Introduction
Gastric cancer is the third most common cause of
cancer-associated mortalities in the world (1,2). The
treatment methods for gastric cancer primarily include surgery,
radiotherapy and chemotherapy (3,4). However,
many patients with gastric cancer often present with advanced
stages of the disease at initial diagnosis. Therefore, the overall
therapeutic effect of advanced cancer remains poor (5,6). The
discovery of new biomarkers for earlier stages of gastric cancer
and testing the sensitivity of these biomarkers is urgently
required for early detection of gastric cancer and for improving
therapy.
miRNAs are endogenous small non-coding RNA
molecules, which have critical roles in multiple biological
processes by regulating mRNAs via cleavage or inhibiting
translation. A number of properties of microRNAs (miRNAs/miRs) make
them attractive as potential biomarkers and therapy targets. An
increasing number of studies report that miRNAs have regulatory
roles in a diverse range of biological processes and that aberrant
expression of miRNAs is involved in numerous diseases (7). miRNAs have been reported to act as
oncogenes or tumor suppressors in a variety of types of cancer,
including lung, pancreatic, breast, hepatic cancer and gastric
cancer (8–13). The deregulation of many miRNAs have
been detected in gastric cancer, including miR-125b, miR-124 and
the miR-106b-25 and miR-221–222 clusters (14). Furthermore, several miRNAs have
critical roles in gastric cancer carcinogenesis and the expression
levels of these miRNAs may predict the outcome of patients with
gastric cancer (15). Consequently,
miRNAs may be important biomarkers for the diagnosis and prevention
of gastric cancer.
miR-142-3p has been reported to be downregulated in
diverse types of cancer, including leukemia, thyroid follicular
carcinomas, cervical cancer, hepatic cancer, glioblastoma,
osteosarcoma and non-small cell lung cancer (16–22).
miR-142-3p has been demonstrated to contribute to carcinogenesis by
regulating cell cycle, cell migration, apoptosis and invasion by
targeting various signaling pathways and targets (19,20,23).
However, aberrant miR-142-3p expression in gastric cancer and the
potential role of miR-142-3p in gastric cancer carcinogenesis are
largely uninvestigated.
In the present study, TaqMan probes were employed to
analyze miR-142-3p expression in 100 pairs of gastric cancer
tissues and the adjacent normal tissues. The results indicated that
miR-142-3p was markedly downregulated in gastric cancer tissues
compared with adjacent non-neoplastic tissues. In addition, a lower
level of miR-142-3p expression in gastric cancer was significantly
associated with higher tumor stages, which indicated its tumor
inhibitory role in gastric cancer carcinogenesis. Furthermore, the
overexpression of miR-142-3p was able to inhibit the proliferation,
invasion and migration of gastric cancer cells, and these effects
may be mediated via downregulating cyclin T2, which is a regulator
of cell cycle.
Materials and methods
Patients and specimens
The human gastric cancer and corresponding adjacent
non-neoplastic tissues were collected from surgical specimens of
100 patients with gastric cancer at the VIP Department, National
Cancer Center, Cancer Hospital, Chinese Academy of Medical Sciences
(Beijing, China) between January 2015 and December 2015. The sex
ratio of the patients and controls was 1:1 and the age range was 30
to 70 years old. Overall staging grouping outlined by AJCC, also
referred to as Roman numeral staging which uses numerals I, II, III
and IV, was used to describe the progression of cancer (24). The present study was approved by the
Ethics Committee of the Department of VIP, National Cancer
Center/Cancer Hospital, Chinese Academy of Medical Sciences. The
tumor and non-cancerous tissues were histologically confirmed by
H&E staining. Briefly, paraffin-embedded tissues were sliced
into 4-µM-thick sections and pretreated at 65°C for 2 h, followed
by deparaffinization using xylene performed twice, for 10 min each.
