Introduction
Primary hepatocellular carcinoma (HCC) is a human
malignancy with a high incidence, mortality and recurrence rate,
worldwide, seriously threatening human health (1,2). China is
one of the areas of high HCC incidence, >45% of the worldwide
cases of HCC were estimated to occur in China in the last decade.
In addition, HCC is the second leading cause of malignant tumor
mortality in males in China, after gastric carcinoma (3,4). For these
reasons, research regarding liver carcinoma treatment is
essential.
The orthotopic transplantation tumor model of HCC is
an ideal model for studying the mechanisms of metastasis and tumor
immunity, and for the development of anti-tumor drugs and novel
therapeutic methods (5,6). Techniques of cell tracking in
vivo may permit the noninvasive monitoring of experimental
animals, which is of great significance for the dynamic study of
tumor diseases. Commonly used in vivo tracing techniques
include radionuclide imaging, magnetic resonance imaging and
optical imaging (7,8). Among these methods, in vivo
optical imaging technology with bioluminescence (bioluminescence
image, BLI) has the advantages of high sensitivity, accurate
quantification with minimal trauma, simple operation and the
capacity for direct observation. At present, it is utilized
extensively in preclinical cancer studies, including stem cell
tracking, progression of tumor metastasis or the kinetics of tumor
growth, to assess the effectiveness of antineoplastic agents in a
tumor xenograft mouse model (9–11).
The murine hepatoma Hepa1-6 cell line, originating
from a BW7756 mouse hepatoma in a C57/L mouse, is commonly used to
establish hepatocarcinogenesis mouse models due to its high
malignancy and low immunogenicity (12). In the present study, the potential
application of the Hepa1-6 cell line transfected with a recombinant
retroviral vector encoding the firefly luciferase (FLuc) gene was
investigated. The resulting Hepa1-6-FLuc cells exhibited similar
cellular morphology and biological characteristics, including
proliferation, migration and invasion rates, to the parental
Hepa1-6 cell line. Furthermore, Hepa1-6-FLuc cells could form tumor
masses subsequent to their subcutaneous transplantation in nude
mice; the bioluminescence signal of the developing tumor masses was
continuously enhanced, reflecting cell proliferation and survival
in vivo. The Hepa1-6-FLuc cell line, with the stable
expression of the FLuc gene, should be an ideal resource to
establish hepatocarcinoma animal models, and longitudinally monitor
tumor proliferation, viability and metastasis, providing a valuable
tool in the study of hepatocarcinoma.
Materials and methods
Cell culture and chemicals
The murine hepatocellular carcinoma cell line
Hepal-6 and human embryonic kidney cell line HEK-293 was purchased
from the American Type Culture Collection (Manassas, VA, USA) and
maintained in complete Dulbecco's modified Eagle's medium (DMEM)
supplemented with 10% fetal bovine serum (FBS; Hyclone; GE
Healthcare Life Sciences, Logan, UT, USA), 100 units/ml penicillin
and 100 µg/ml streptomycin at 37°C and 5% CO2. Cells
were subcultured at 90% confluence.
Establishment of Hepa1-6 cell lines
containing the FLuc gene
A retroviral vector, expressing FLuc and a
blasticidin selection marker, and a pCLAmpho mammalian expression
vector (Novus Biologicals, LLC, Littleton, CO, USA) were
co-transfected into 293 cells with Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA),
according to the manufacture's instructions, to package the
recombinant-retrovirus. Hepa1-6 cells were seeded in T-25 flasks
and infected with the retrovirus for 7 days, then selected in the
presence of 3 µg/ml blasticidin S (Invitrogen; Thermo Fisher
Scientific, Inc.) for 14 days. The surviving cells were passaged
and designated as Hepa1-6-FLuc.
The in vitro luciferase activity of the
Hepa1-6-FLuc cells was assessed by using the Firefly Luciferase
Assay kit (Promega Corporation, Madison, WI, USA). A total of
~2×105 of cells were incubated in 24-well plates for 3
days and lysed in 1X passive lysis buffer (PLB). Cell lysate (20
µl) and luciferase assay buffer (100 µl) were mixed, and the
absorbance at 560 nm was read immediately in the GloMax®
20/20 luminometer (Promega Corporation). The experiment was
performed in triplicate.
Cell proliferation and viability
assay
An MTT assay and crystal violet staining were used
to detect the cell proliferation and viability, as previously
described (13). Briefly, 200 µl cell
suspensions (~5,000 cells) were seeded into each well of 96-well
plates and incubated overnight. At 1, 2, 3, 4 and 5 days later, 20
µl freshly prepared 5 mg/ml MTT was added to each well. Following a
further 4-h incubation, the medium was carefully removed and 150 µl
dimethyl sulfoxide was added to dissolve the MTT-formazan crystals.
The plate was covered with tinfoil and agitated on an orbital
shaker for 15 min, and the absorbance was read at 490 nm.
For crystal violet staining, fixed cells in 24-well
plates were stained with 0.05% crystal violet solution for 30 min
and images were captured using a digital camera at ×1 magnification
(D7000; Nikon, Tokyo, Japan) after washing three times by PBS.
Following treatment with 500 µl 33% acetic acid, mission spectra
were measured at an excitation wavelength of 570 nm using a
multimode microplate reader (Thermo Fisher Scientific, Inc.). A
total of three independent experiments were performed in duplicate,
from which the means and standard deviations (SDs) were
calculated.
Colony formation assay
Oncogenic transformation was evaluated with a colony
formation assay, as previously described (14,15). A
total of 400 cells were seeded onto 6-well plates, and cultured in
complete DMEM with 10% FBS, which was replaced every 3 days. After
14 days, cells were stained with Giemsa stain. The number of the
colonies containing >50 cells was counted under an inverted
phase microscope (TE2000-S; Nikon) at ×40 magnification and the
plate clone-forming efficiency was calculated as follows: Number of
colonies/number of cells seeded × 100%.
