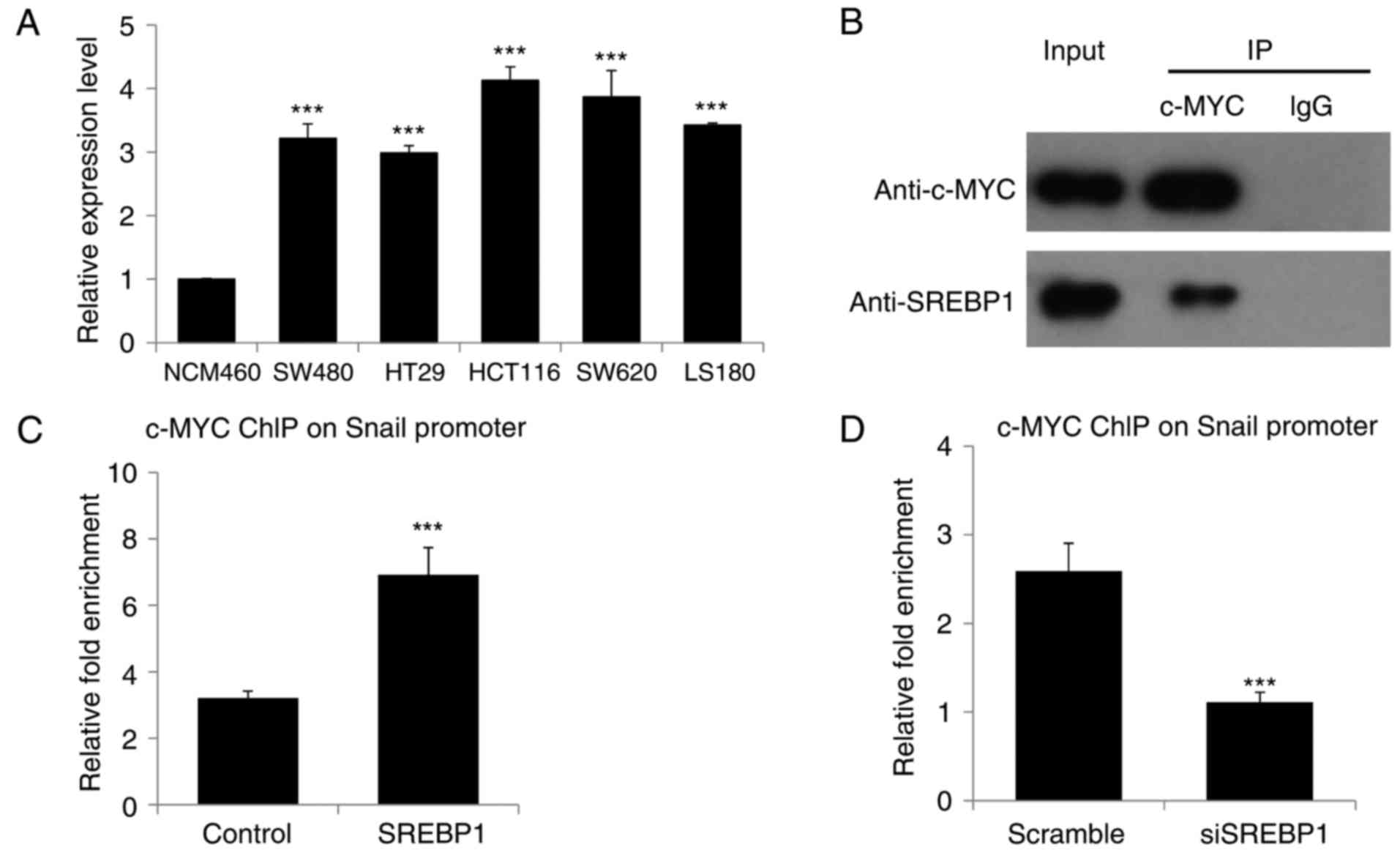
Due to the fact that c-Myc is highly expressed in
CRC and that SNAIL is one of the crucial targets of c-Myc in EMT
(12–14), the present study investigated whether
or not c-Myc is responsible for SREBP1-driven SNAIL expression and
EMT in CRC. To begin with, the expression of c-Myc was measured in
different CRC cell lines and it was revealed that c-Myc is highly
expressed in all these CRC cell lines (Fig. 4A), consistent with the results of
previous studies (12–14). As SREBP1 has been reported to interact
with c-Myc to enhance the function of c-Myc in somatic cell
reprogramming (15), we hypothesized
that there may be an associated between SREBP1 and c-Myc in CRC. In
the present study, Co-IP was performed, revealing that SREBP1
interacts with c-Myc in CRC cells (Fig.