Subsequently the tissues were rehydrated in absolute alcohol twice,
for 5 min each, followed by 95% ethanol for 2 min and 70% ethanol
for 2 min. The sections were washed briefly in distilled water
twice and stained using Harris hematoxylin solution for 8 min at
room temperature followed by washing in running tap water for 5
min, then differentiating in 1% acid ethanol for 30 sec. Bluing in
0.2% ammonia water or saturated lithium carbonate solution for 30
sec to 1 min at room temperature was conducted followed by washing
in running tap water for 5 min. Finally, the sections were
dehydrated using 95% ethanol, followed by absolute ethanol twice
for 5 min each, followed by xylene twice for 5 min each and mounted
with a xylene based mounting medium. The sections were photographed
under a light microscope (Nikon TE2000, Japan) equipped with a
digital camera. All clinical samples were snapped frozen in liquid
nitrogen immediately and stored at −80°C until RNA extraction.
Cell culture and transfection
The human gastric cancer cell lines HGC-27, MGC-803
and 293T cells (American Type Culture Collection, Manassas, VA,
USA) were cultured in Dulbecco's modified Eagle's medium
(Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS) at 37°C in 5%
CO2. Scrambled control mimic (cat. no. CN-001000-01-05),
miR-142-3p mimics (cat. no. MIMAT0000434), cyclin T2 (CCNT2)
small-interfering siRNA (cat. no. L-003221-00-0005) and siRNA
control (cat. no. D-001810-01-05) were purchased from GE Healthcare
Dharmacon, Inc., (Lafayette, CO, USA) and transfected into HGC-27,
MGC-803 and 293T cells at 50 nM using DharmFECT1 (GE Healthcare
Dharmacon, Inc.).
RNA extraction, cDNA synthesis and
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) assays
Total RNA was extracted using Trizol from human
tissues and gastric cancer cells according to the manufacturer's
instructions. cDNA was synthesized using 1–5 µg total RNA and a
high-capacity cDNA reverse transcription kit (Thermo Fisher
Scientific, Inc.), according to the manufacturer's instructions.
For detection of miR-142-3p expression, a miR-142-3p specific RT
primer was used for reverse transcription. RT-qPCR using SYBR-Green
qPCR Master mix (Takara Bio, Inc., Otsu, Japan) was performed in a
Bio-Rad CFX96 real-time PCR system using TaqMan probes with U6
small nuclear RNA as an endogenous control. The PCR parameters were
as follows: 95°C for 30 sec, followed by 40 cycles of 95°C for 5
sec and 60°C for 34 sec. The relative expression of miRNAs and
coding genes was calculated using the 2−ΔΔCq method
(25). The primer sequences used in
the present study are listed in Table
I.
 | Table I.Sequence of primers used in reverse
transcription-quantitative polymerase chain reaction and
constructs. |
Table I.
Sequence of primers used in reverse
transcription-quantitative polymerase chain reaction and
constructs.
| Primer | Sequence
(5′-3′) |
|---|
| miR-142-3p |
|
| RT |
GTCGTATCCAGTGCAGGGTCCGAGGTA |
|
|
TTCGCACTGGATACGACTCCATAA |
|
Forward |
CTGTGTAGTGTTTCCTACTTTA |
|
Reverse |
GTGCAGGGTCCGAGGT |
|
Probe |
FAM-GTAGTGTTTCCTACTTTATGG-MGB |
| U6 |
|
| RT |
AAAATATGGAACGCTTCACGAATTTG |
|
Forward |
CTCGCTTCGGCAGCACATATACT |
|
Reverse |
ACGCTTCACGAATTTGCGTGTC |
|
Probe |
FAM-CCATGCTAATCTTCTCTGTA-MGB |
| CCNT2-UTR |
|
|
Forward |
ACCAGATTGGCTGCTGAA |
|
Reverse |
CCCTACAAAGAGGCTCCTAAGT |
Cell proliferation and colony
formation assay
To determine the possible effect of miRNA mimics or
CCNT2 siRNA on cell proliferation, the transfected gastric cancer
cells were incubated with 10% Cell Counting Kit-8 (CCK-8) (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) at 37°C for 1 h and
analyzed at 450 nm. Growth curve of gastric cancer cells were
constructed by detecting absorbance at day 1, 2, 3 and 4
post-transfection. Subsequently, the colony formation ability was
detected. Gastric cancer cells were trypsinized and replated at 200
cells per well in 6-well plates, maintained for a week and then
stained with 0.1% crystal violet in 20% methanol for 15 min. The
number of colonies was counted., and was performed. All the
experiments were repeated three times. The aforementioned control
mimic and siRNA control were used as negative controls for miRNA
mimic and CCNT2 siRNA, respectively.
