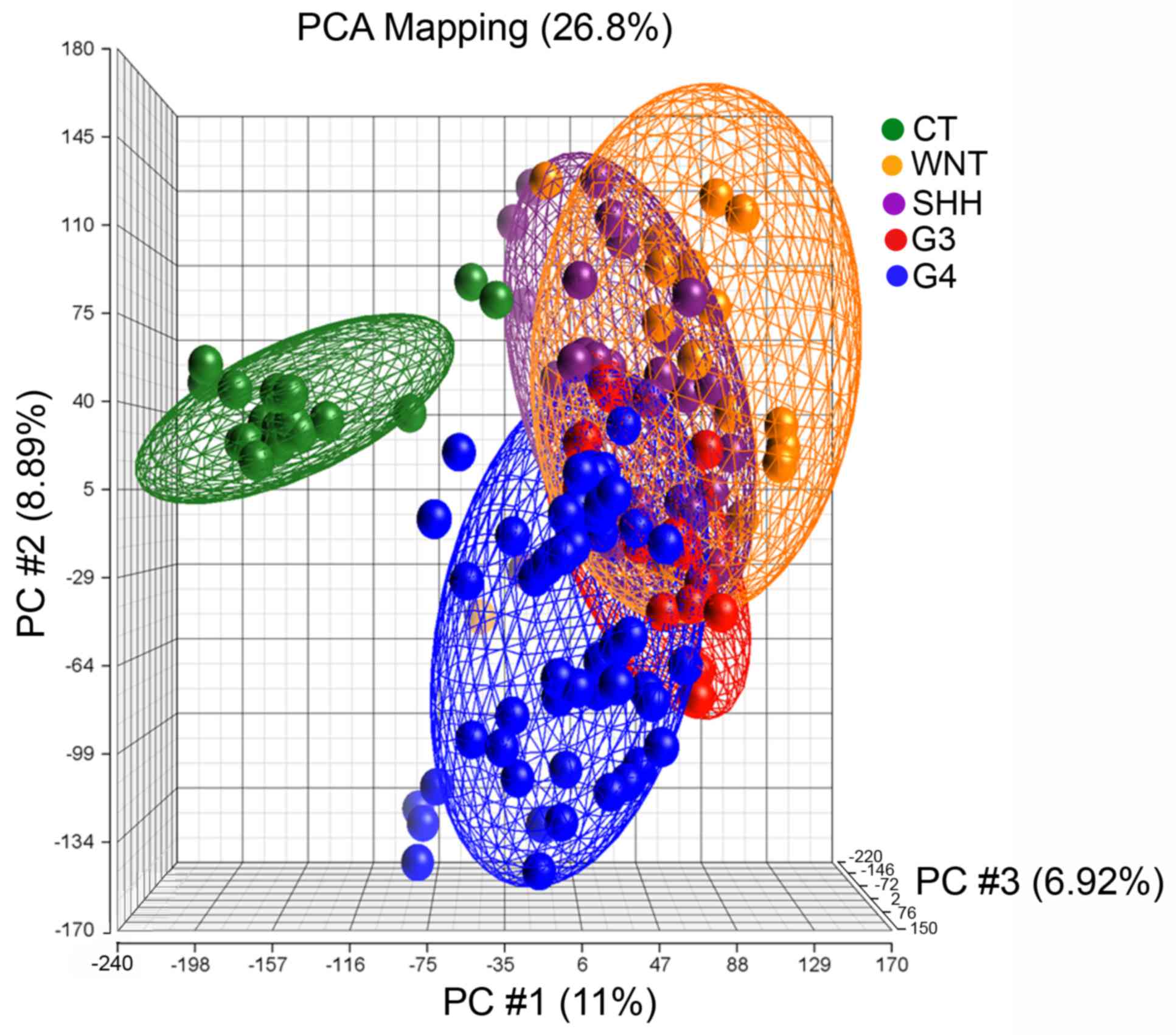
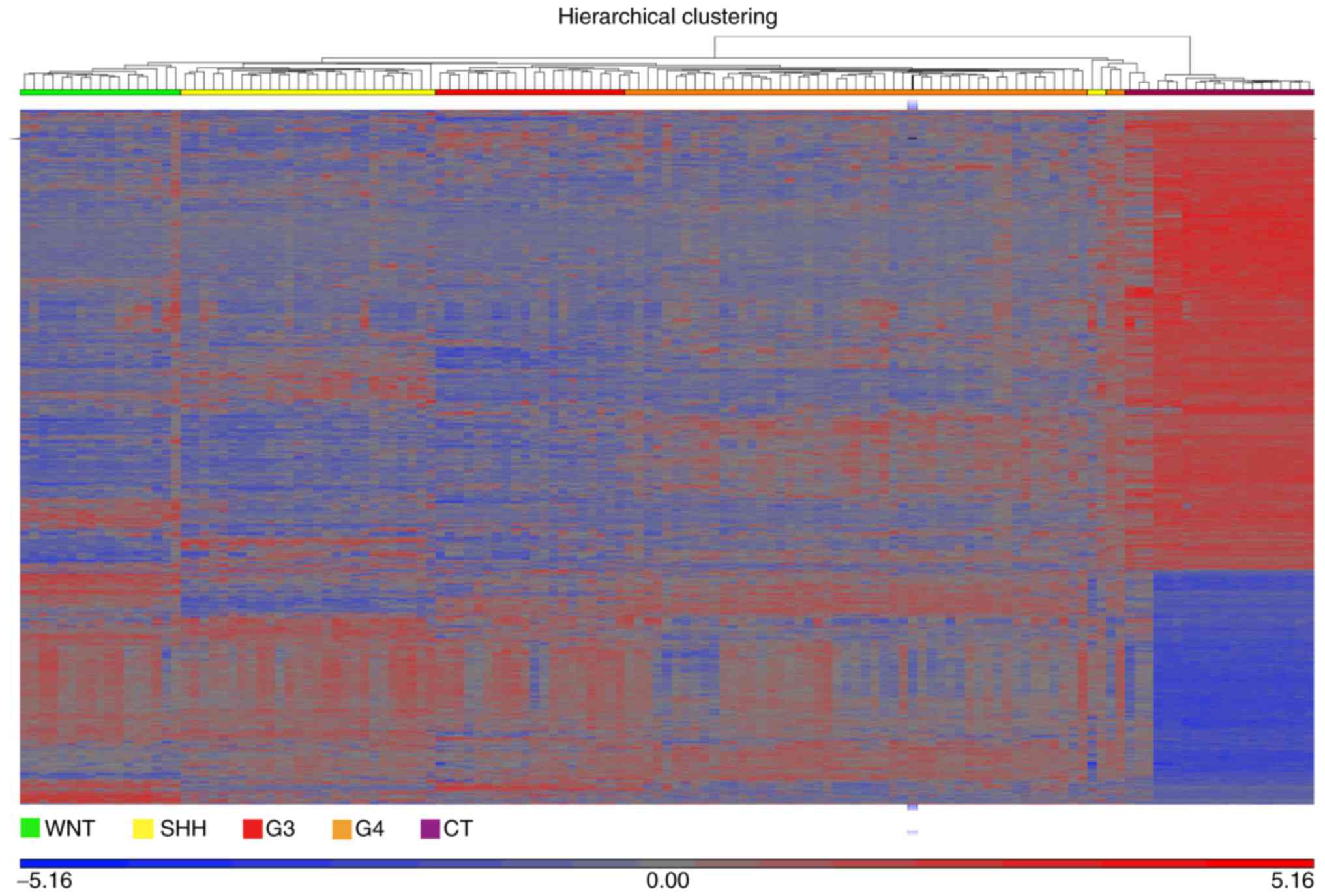
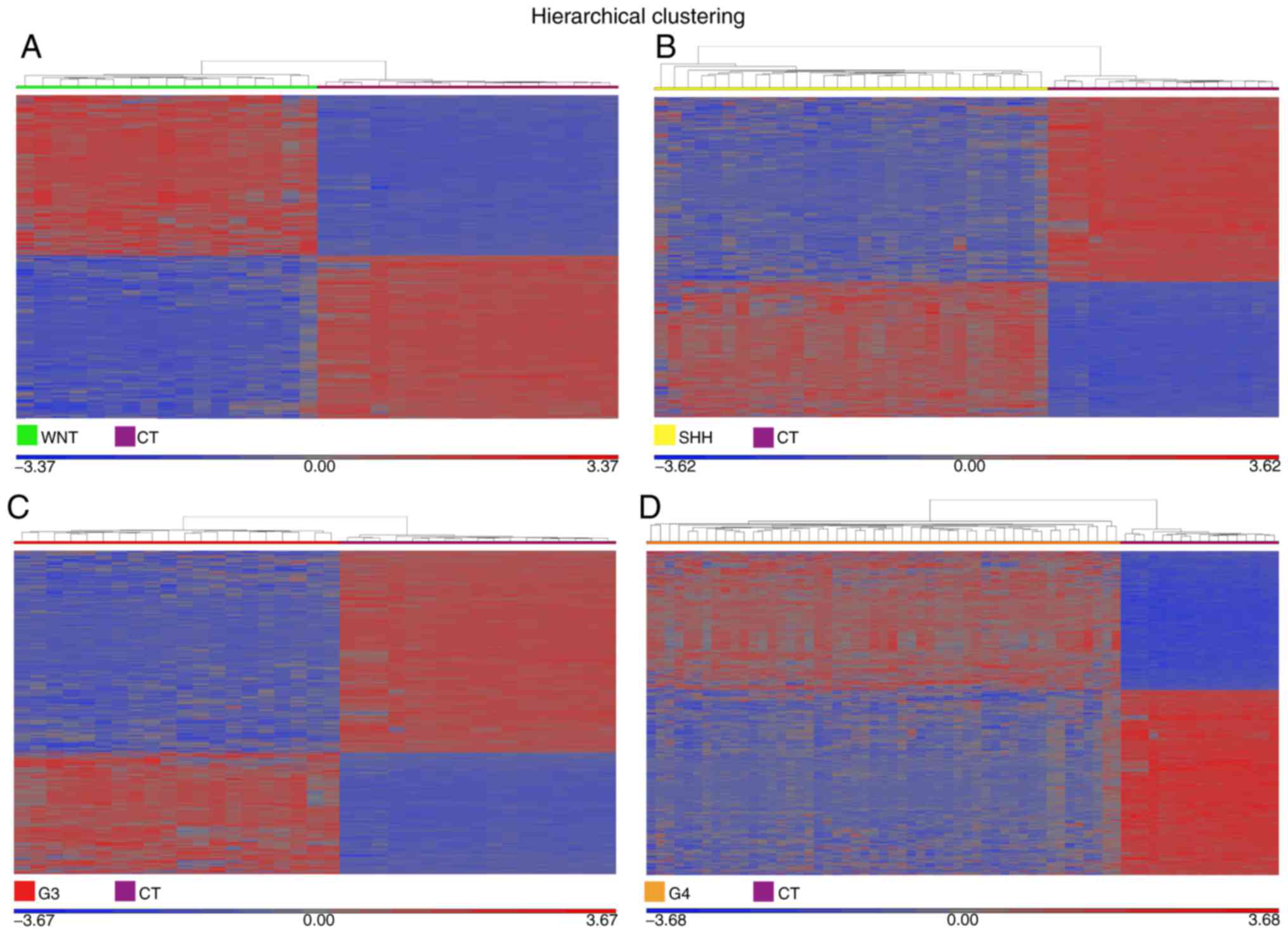
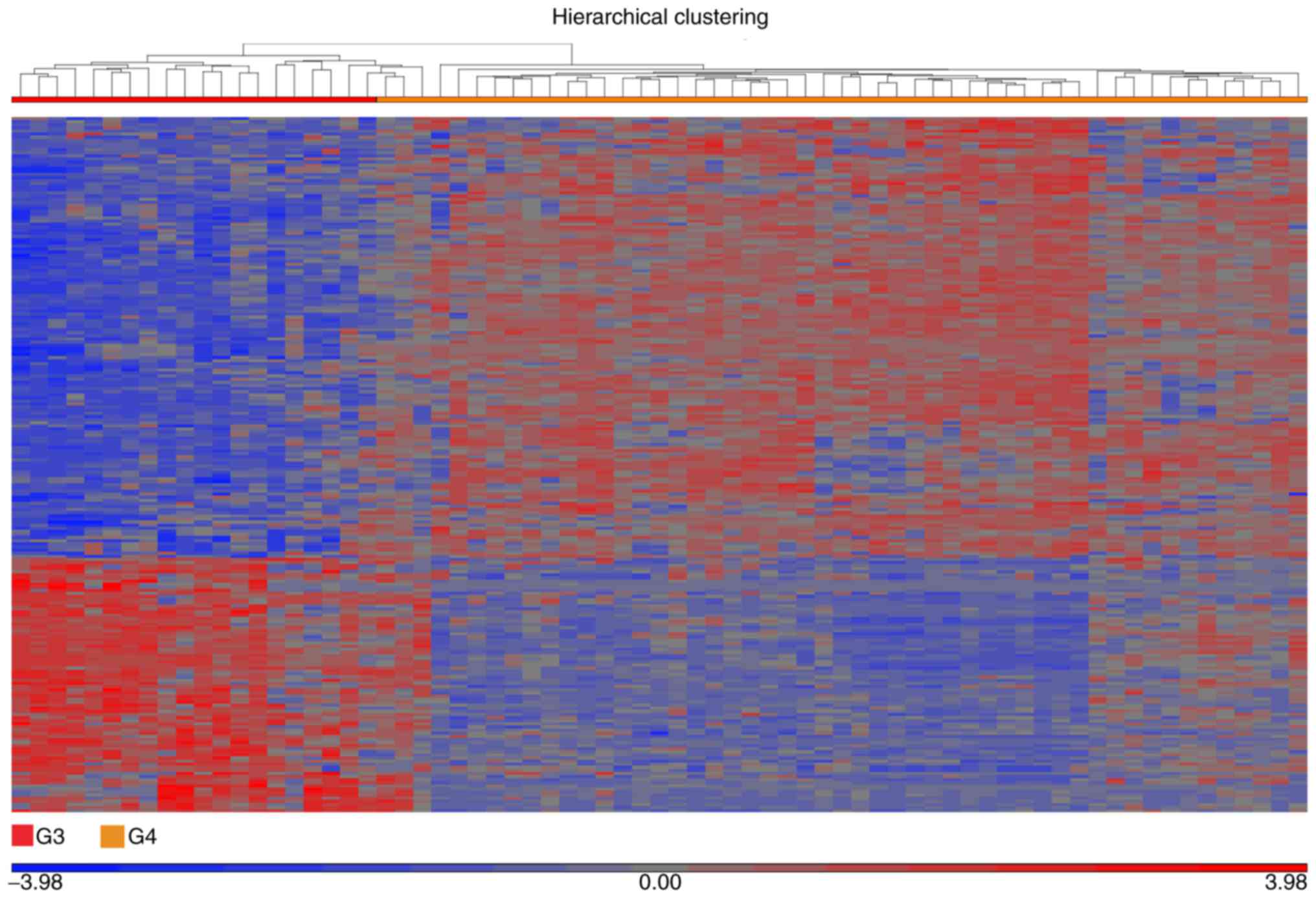