Monolayer wound healing cell migration
assay
The scratch wound healing assay was performed to
detect cell migration in vitro, as previously described
(15). Approximately 5×105
cells were seeded into 6-well plates in DMEM with 1% FBS. Following
the formation of confluent monolayer, a gap in the surface of the
confluent cells was created with a pipette tip. Bright field images
at ×40 magnification of the wounds were captured at 0, 1, 2, 3, 4
and 5 days to assess the cell migration across the gap. Each assay
was performed in triplicate.
Cell migration and invasion (Matrigel)
assay
For the cell migration and invasion assays, a Cell
Invasion Assay kit (Cell Biolabs, Inc., San Diego, CA, USA) was
used according to the manufacturer's instructions. Briefly, a total
of ~1×104 cells were seeded into the Transwell insert in
serum-free DMEM, whereas DMEM with 10% FBS was added to the lower
well. At 48 h, cells were fixed and cells at the top of chamber
were removed. Cells on the lower side of the chamber were stained
with crystal violet, and visualized with a light microscope. The
stain was dissolved with 33% acetic acid and absorbance of each
well was measured at 570 nm with a microplate reader (Thermo Fisher
Scientific, Inc.). The procedure was repeated independently three
times, with triplicate chambers for each group.
Cell implantation and in vivo
imaging
The use and care of animals was approved by the
Institutional Animal Care and Use Committee of the Children's
Hospital of Chongqing Medical University (Chongqing, China). A
total of nine female BALB/c nude mice (6–8 weeks, weight ~20 g;
Tengxin Biotechnology Co., Ltd, Chongqing, China) were housed in
specific pathogen-free laboratory with a 12/12 h light/dark cycle
under a controlled temperature of 22±2°C and humidity of 50±10%
with ad libitum access to food and water. Hepa1-6 cells were
infected with adenovirus AdFLuc (Molecular Oncology Laboratory, The
University of Chicago Medical Center, Chicago, IL, USA) for 24 h
and termed as Hepa1-6-AdFLuc. Subconfluent Hepa1-6, Hepa1-6-FLuc or
Hepa1-6-AdFLuc cells were collected and subcutaneously injected
into the front and rear notum on the left and/or right side(s) of
the nude mice (1×106 cells/injection) (16). At 1 day, 1 week and 2 weeks after
implantation, mice were intraperitoneally injected with 2 mg/ml 0.1