Cell migration and invasion
assays
To determine the migratory ability of gastric cancer
cells, artificial scratches were created by tips 24 h after
transfection. The images were captured under a light microscope
(Nikon TE2000; Nikon Corporation, Tokyo, Japan) equipped with a
digital camera at 0, 24 and 48 h following the scratches, and the
percentage of open wound area was calculated [(open wound
area/initial wound area) ×100].
For the invasion assay, 1×105 gastric
cancer cells were plated onto a Matrigel-coated Transwell chamber
and inserted in wells of a 24-well plate in serum-free DMEM medium.
In addition, 10% FBS was added below the chamber as a
chemoattractant. After culturing at 37°C for 24 h, the cells that
invaded the lower surface of the chamber were stained with 0.1%
crystal violet for 10 min at room temperature and washed with PBS
and counted under a microscope (Nikon TE2000; Nikon Corporation,)
equipped with a digital camera.
Constructs and luciferase assay
A number of potential targets of miR-142-3p were
predicted using online microRNA target predicting tools: PicTar
(http://www.pictar.org/) and TargetScan
(http://www.targetscan.org/vert_71/).
The 3′untranslated region (UTR) of human CCNT2 containing
miR-142-3p binding sites was inserted downstream of the firefly
luciferase reporter in the pMIR-reporter (Promega Corporation,
Madison, WI, USA). Mutations at the miRNA binding site in the 3′UTR
sequence were created using bridging PCR and then the mutated CCNT2
3′-UTR was also cloned into the pMIR-reporter. For luciferase
assay, 0.4 µg reporter construct, 0.02 µg pRL-TK vector and 5 pmol
miRNA mimic or scrambled controls were co-transfected using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.)
into 293T cells. The cells were harvested and lysed 48 h
post-transfection, the luciferase activity was detected using a
dual luciferase assay (Promega Coporation) in triplicate. The
relative luciferase activity was normalized with Renilla luciferase
activity and a pMIR-reporter containing a complete complementary
sequence to miR-142-3p as positive control.
Western blotting
The whole-cell lysate of MGC-803 cells and HGC-27
cells was extracted with the lysis buffer (0.05 M Tris, pH, 7.5,
0.15 M NaCl and 2% NP-40) containing 200 mM NaF, 200 lm
Na3VO4, 0.5 M EDTA and proteinase inhibitors,
for 30 min on ice and then quantified using a BCA protein assay
kit. 10 µg protein lysates were then separated by 12% SDS-PAGE and
transferred onto a polyvinylidene difluoride membrane for the
detection of CCNT2 and GAPDH. The membraned were blocked in 5% skim
milk. The following antibodies were used at a 1:1,000 dilution:
Anti-CCNT2 (catalog no. 21860-1-AP; ProteinTech Group, Inc.,
Chicago, IL, USA) and anti-GAPDH (catalog no. 10494-1-AP;
ProteinTech Group, Inc.). Horseradish peroxidase-conjugated goat
anti-rabbit IgG secondary antibody (catalog no. SA00001-1;
ProteinTech Group, Inc.) was used at a 1:10,000 dilution and 5%
milk was used as blocking reagent. Immobilon Western HRP substrate
(Merck KGaA, Darmstadt, Germany) was used as a visualization
reagent.
Statistical analysis
Spearman's correlation was used to analyze the
association between clinicopathological characteristics and
miR-142-3p expression. A Student's t-test (two-tailed) was used to
analyze the data. All statistical analysis was conducted using SPSS
19.0 (IBM Corp., Armonk, NY, USA). Data are presented as the mean ±
standard deviation. P≤0.05 was considered to indicate a
statistically significant difference.