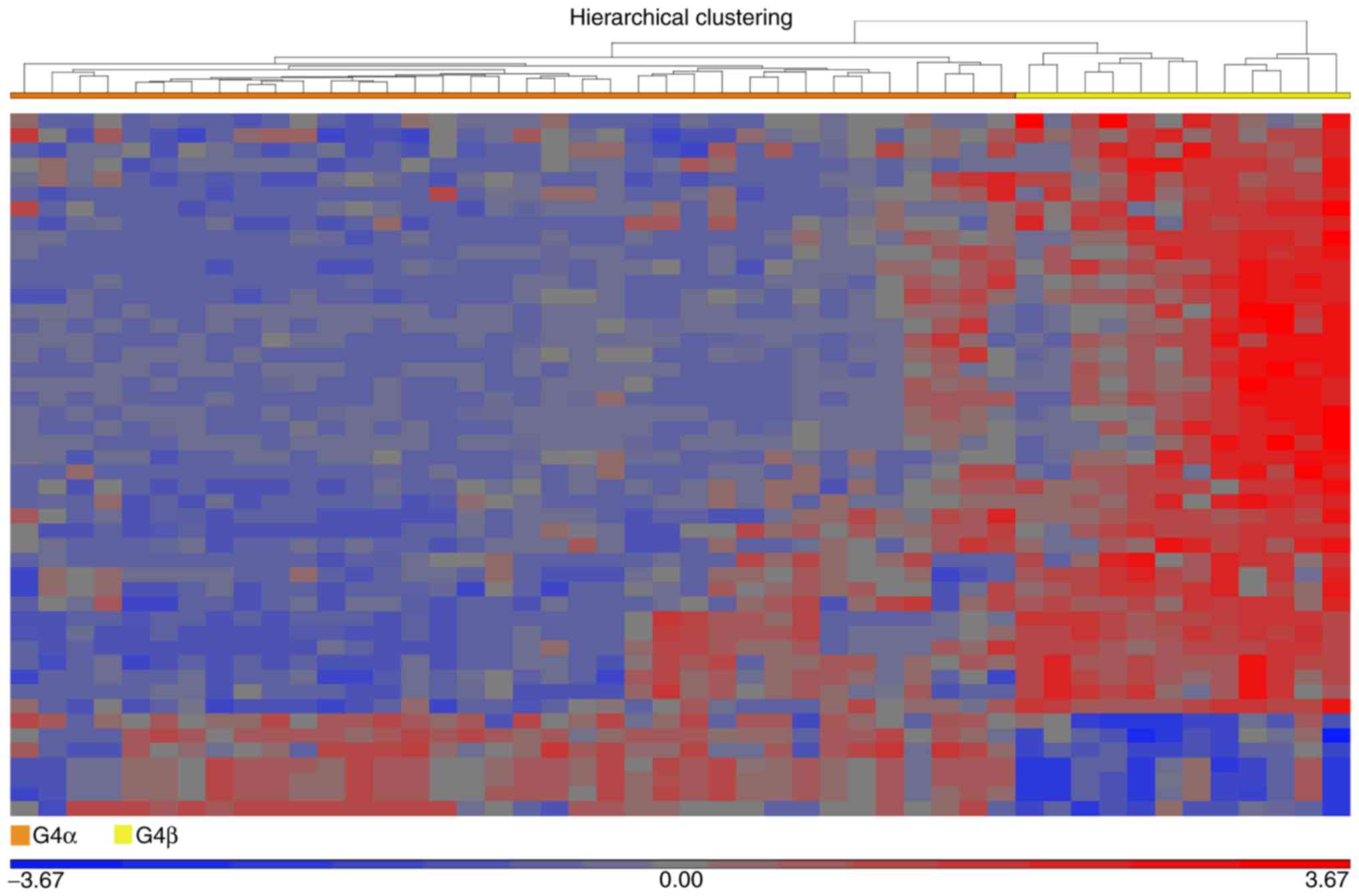
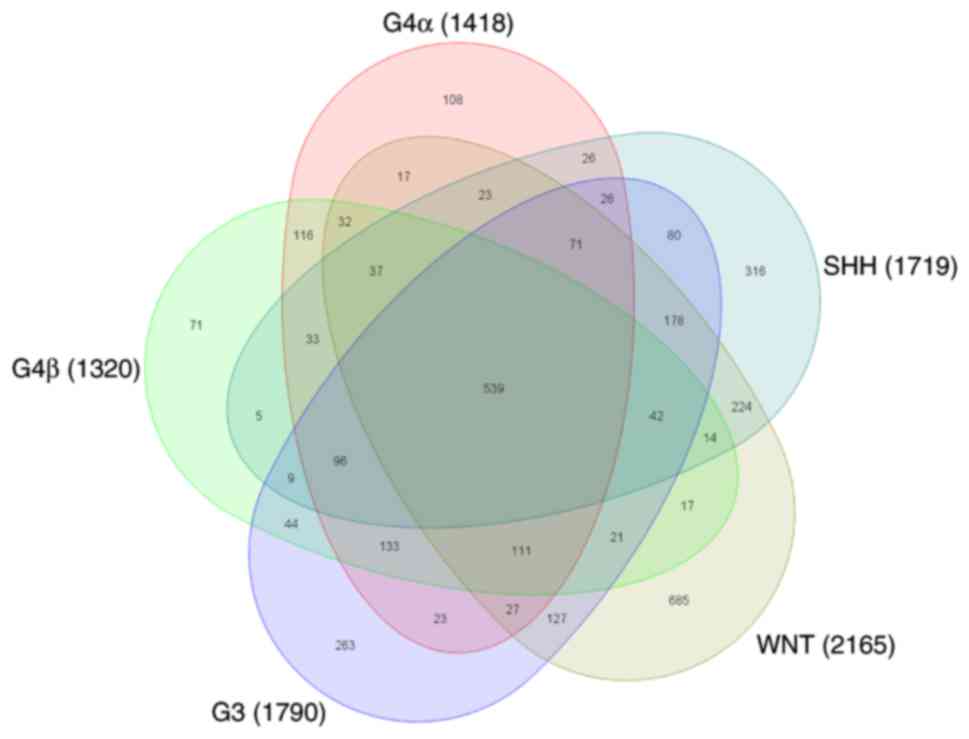
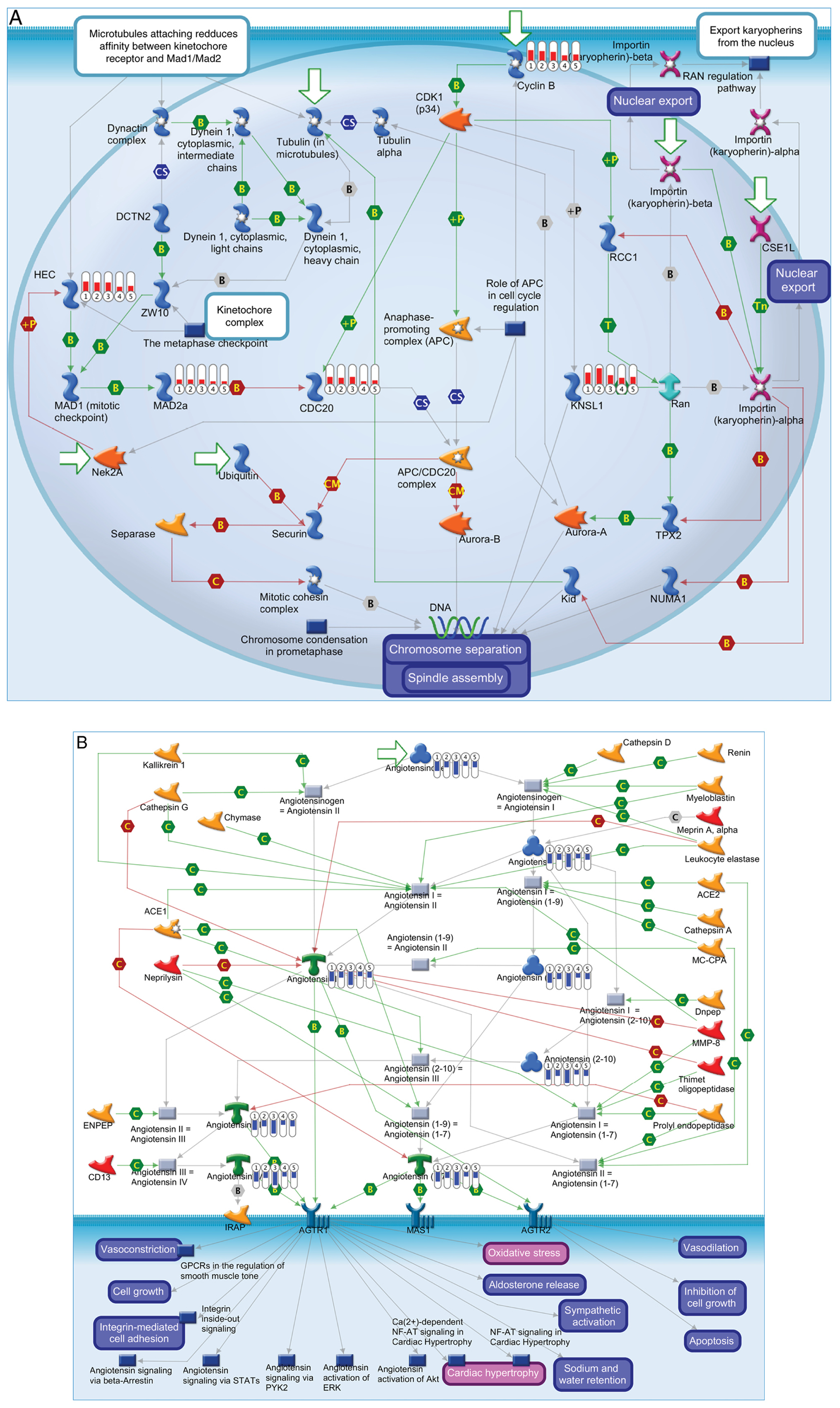
|
1
|
Smoll NR: Relative survival of childhood
and adult medulloblastomas and primitive neuroectodermal tumors
(PNETs). Cancer. 118:1313–1322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Smoll NR and Drummond KJ: The incidence of
medulloblastomas and primitive neurectodermal tumours in adults and
children. J Clin Neurosci. 19:1541–1544. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 world health organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scotting PJ, Walker DA and Perilongo G:
Childhood solid tumours: A developmental disorder. Nat Rev Cancer.