ml D-luciferin (Gold Biotechnology, Inc., Olivette, MO, USA) and
visualized using an IVIS-200 optical in vivo imaging system
(Xenogen Corporation, Alameda, CA, USA) to quantify cell
survival.
Assessment of tumor size and
histochemical stain
Mice were sacrificed by CO2 asphyxiation
at 14 days after cell implantation. Tumor tissues were harvested,
the size of the tumors was measured and images were captured using
a digital camera. The specimens were fixed with 10% formalin at
room temperature for 30 min, embedded in paraffin and serially
cutinto 5-µm thick sections. The sections were stained with 1%
hematoxylin and 0.2% eosin (H&E) at room temperature for 10
min, and then photographed with a microscope (Nikon).
Statistical analysis
Data are presented as the means ± SD and analyzed by
SPSS 18.0 software (SPSS, Inc., Chicago, IL, USA). A two-tailed
student's t-test was used to evaluate the difference between two
groups, and a one-way analysis of variance with a
Student-Newman-Keuls post hoc test was used to evaluate the
differences among three or more groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
Establishment of the Hepa1-6-FLuc
stable cell line
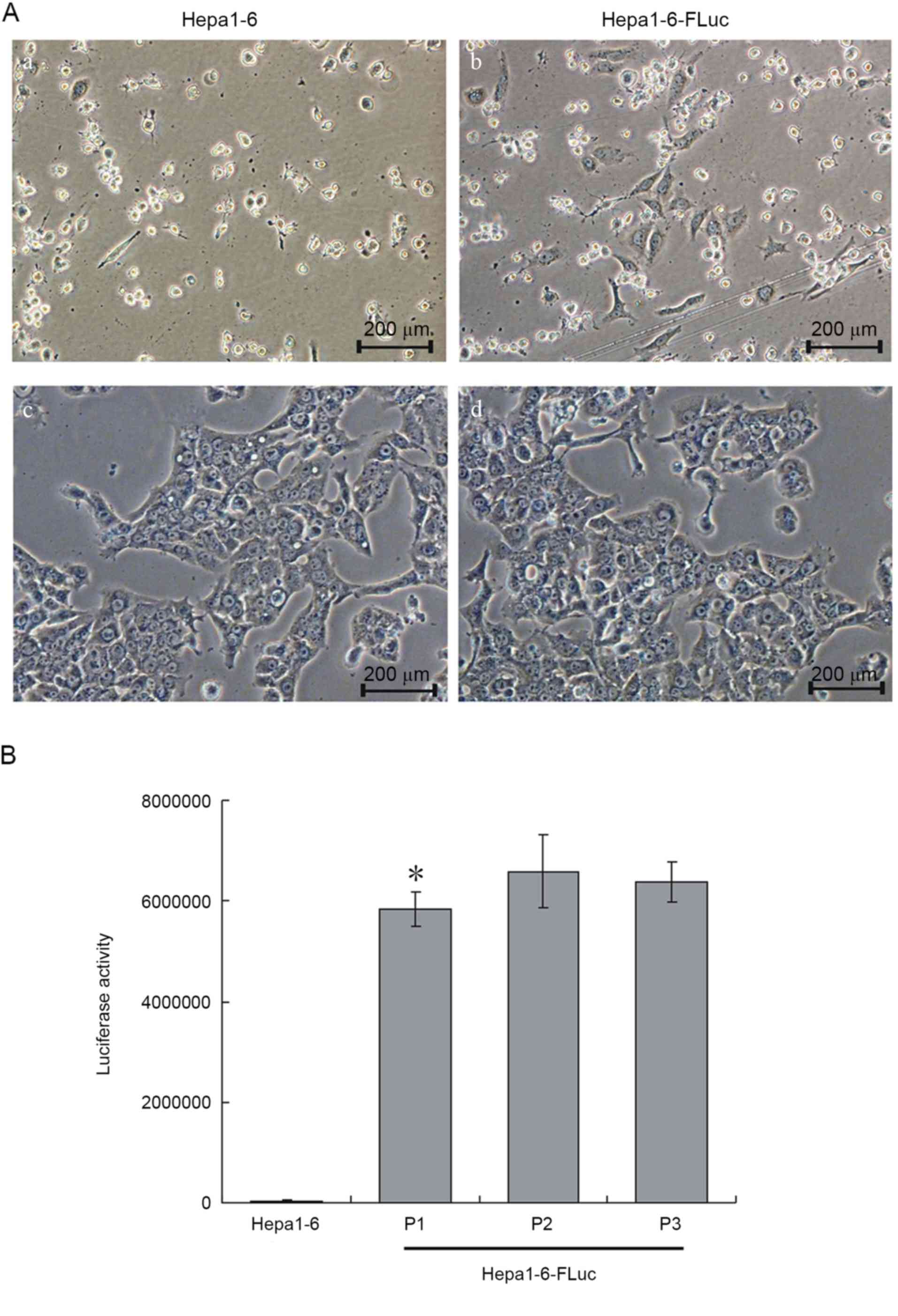
Following 14 days of blasticidin S selection, all
Hepa1-6 cells were dead (Fig. 1A-a)
and only 5–7% of Hepa1-6-FLuc cells had survived (Fig. 1A-b). The 5th passage of Hepa1-6-FLuc
cells was uniform and exhibited the same cell shape as the Hepa1-6
progenitor cells (Fig. 1A-c and -d).
The luciferase activity assay revealed significantly increased
luciferase activity in the Hepa1-6-FLuc cells compared with the
parental Hepa1-6 cells (Fig. 1B;
P<0.05). Thus, a stable Hepa1-6-FLuc cell line, with blasticidin
resistance and expressing FLuc, was successfully constructed.
Hepa1-6-FLuc cell line has similar
characteristics to the Hepa1-6 cell line
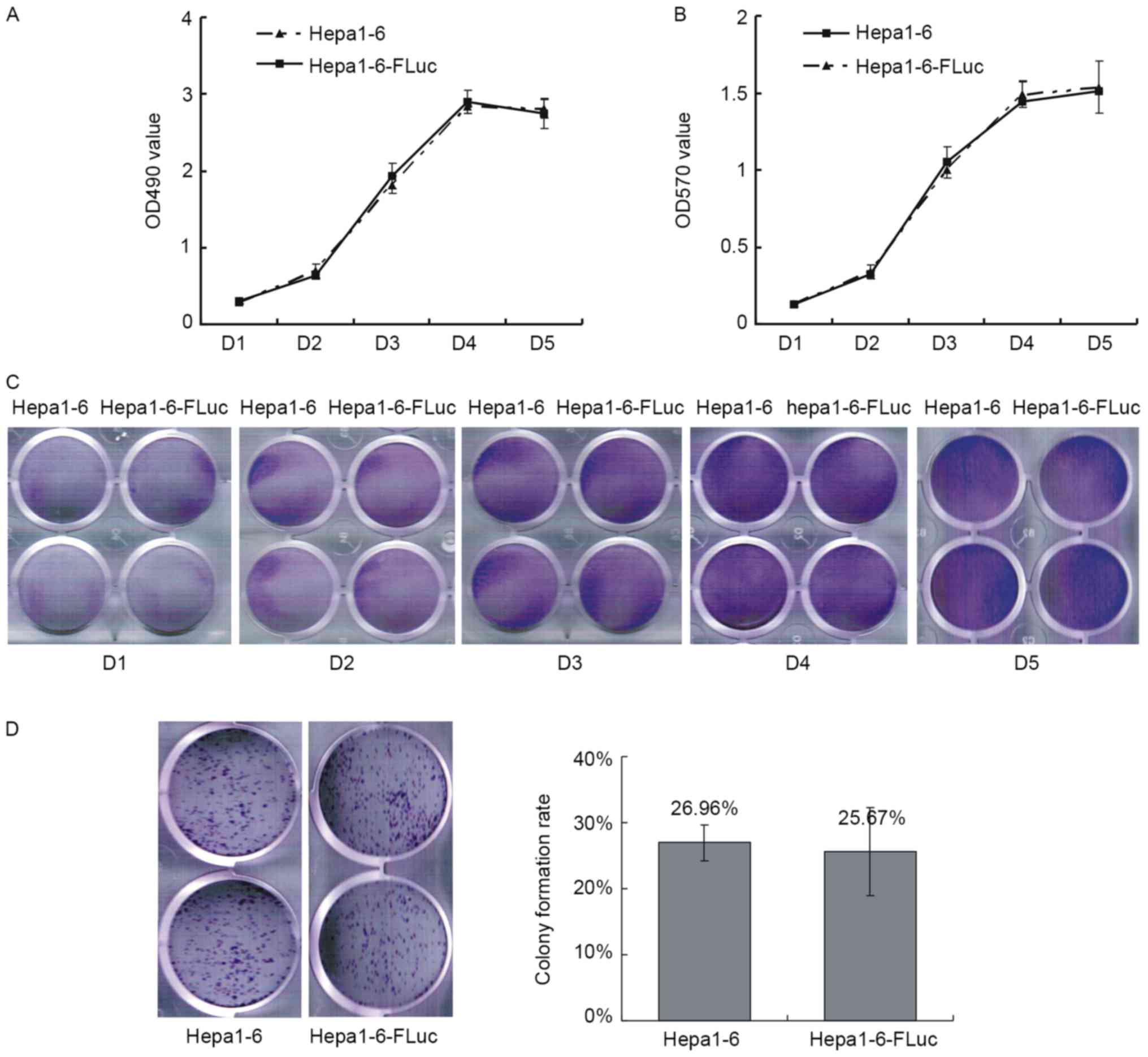
Growth curves were produced by MTT and crystal
violet staining assays. The growth curve of the Hepa1-6 cells
exhibited a typical sigmoid shape, and the stable expression of Luc
in Hepa1-6-FLuc cells did not affect their proliferation (Fig. 2A-C). Colony formation reflects the
proliferation and migration ability of cells; the colony formation
rates of Hepa1-6 and Hepa1-6-FLuc cells were 26.59±2.67 and
25.67±6.68%, respectively, which were not statistically different
(Fig. 2D; P>0.05).