Results
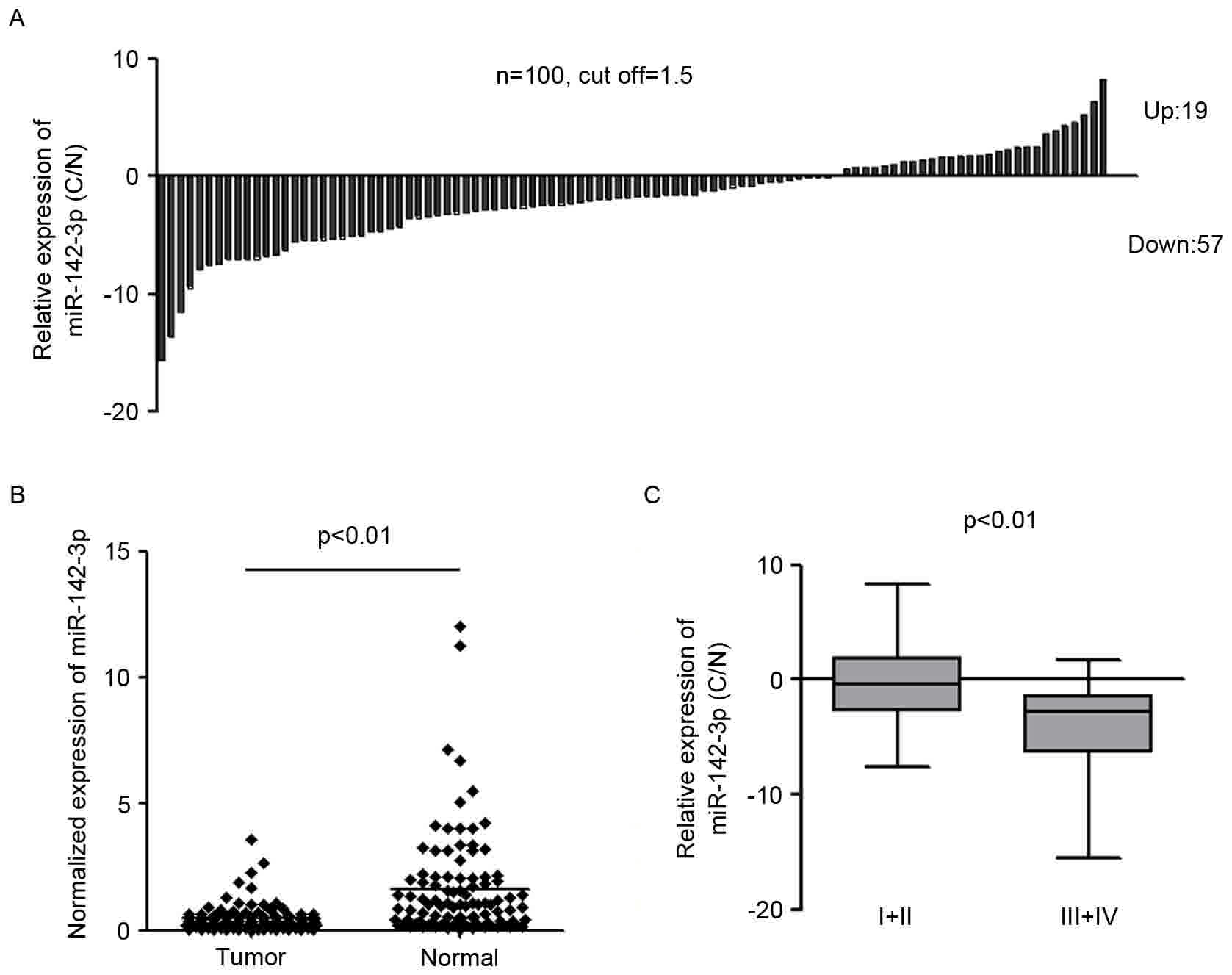
miR-142-3p is significantly
downregulated in gastric cancer tissues
To investigate the expression of miR-142-3p in
gastric cancer, miR-142-3p expression was examined in 100 pairs of
gastric cancer tissues and the adjacent normal tissues by qPCR
using TaqMan probes. The threshold for miR-142-3p expression change
was set as ±1.5. The results indicated that miR-142-3p expression
was decreased in 57% (57/100) of cancer tissues compared with the
adjacent non-neoplastic tissues in patients with gastric cancer
(Fig. 1A). miR-142-3p expression was
upregulated in 19% (19/100) of cancer tissues compared with
corresponding normal tissues in patients with gastric cancer.