5:481–488. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gilbertson RJ and Ellison DW: The origins
of medulloblastoma subtypes. Annu Rev Pathol. 3:341–365. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Childhood Central Nervous System Embryonal
Tumors Treatment (PDQ®), . Health Professional Version: PDQ
pediatric treatment editorial boardPDQ Cancer Information
Summaries[Internet]. Bethesda (MD): National Cancer Institute (US);
2002
|
|
7
|
Crawford JR, MacDonald TJ and Packer RJ:
Medulloblastoma in childhood: New biological advances. Lancet
Neurol. 6:1073–1085. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Paulino AC, Lobo M, Teh BS, Okcu MF, South
M, Butler EB, Su J and Chintagumpala M: Ototoxicity after
intensity-modulated radiation therapy and cisplatin-based
chemotherapy in children with medulloblastoma. Int J Radiat Oncol
Biol Phys. 78:1445–1450. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kool M, Korshunov A, Remke M, Jones DT,
Schlanstein M, Northcott PA, Cho YJ, Koster J, Schouten-van
Meeteren A, van Vuurden D, et al: Molecular subgroups of
medulloblastoma: An international meta-analysis of transcriptome,
genetic aberrations, and clinical data of WNT, SHH, Group 3, and
Group 4 medulloblastomas. Acta Neuropathol. 123:473–484. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kijima N and Kanemura Y: Molecular
classification of medulloblastoma. Neurol Med Chir (Tokyo).
56:687–697. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Skowron P, Ramaswamy V and Taylor MD:
Genetic and molecular alterations across medulloblastoma subgroups.
J Mol Med (Berl). 93:1075–1084. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang H, Mahler-Araujo BM, Sankila A,
Chimelli L, Yonekawa Y, Kleihues P and Ohgaki H: APC mutations in
sporadic medulloblastomas. Am J Pathol. 156:433–437. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dahmen RP, Koch A, Denkhaus D, Tonn JC,
Sörensen N, Berthold F, Behrens J, Birchmeier W, Wiestler OD and
Pietsch T: Deletions of AXIN1, a component of the WNT/wingless
pathway, in sporadic medulloblastomas. Cancer Res. 61:7039–7043.
2001.PubMed/NCBI
|
|
14
|
Clifford SC, Lusher ME, Lindsey JC,
Langdon JA, Gilbertson RJ, Straughton D and Ellison DW:
Wnt/wingless pathway activation and chromosome 6 loss characterize
a distinct molecular sub-group of medulloblastomas associated with
a favorable prognosis. Cell cycle. 5:2666–2670. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Evans DG, Farndon PA, Burnell LD,
Gattamaneni HR and Birch JM: The incidence of gorlin syndrome in
173 consecutive cases of medulloblastoma. Br J Cancer. 64:959–961.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fujii K and Miyashita T: Gorlin syndrome
(nevoid basal cell carcinoma syndrome): Update and literature
review. Pediatr Int. 56:667–674. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He X, Zhang L, Chen Y, Remke M, Shih D, Lu
F, Wang H, Deng Y, Yu Y, Xia Y, et al: The G protein α subunit Gαs
is a tumor suppressor in sonic hedgehog-driven medulloblastoma. Nat
Med. 20:1035–1042. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ellison DW, Dalton J, Kocak M, Nicholson
SL, Fraga C, Neale G, Kenney AM, Brat DJ, Perry A, Yong WH, et al:
Medulloblastoma: Clinicopathological correlates of SHH, WNT, and
non-SHH/WNT molecular subgroups. Acta Neuropathol. 121:381–396.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kool M, Koster J, Bunt J, Hasselt NE,
Lakeman A, van Sluis P, Troost D, Meeteren NS, Caron HN, Cloos J,
et al: Integrated genomics identifies five medulloblastoma subtypes
with distinct genetic profiles, pathway signatures and
clinicopathological features. PLoS One. 3:e30882008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pöschl J, Stark S, Neumann P, Gröbner S,
Kawauchi D, Jones DT, Northcott PA, Lichter P, Pfister SM, Kool M
and Schüller U: Genomic and transcriptomic analyses match
medulloblastoma mouse models to their human counterparts. Acta
Neuropathol. 128:123–136. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rack PG, Ni J, Payumo AY, Nguyen V,
Crapster JA, Hovestadt V, Kool M, Jones DT, Mich JK, Firestone AJ,
et al: Arhgap36-dependent activation of Gli transcription factors.
Proc Natl Acad Sci USA. 111:pp. 11061–11066. 2014; View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Robinson G, Parker M, Kranenburg TA, Lu C,
Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al: Novel
mutations target distinct subgroups of medulloblastoma. Nature.
488:43–48. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lambert SR, Witt H, Hovestadt V, Zucknick
M, Kool M, Pearson DM, Korshunov A, Ryzhova M, Ichimura K, Jabado
N, et al: Differential expression and methylation of brain
developmental genes define location-specific subsets of pilocytic
astrocytoma. Acta Neuropathol. 126:291–301. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kool M, Jones DT, Jäger N, Northcott PA,
Pugh TJ, Hovestadt V, Piro RM, Esparza LA, Markant SL, Remke M, et
al: Genome sequencing of SHH medulloblastoma predicts
genotype-related response to smoothened inhibition. Cancer Cell.