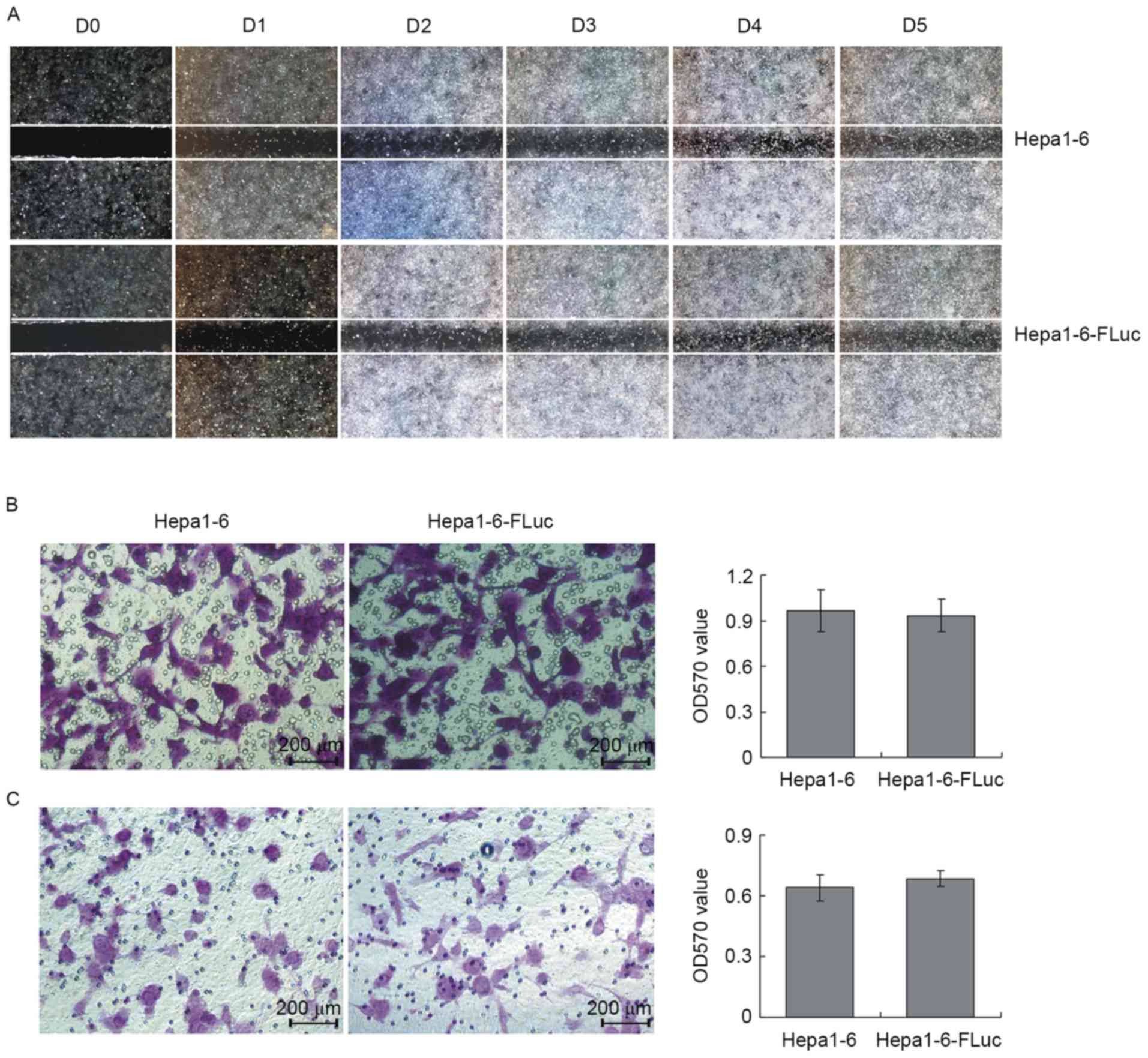
The results of the wound healing assays revealed the
migration ability of Hepa1-6 and Hepa1-6-FLuc cells was similar
(Fig. 3A). In the Transwell cell
migration and invasion assays, the migrating cell number for the
Hepa1-6 and Hepa1-6-FLuc groups was consistent (Fig. 3B; P>0.05), which was also true for
the invading cell number (Fig. 3C;
P>0.05). Therefore, the present study demonstrated that the
Hepa1-6 and Hepa1-6-FLuc cell lines exhibited similar
proliferation, migration and invasion abilities.