24/100 (24%) of patients exhibited no change in miR-142-3p
expression (Fig. 1A). The results
also indicated that miR-142-3p expression in gastric cancer samples
was markedly lower compared with non-neoplastic tissues (P<0.01)
(Fig. 1B). To further investigate the
association between the clinicopathological characteristics and the
expression level of miR-142-3p, the relative expression of
miR-142-3p in 100 pairs of gastric cancer and normal tissues were
analyzed. Clinical correlation analysis by Spearman's correlation
demonstrated that a lower level of miR-142-3p expression in gastric
cancer was associated with higher stages of the disease (stage I/II
vs. III/IV, P<0.01; Fig. 1C). No
significant associations were observed between miR-142-3p
expression and sex, age, position or Borrmann typing (data not
shown). The significant downregulation of miR-142-3p in gastric
cancer tissues and its association with the malignant phenotype
strongly indicated miR-142-3p is able to regulate the development
of gastric cancer.
Overexpression of miR-142-3p is able
to inhibit the growth of gastric cancer cells
To investigate the potential role of miR-142-3p in
gastric cancer carcinogenesis, miR-142-3p was overexpressed in two
gastric cancer cell lines: HGC-27 and MGC-803 (Fig. 2). miR-142-3p was successfully
overexpressed in the two cell lines as detected by qPCR (Fig. 2A and C). CCK-8 assay was used to
determine the cell proliferation rate at day 1, 2, 3 and 4
post-transfection. Cell growth curve indicated that the
overexpression of miR-142-3p in HGC-27 and MGC-803 cell lines was