4B). Due to the fact that c-Myc is involved in EMT through
direct targeting of the promoter of SNAIL (16,17),
ChIP-qPCR was performed in order to analyze the binding of c-Myc to
the SNAIL promoter in CRC cells transfected with SREBP1 or siSREBP1
and corresponding control vectors. ChIP-qPCR revealed a higher
enrichment of c-Myc on the SNAIL promoter when SREBP1 was
overexpressed in CRC cells (Fig. 4C).
Knockdown of SREBP1 led to a decreased binding of c-MYC to the
SNAIL promoter (Fig. 4D). Therefore,
the results of the present study demonstrated that SREBP1
facilitates the binding of c-Myc to the SNAIL promoter in CRC
cells, thereby accelerating SNAIL expression, EMT and
migration.
Discussion
Metastasis is the primary cause of mortality in
patients with CRC, the mechanism of which remains to be fully
elucidated (3,18). Further investigation into the
underlying mechanisms of CRC metastasis is required. Emerging
evidence has indicated that SREBP1, a master transcription factor
that controls metabolic reprogramming in cancer cells, is also
involved in the invasion and migration of cancer cells (19). However, the roles of SREBP1 in CRC
remain unclear. The present study demonstrated that SREBP1 is
highly expressed in CRC tissues and cell lines, suggesting a
potential role for SREBP1 in CRC. Furthermore, it was revealed that
SREBP1 regulates CRC cell proliferation, migration and invasion
through increasing SNAIL expression and accelerating EMT. Finally,
mechanistic studies revealed that SREBP1-driven EMT depends upon
c-Myc, as SREBP1 interacts with c-Myc to enhance its binding to the
SNAIL promoter, thereby increasing SNAIL expression and subsequent
EMT and cell migration.
Metabolic reprogramming is one of the key features
of cancer cells (18). SREBP1 has
become an area of interest in cancer biology because it is a key
regulator of lipid metabolism (19).
Recently, the high expression level of SREBP1 in cancer cells has
encouraged novel research (4,5,7,20). The majority of these studies have
focused on the impact of SREBP1 on cancer cell metabolism,
particularly lipid synthesis and desaturation (4,5,7,20). Few
previous studies have demonstrated that SREBP1 is involved in cell
migration and invasion (6), and the
underlying mechanism of this remains unclear. However, the present
study demonstrated that SREBP1 regulates the migration and invasion
of CRC cells. Furthermore, the mechanism through which SREBP1
promoting EMT was also investigated, and it was revealed that
SREBP1 interacts with c-Myc to increase the expression of SNAIL,
thereby promoting EMT in CRC.
The proto-oncogene, c-Myc, serves crucial roles in
various types of cancer and is known to be highly expressed in the
majority of cases of CRCs (12–14,21–23).
It has been reported that c-Myc is primarily involved in regulation
at the transcriptional and post-transcriptional levels in multiple
types of cancer (14,21–23).
However, the regulatory mechanisms of c-Myc in CRC remain unclear.
The present study not only revealed a synergistic as association
between SREBP1 and c-Myc, which was consistent with the
observations of previous studies (15), but also provided a novel regulatory
model for the regulation of c-Myc in CRC.
To conclude, the results of the present study
demonstrated that SREBP1 interacts with c-Myc to enhance the
EMT-promoting function of c-Myc. The present study identified a
novel role for SREBP1 in the metastasis of CRC cells and provided
insight on the regulatory mechanism of c-Myc.
Acknowledgements
The authors would like to thank the Department of
General Surgery, Zhujiang Hospital of Southern Medical University
(Guangzhou, China) of Professor Jinglong Yu for providing technical
support.
References
|
1
|
Weitz J, Koch M, Debus J, Hohler T, Galle
P and Buchler MW: Colorectal cancer. Lancet. 365:153–165. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hu CE and Gan J: TRIM37 promotes
epithelial-mesenchymal transition in colorectal cancer. Mol Med
Rep. 15:1057–1062. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Boutin AT, Liao WT, Wang M, Hwang SS,
Karpinets TV, Cheung H, Chu GC, Jiang S, Hu J, Chang K, et al:
Oncogenic Kras drives invasion and maintains metastases in
colorectal cancer. Genes Dev. 31:370–382. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Williams KJ, Argus JP, Zhu Y, Wilks MQ,
Marbois BN, York AG, Kidani Y, Pourzia AL, Akhavan D, Lisiero DN,
et al: An essential requirement for the SCAP/SREBP signaling axis
to protect cancer cells from lipotoxicity. Cancer Res.