Hepa1-6-Fluc cells reflect Hepa1-6
cell proliferation and survival in vivo
The Xenogen IVIS imaging system a highly sensitive
in vivo imaging system that can be used to track cells in
real time if cells are tagged with a gene encoding a luciferase
enzyme; its non-invasive visualization allows the monitoring of
cell dynamics in vivo (16,17). In
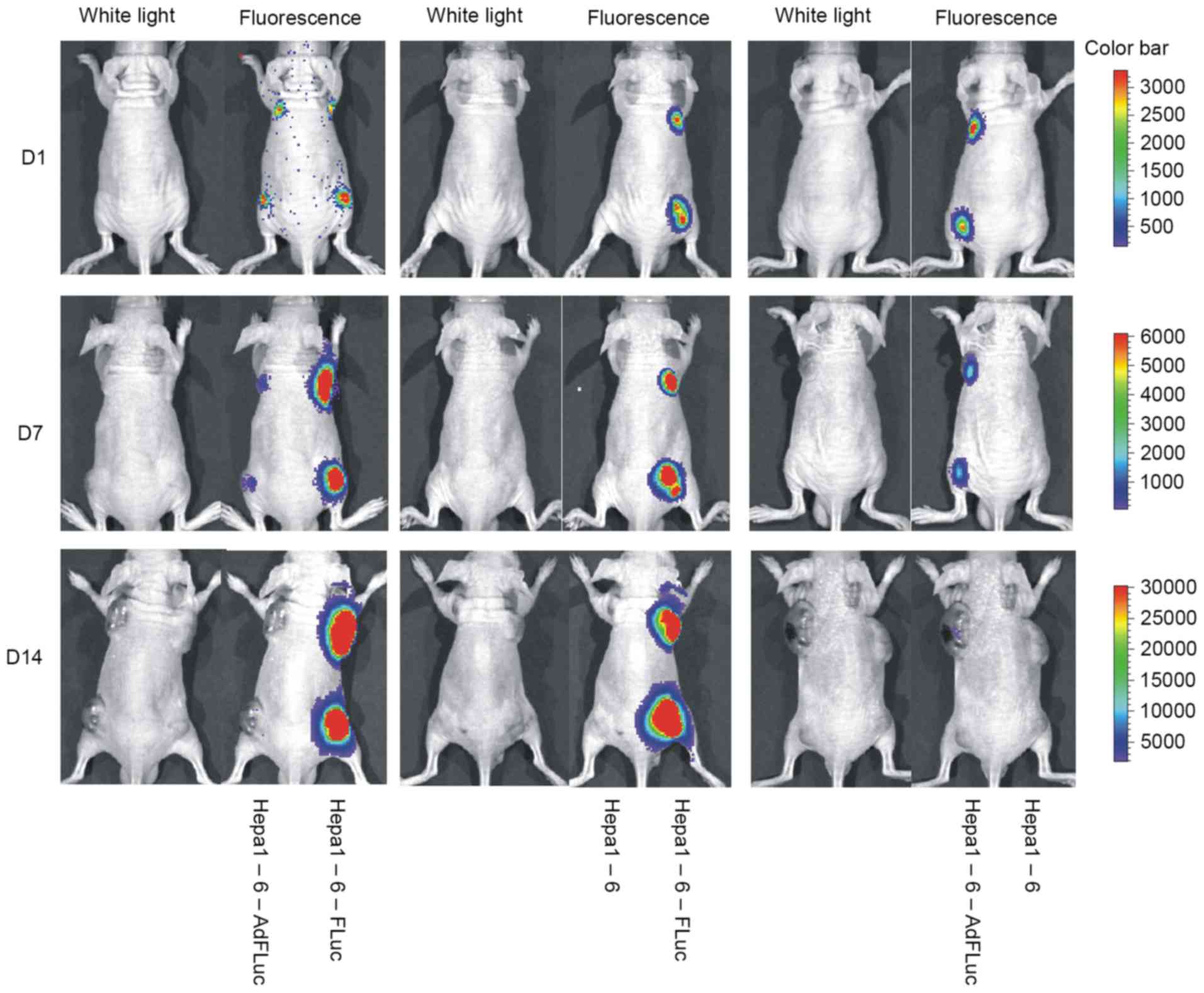
the present study, subcutaneous Hepa1-6 tumors were monitored,
which exhibited stable FLuc expression or temporary
adenovirus-mediated FLuc expression. Hepa1-6, Hepa1-6-FLuc and
Hepa1-6-AdFLuc cells were able to form tumor masses. No signal was
observed at the Hepa1-6 implantation sites, and the signals of
Hepa1-6-FLuc and Hepa1-6-AdFLuc cells were similar at day 1 after
implantation (Fig. 4). With increases
in tumor size, the luciferase signal of the Hepa1-6-FLuc tumors
gradually strengthened, whereas the signal for the Hepa1-6-AdFLuc
cells became weaker and eventually disappeared after four weeks.