able to markedly inhibit the proliferation of cancer cells compared
with the scrambled control. (Fig. 2B and
D). Furthermore, the colony formation ability of
miR-142-3p-overexpressed gastric cells was also examined. The
transfected cells were re-plated at a relative low density and
maintained for 7 days to allow the colonies to form. The
overexpression of miR-142-3p was able to significantly decrease the
number of HGC-27 and MGC-803 colonies compared with the scrambled
control. By contrast, the scrambled control exerted little effects
on the number of colonies formed compared with the untreated cells
(Fig. 2E and F). Collectively, the
results suggested that miR-142-3p may perform a tumor suppressive
role in gastric cancer.
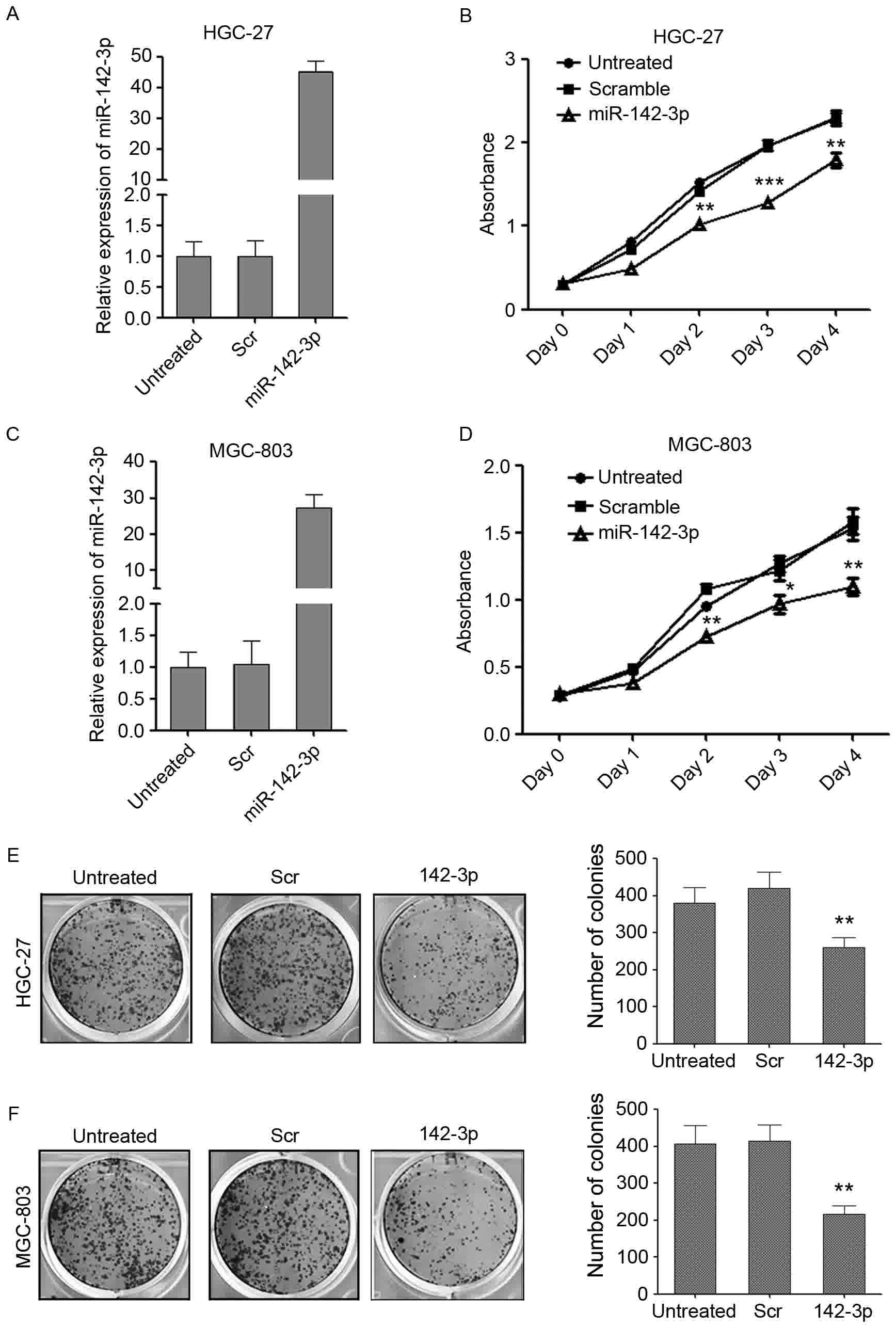 | Figure 2.miR-142-3p is able to suppress the
ability of the gastric cancer cells to proliferate and form
colonies. (A) The enforced expression of miR-142-3p in HGC-27 cells
was confirmed by RT-qPCR. (B) CCK-8 proliferation assays performed
at day 0, 1, 2, 3 and 4 post-transfection of HGC-27 cells. Data are
expressed as the mean ± standard deviation (n=3). (C) The enforced
expression of miR-142-3p in MGC-803 cells was confirmed by RT-qPCR.
(D) CCK-8 proliferation assays performed at day 0, 1, 2, 3 and 4
post-transfection of MGC-803 cells. Data are expressed as the mean
± standard deviation (n=3). The number of colonies/well in 6-well
plates: (E) HGC-27 and (F) MGC-803. The cells were cultured for 7
days. *P<0.05; **P<0.01, ***P<0.001 vs. scrambled control.