73:2850–2862. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Griffiths B, Lewis CA, Bensaad K, Ros S,
Zhang Q, Ferber EC, Konisti S, Peck B, Miess H, East P, et al:
Sterol regulatory element binding protein-dependent regulation of
lipid synthesis supports cell survival and tumor growth. Cancer
Metab. 1:32013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bao J, Zhu L, Zhu Q, Su J, Liu M and Huang
W: SREBP-1 is an independent prognostic marker and promotes
invasion and migration in breast cancer. Oncol Lett. 12:2409–2416.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li W, Tai Y, Zhou J, Gu W, Bai Z, Zhou T,
Zhong Z, McCue PA, Sang N, Ji JY, et al: Repression of endometrial
tumor growth by targeting SREBP1 and lipogenesis. Cell cycle.
11:2348–2358. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jackstadt R, Röh S, Neumann J, Jung P,
Hoffmann R, Horst D, Berens C, Bornkamm GW, Kirchner T, Menssen A
and Hermeking H: AP4 is a mediator of epithelial-mesenchymal
transition and metastasis in colorectal cancer. J Exp Med.
210:1331–1350. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kurita K, Maeda M, Mansour MA, Kokuryo T,
Uehara K, Yokoyama Y, Nagino M, Hamaguchi M and Senga T: TRIP13 is
expressed in colorectal cancer and promotes cancer cell invasion.
Oncol Lett. 12:5240–5246. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Long J, Xie Y, Yin J, Lu W and Fang S:
SphK1 promotes tumor cell migration and invasion in colorectal
cancer. Tumour Biol. 37:6831–6836. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Boudjadi S, Carrier JC, Groulx JF and
Beaulieu JF: Integrin α1β1 expression is controlled by c-MYC in
colorectal cancer cells. Oncogene. 35:1671–1678. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang L, Liu X, Zuo Z, Hao C and Ma Y:
Sphingosine kinase 2 promotes colorectal cancer cell proliferation
and invasion by enhancing MYC expression. Tumour Biol.
37:8455–8460. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Z, Jiang Y, Hou Y, Hu Y, Cao X, Tao Y,
Xu C, Liu S, Wang S, Wang L, et al: The IκB family member Bcl-3
stabilizes c-Myc in colorectal cancer. J Mol Cell Biol. 5:280–282.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu Y, Chen K and Liu X, Huang L, Zhao D,
Li L, Gao M, Pei D, Wang C and Liu X: Srebp-1 Interacts with c-Myc
to enhance somatic cell reprogramming. Stem cells. 34:83–92. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Smith AP, Verrecchia A, Fagà G, Doni M,
Perna D, Martinato F, Guccione E and Amati B: A positive role for
Myc in TGFbeta-induced Snail transcription and
epithelial-to-mesenchymal transition. Oncogene. 28:422–430. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cho KB, Cho MK, Lee WY and Kang KW:
Overexpression of c-myc induces epithelial mesenchymal transition
in mammary epithelial cells. Cancer Lett. 293:230–239. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yao C, Su L, Shan J, Zhu C, Liu L, Liu C,
Xu Y, Yang Z, Bian X, Shao J, et al: IGF/STAT3/NANOG/slug signaling
axis simultaneously controls epithelial-mesenchymal transition and
stemness maintenance in colorectal cancer. Stem cells. 34:820–831.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Beloribi-Djefaflia S, Vasseur S and
Guillaumond F: Lipid metabolic reprogramming in cancer cells.
Oncogenesis. 5:e1892016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sun Y, He W, Luo M, Zhou Y, Chang G, Ren
W, Wu K, Li X, Shen J, Zhao X and Hu Y: SREBP1 regulates
tumorigenesis and prognosis of pancreatic cancer through targeting
lipid metabolism. Tumour Biol. 36:4133–4141. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chuan YC, Iglesias-Gato D, Fernandez-Perez
L, Cedazo-Minguez A, Pang ST, Norstedt G, Pousette A and
Flores-Morales A: Ezrin mediates c-Myc actions in prostate cancer
cell invasion. Oncogene. 29:1531–1542. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhu W, Cai MY, Tong ZT, Dong SS, Mai SJ,
Liao YJ, Bian XW, Lin MC, Kung HF, Zeng YX, et al: Overexpression
of EIF5A2 promotes colorectal carcinoma cell aggressiveness by
upregulating MTA1 through C-myc to induce
epithelial-mesenchymaltransition. Gut. 61:562–575. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mansour MA, Hyodo T, Akter KA, Kokuryo T,
Uehara K, Nagino M and Senga T: SATB1 and SATB2 play opposing roles
in c-Myc expression and progression of colorectal cancer.
Oncotarget. 7:4993–5006. 2016. View Article : Google Scholar : PubMed/NCBI
|