25:393–405. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Villegas-Ruiz V, Moreno J, Jacome-Lopez K,
Zentella-Dehesa A and Juarez-Mendez S: Quality control usage in
high-density microarrays reveals differential gene expression
profiles in ovarian cancer. Asian Pac J Cancer Prev. 17:2519–2525.
2016.PubMed/NCBI
|
|
26
|
Villegas-Ruiz V and Juarez-Mendez S: Data
mining for identification of molecular targets in ovarian cancer.
Asian Pac J Cancer Prev. 17:1691–1699. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
McCall MN, Murakami PN, Lukk M, Huber W
and Irizarry RA: Assessing affymetrix Genechip microarray quality.
BMC Bioinformatics. 12:1372011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Northcott PA, Korshunov A, Witt H,
Hielscher T, Eberhart CG, Mack S, Bouffet E, Clifford SC, Hawkins
CE, French P, et al: Medulloblastoma comprises four distinct
molecular variants. J Clin Oncol. 29:1408–1414. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li KK, Lau KM and Ng HK: Signaling pathway
and molecular subgroups of medulloblastoma. Int J Clin Exp Pathol.
6:1211–1222. 2013.PubMed/NCBI
|
|
30
|
Thompson MC, Fuller C, Hogg TL, Dalton J,
Finkelstein D, Lau CC, Chintagumpala M, Adesina A, Ashley DM,
Kellie SJ, et al: Genomics identifies medulloblastoma subgroups
that are enriched for specific genetic alterations. J Clin Oncol.
24:1924–1931. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cho YJ, Tsherniak A, Tamayo P, Santagata
S, Ligon A, Greulich H, Berhoukim R, Amani V, Goumnerova L,
Eberhart CG, et al: Integrative genomic analysis of medulloblastoma
identifies a molecular subgroup that drives poor clinical outcome.
J Clin Oncol. 29:1424–1430. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hooper CM, Hawes SM, Kees UR, Gottardo NG
and Dallas PB: Gene expression analyses of the spatio-temporal
relationships of human medulloblastoma subgroups during early human
neurogenesis. PLoS One. 9:e1129092014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dittmer S, Kovacs Z, Yuan SH, Siszler G,
Kögl M, Summer H, Geerts A, Golz S, Shioda T and Methner A: TOX3 is
a neuronal survival factor that induces transcription depending on
the presence of CITED1 or phosphorylated CREB in the
transcriptionally active complex. J Cell Sci. 124(Pt 2): 1–260.
2011.PubMed/NCBI
|
|
34
|
Yuan SH, Qiu Z and Ghosh A: TOX3 regulates
calcium-dependent transcription in neurons. Proc Natl Acad Sci USA.
106:pp. 2909–2914. 2009; View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tessema M, Yingling CM, Grimes MJ, Thomas
CL, Liu Y, Leng S, Joste N and Belinsky SA: Differential epigenetic
regulation of TOX subfamily high mobility group box genes in lung
and breast cancers. PLoS One. 7:e348502012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Morohashi Y, Hatano N, Ohya S, Takikawa R,
Watabiki T, Takasugi N, Imaizumi Y, Tomita T and Iwatsubo T:
Molecular cloning and characterization of CALP/KChIP4, a novel
EF-hand protein interacting with presenilin 2 and voltage-gated
potassium channel subunit Kv4. J Biol Chem. 277:14965–14975. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Northcott PA, Shih DJ, Peacock J, Garzia
L, Morrissy AS, Zichner T, Stütz AM, Korshunov A, Reimand J,
Schumacher SE, et al: Subgroup-specific structural variation across
1,000 medulloblastoma genomes. Nature. 488:49–56. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Engelender S, Kaminsky Z, Guo X, Sharp AH,
Amaravi RK, Kleiderlein JJ, Margolis RL, Troncoso JC, Lanahan AA,
Worley PF, et al: Synphilin-1 associates with alpha-synuclein and
promotes the formation of cytosolic inclusions. Nat Genet.
22:110–114. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rudin CM, Hann CL, Laterra J, Yauch RL,
Callahan CA, Fu L, Holcomb T, Stinson J, Gould SE, Coleman B, et
al: Treatment of medulloblastoma with hedgehog pathway inhibitor
GDC-0449. N Engl J Med. 361:1173–1178. 2009. View Article : Google Scholar : PubMed/NCBI
|