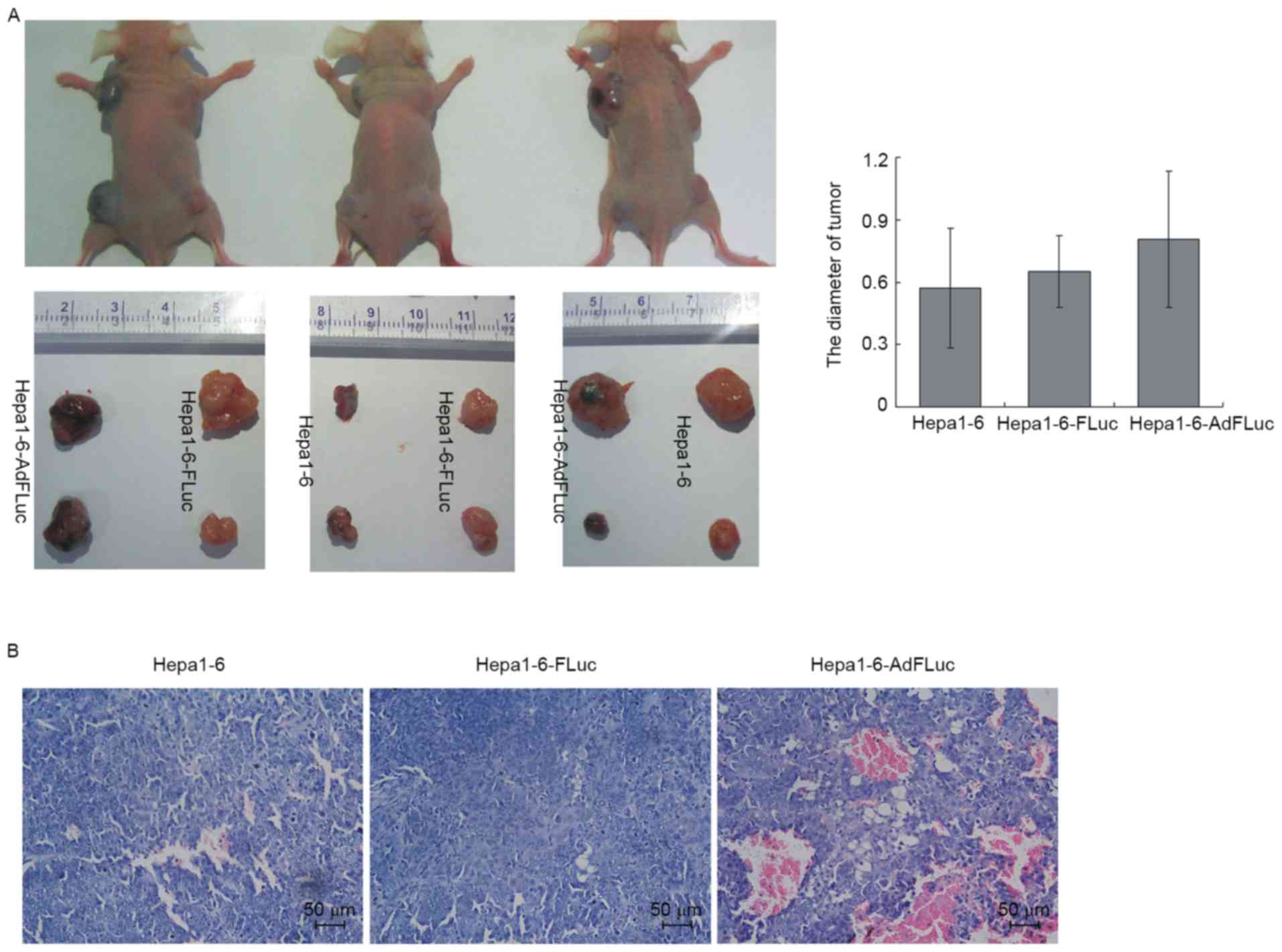
There was no significant difference in tumor sizes observed among
the three groups, although the Hepa1-6 AdFLuc tumors were trending
towards a larger mean size (Fig. 5A;
P>0.05). Based on H&E staining (Fig. 5B), it was identified that the Hepa1-6
and Hepa1-6-FLuc cells exhibited polygonal, irregular shapes and
different cell sizes, and were arranged densely with deeply stained
nuclei, reduced cytoplasm and hyperchromatic mitosis. A number of
cells presented with nuclear pyknosis, deep staining or mitotic
characteristics. The Hepa1-6-AdFLuc cells formed a tumor mass that
exhibited similar cell morphology to the other groups, but with a
substantial amount of neovascularization; a number of vessel walls
were infiltrated, tumor cells appeared in the vascular lumen and
were mixed with blood cells. Therefore, these results suggested
that the stable expression of FLuc in Hepa1-6 cells, and not
transient adenovirus-mediated expression, did not affect its tumor
formation ability in vivo; additionally, this stable
expression could be used to monitor cell proliferation and survival
in vivo using a cell tracing technique.
Discussion
The pathogenesis of hepatocellular carcinoma remains
incompletely understood at the cellular and molecular levels. The
accurate and sensitive evaluation of the effect of therapeutics on
in vivo tumor development is also required, as a tool for
the longitudinal monitoring of tumor proliferation, viability and
metastasis (18–20). Traditional cell tracking methods
usually involve histopathological techniques to observe the labeled
cells in vivo, which require the isolation of samples from
patients or animals at different time-points. Although this method
allows the collection of data from the transplanted cell tumors at
the time of sacrifice, it does not allow the collection of
real-time dynamic information of cell location, viability,
migration, activation and differentiation in vivo (21,22). BLI
is a novel non-invasive technique for obtaining biomedical images
of living tissues at the cellular and molecular levels, which may
be utilized to constantly monitor the physiological, biochemical
and pathological processes of diseases in vivo. Compared
with other techniques, BLI is preferable because of its
non-invasion, high sensitivity and dynamic monitoring (23–25), and
serves an important role in quantitatively assessing in vivo
tumor cell proliferation and invasion over time. In the study of
pre-clinical oncology, BLI is a versatile and sensitive tool that
is based on the detection of light emission from cells or tissues.
Live animal imaging of small animal tumor models using BLI involves
the production of light by luciferase-expressing cells in the
animal in the presence of substrate (24,26–28).
The FLuc gene has been isolated from the cDNA
library of Photinus pyralis. Luciferases emit light in the
presence of D-luciferin substrate, ATP, magnesium and oxygen, which
is a valuable tool for noninvasively monitoring cells in
vivo (24,29). A number of studies (30–32) have
demonstrated the use of recombinant adenoviruses as a gene delivery
vector to express the FLuc gene in different animal cells. However,
in the present study it was identified that adenovirus medicated
FLuc expression was not long-lasting due to its low and transient
level of transgenic expression, potentially as a result of cellular
immunity. Most importantly, in a successful hepatocarcinoma model,
the transplanted cells should proliferate in vivo and
gradually form a mass, but the signals of the AdFLuc-labeled cells
in the present study were not consistent with the growth of the
tumors. Adenoviruses do not integrate the reporter gene into the
host cell genome, preventing the tracing of the daughter cells
originating from the transplanted tumor cells in vivo
(33,34).
In the present study, a Hepa1-6-FLuc cell line with
FLuc gene expression was constructed by retroviral infection. The
passaged Hepa1-6-FLuc cells stably expressed FLuc activity, and
exhibited similar morphology, proliferation, migration and invasion
characteristics compared with the Hepa1-6 cells. The FLuc gene was
replicated with Hepa1-6-FLuc cell proliferation in vivo
following the implantation in nude mice, therefore
luciferin-mediated BLI traced the implanted cell accurately. No
difference in tumor mass volume between the Hepa1-6-FLuc and
Hepa1-6 cell masses was observed, but the volume of the
AdFLuc-infected Hepa1-6 cell tumor mass was non-significantly
increased compared with that of the Hepa1-6 cell tumors. In
addition, hemorrhage and blood cells were present in the gross
specimens and histological sections of Hepa1-6 AdFLuc tumors,
indicating that adenovirus infection may promote the
neovascularization and development of Hepa1-6 tumors (35). Therefore, compared with
adenovirus-based methods, the retrovirus-mediated stable expression
of exogenous FLuc gene may more accurately label and trace cell
survival and proliferation in vivo.
In conclusion, the present study describes a
hepatocarcinoma cell line that stably expressed the FLuc gene. The
Hepa1-6-FLuc cells exhibited the same cellular characteristics as
the Hepa1-6 progenitor cells, were able to replace Hepa1-6 cells in
the establishment of a hepatocarcinoma animal model, and may be
useful for the future study of tumor pathogenesis and the screening
of novel anticancer drugs for the treatment of hepatocarcinoma.
Acknowledgements
Not applicable.
Funding
The present study was supported by a research grant
from the Natural Science Foundation of Chongqing City (grant no.
csct2016jcyjA0228) and the National Natural Science Foundation of
China (grant no. 81100309).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YL carried out the animal experiments. MNL and JJC
carried out the cell experiments. KY and LZ did pathological
histochemistry and helped to evaluate bioluminescence imaging. YW
and MJG participated in cell culture. YH executed statistical
analyses. TCH and YB designed the research and wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The use and care of animals within the study was
approved by the Institutional Animal Care and Use Committee of the
Children's Hospital of Chongqing Medical University (Chongqing,
China).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang CH, Wey KC, Mo LR, Chang KK, Lin RC
and Kuo JJ: Current trends and recent advances in diagnosis,
therapy, and prevention of hepatocellular carcinoma. Asian Pac J
Cancer Prev. 16:3595–3604. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wallace MC, Preen D, Jeffrey GP and Adams
LA: The evolving epidemiology of hepatocellular carcinoma: A global
perspective. Expert Rev Gastroenterol Hepatol. 9:765–779.
2015.PubMed/NCBI
|
|
3
|
Zhu RX, Seto WK, Lai CL and Yuen MF:
Epidemiology of hepatocellular carcinomain the Asia-pacific region.
Gut Liver. 10:332–339. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu Q, Li N, Zeng X, Han Q, Li F, Yang C,
Lv Y, Zhou Z and Liu Z: Hepatocellular carcinoma in a large medical
center of China over a 10-year period: Evolving therapeutic option
and improving survival. Oncotarget. 6:4440–4450. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao GJ, Xu LX, Chu ES, Zhang N, Shen JY,
Damirin A and Li XX: Establishment of an orthotopic transplantation
tumor model of hepatocellular carcinoma in mice. World J
Gastroenterol. 18:7087–7092. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Heindryckx F, Colle I and Van Vlierberghe
H: Experimental mouse models for hepatocellular carcinoma research.
Int J Exp Pathol. 90:367–386. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kircher MF, Gambhir SS and Grimm J:
Noninvasive cell-tracking methods. Nat Rev Clin Oncol. 8:677–688.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Masuda H, Okano HJ, Maruyama T, Yoshimura
Y, Okano H and Matsuzaki Y: In vivo imaging in humanized mice. Curr
Top Microbiol Immunol. 324:179–196. 2008.PubMed/NCBI
|
|
9
|
Kim JE, Kalimuthu S and Ahn BC: In vivo
cell tracking with bioluminescence imaging. Nucl Med Mol Imaging.
49:3–10. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Huang NF, Okogbaa J, Babakhanyan A and
Cooke JP: Bioluminescence imaging of stem cell-based therapeutics
for vascular regeneration. Theranostics. 2:346–354. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Madero-Visbal RA, Colon JF, Hernandez IC,
Limaye A, Smith J, Lee CM, Arlen PA, Herrera L and Baker CH:
Bioluminescence imaging correlates with tumor progression in an
orthotopic mouse model of lung cancer. Surg Oncol. 21:23–29. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Q, Luan W, Goz V, Burakoff SJ and
Hiotis SP: Non-invasive in vivo imaging for liver tumour
progression using an orthotopic hepatocellular carcinoma model in
immunocompetent mice. Liver Int. 31:1200–1208. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
He Y, Zhou JW, Xu L, Gong MJ, He TC and Bi
Y: Comparison of proliferation and differentiation potential
between mouse primary hepatocytes and embryonic hepatic progenitor
cells in vitro. Int J Mol Med. 32:476–484. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu J, Yong M, Li J, Dong X, Yu T, Fu X and
Hu L: High level of CFTR expression is associated with tumor
aggression and knockdown of CFTR suppresses proliferation of
ovarian cancer in vitro and in vivo. Oncol Rep. 33:2227–2234. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cui J, Gong M, He Y, Li Q, He T and Bi Y:
All-trans retinoic acid inhibits proliferation, migration, invasion
and induces differentiation of hepa1-6 cells through reversing EMT
in vitro. Int J Oncol. 48:349–357. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bi Y, He Y, Huang J, Su Y, Zhu GH, Wang Y,
Qiao M, Zhang BQ, Zhang H, Wang Z, et al: Functional
characteristics of reversibly immortalized hepatic progenitor cells
derived from mouse embryonic liver. Cell Physiol Biochem.
34:1318–1338. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Henriques C, Henriques-Pons A,
Meuser-Batista M, Ribeiro AS and de Souza W: In vivo imaging of
mice infected with bioluminescent Trypanosoma cruzi unveils novel
sites of infection. Parasit Vector. 7:892014. View Article : Google Scholar
|
|
18
|
Chen X, Yin S, Hu C, Chen X, Jiang K, Ye
S, Feng X, Fan S, Xie H, Zhou L and Zheng S: Comparative study of
nanosecond electric fields in vitro and in vivo on hepatocellular
carcinoma indicate macrophage infiltration contribute to tumor
ablation in vivo. PLoS One. 9:e864212014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hossain MA, Kim DH, Jang JY, Kang YJ, Yoon
JH, Moon JO, Chung HY, Kim GY, Choi YH, Copple BL and Kim ND:
Aspirin enhances doxorubicin-induced apoptosis and reduces tumor
growth in human hepatocellular carcinoma cells in vitro and in
vivo. Int J Oncol. 40:1636–1642. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kwak MS, Yu SJ, Yoon JH, Lee SH, Lee SM,
Lee JH, Kim YJ, Lee HS and Kim CY: Synergistic anti-tumor efficacy
of doxorubicin and flavopiridol in an in vivo hepatocellular
carcinoma model. J Cancer Res Clin Oncol. 141:2037–2345. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gu E, Chen WY, Gu J, Burridge P and Wu JC:
Molecular imaging of stem cells: Tracking survival,
biodistribution, tumorigenicity, and immunogenicity. Theranostics.
2:335–345. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Alam F and Yadav N: Potential applications
of quantum dots in mapping sentinel lymph node and detection of
micrometastases in breast carcinoma. J Breast Cancer. 16:1–11.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heffern MC, Park HM, Au-Yeung HY, Van de
Bittner GC, Ackerman CM, Stahl A and Chang CJ: In vivo
bioluminescence imaging reveals copper deficiency in a murine model
of nonalcoholic fatty liver disease. Proc Natl Acad Sci USA.
113:pp. 14219–14224. 2016; View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun A, Hou L, Prugpichailers T, Dunkel J,
Kalani MA, Chen X, Kalani MY and Tse V: Firefly luciferase-based
dynamic bioluminescence imaging: A noninvasive technique to assess
tumor angiogenesis. Neurosurgery. 66:751–757. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang R, Zhang K, Tao H, Du W, Wang D,
Huang Z, Zhou M, Xu Y, Wang Y, Liu N, et al: Molecular imaging of
tumor angiogenesis and therapeutic effects with dual
bioluminescence. Curr Pharm Biotechnol. 18:422–428. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Brutkiewicz S, Mendonca M, Stantz K,
Comerford K, Bigsby R, Hutchins G, Goebl M and Harrington M: The
expression level of luciferase within tumour cells can alter tumour
growth upon in vivo bioluminescence imaging. Luminescence.
22:221–228. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mezzanotte L, Fazzina R, Michelini E,
Tonelli R, Pession A, Branchini B and Roda A: In vivo
bioluminescence imaging of murine xenograft cancer models with a
red-shifted thermostable luciferase. Mol Imaging Biol. 12:406–414.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hemmati R, Hosseinkhani S, Sajedi RH, Azad
T, Tashakor A, Bakhtiari N and Ataei F: Luciferin-regenerating
enzyme mediates firefly luciferase activation through direct
effects of D-cysteine on luciferase structure and activity.
Photochem Photobiol. 91:828–836. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu JC, Sundaresan G, Iyer M and Gambhir
SS: Noninvasive optical imaging of firefly luciferase reporter gene
expression in skeletal muscles of living mice. Mol Ther. 4:297–306.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang F, Wang Z, Tian H, Qi M, Zhai Z, Li
S, Li R, Zhang H, Wang W, Fu S, et al: Biodistribution and safety
assessment of bladder cancer specific recombinant oncolytic
adenovirus in subcutaneous xenografts tumor model in nude mice.
Curr Gene Ther. 12:67–76. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cao L, Zeng Q, Xu C, Shi S, Zhang Z and
Sun X: Enhanced antitumor response mediated by the codelivery of
paclitaxel and adenoviral vector expressing IL-12. Mol Pharm.
10:1804–1814. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Man K, Ng KT, Xu A, Cheng Q, Lo CM, Xiao
JW, Sun BS, Lim ZX, Cheung JS, Wu EX, et al: Suppression of liver
tumor growth and metastasis by adiponectin in nude mice through
inhibition of tumor angiogenesis and downregulation of Rho
kinase/IFN-inducible protein 10/matrix metalloproteinase 9
signaling. Clin Cancer Res. 16:967–977. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Volpers C and Kochanek S: Adenoviral
vectors for gene transfer and therapy. J Gene Med. 6 Suppl
1:S164–S171. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hall K, Blair Zajdel ME and Blair GE:
Unity and diversity in the human adenoviruses: Exploiting
alternative entry pathways for gene therapy. Biochem J.
431:321–336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Suzuki K, Sun R, Origuchi M, Kanehira M,
Takahata T, Itoh J, Umezawa A, Kijima H, Fukuda S and Saijo Y:
Mesenchymal stromal cells promote tumor growth through the
enhancement of neovascularization. Mol Med. 17:579–587. 2011.
View Article : Google Scholar : PubMed/NCBI
|