CCK-8, Cell Counting Kit-8; miRNA, microRNA; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction. Scr,
scrambled control; 142-3p, miR-142-3p mimic. |
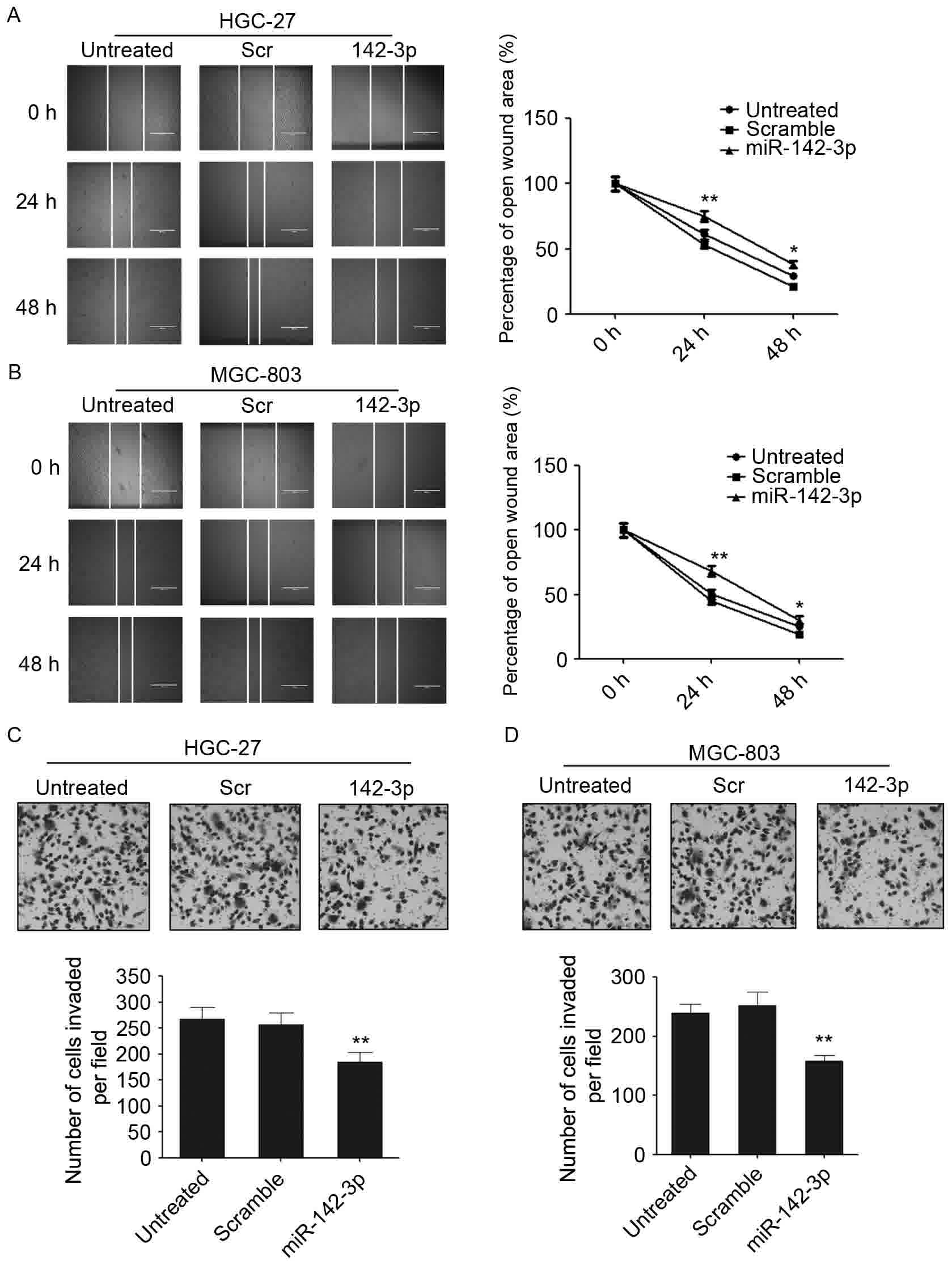
miR-142-3p is able to suppress the
migration and invasion of gastric cancer cells
To investigate whether miR-142-3p regulates the
aggressive properties of gastric cancer cells, the migratory and
invasive abilities of gastric cancer cells that were overexpressed
with miR-142-3p were assessed. A wound-healing/scratch assay was
used to evaluate the role of miR-142-3p in regulating the migration
of gastric cancer cells. The overexpression of miR-142-3p was able
to decrease the migration rate in the two gastric cancer cell lines
(Fig. 3A and B). Wound healing was
markedly decreased in miR-142-3p mimic-transfected gastric cancer
cells at different time points compared with the scrambled control
(Fig. 3A and B). In addition, a
Matrigel cell invasion assay was employed to investigate the
potential effect of miR-142-3p on the invasive ability of gastric
cancer cells. The overexpression of miR-142-3p was able to
significantly decrease the number of invaded cells compared with
the scrambled control and untreated cells (Fig. 3C and D). These results suggested that
miR-142-3p is able to regulate the migration and invasion of
gastric cancer cells.
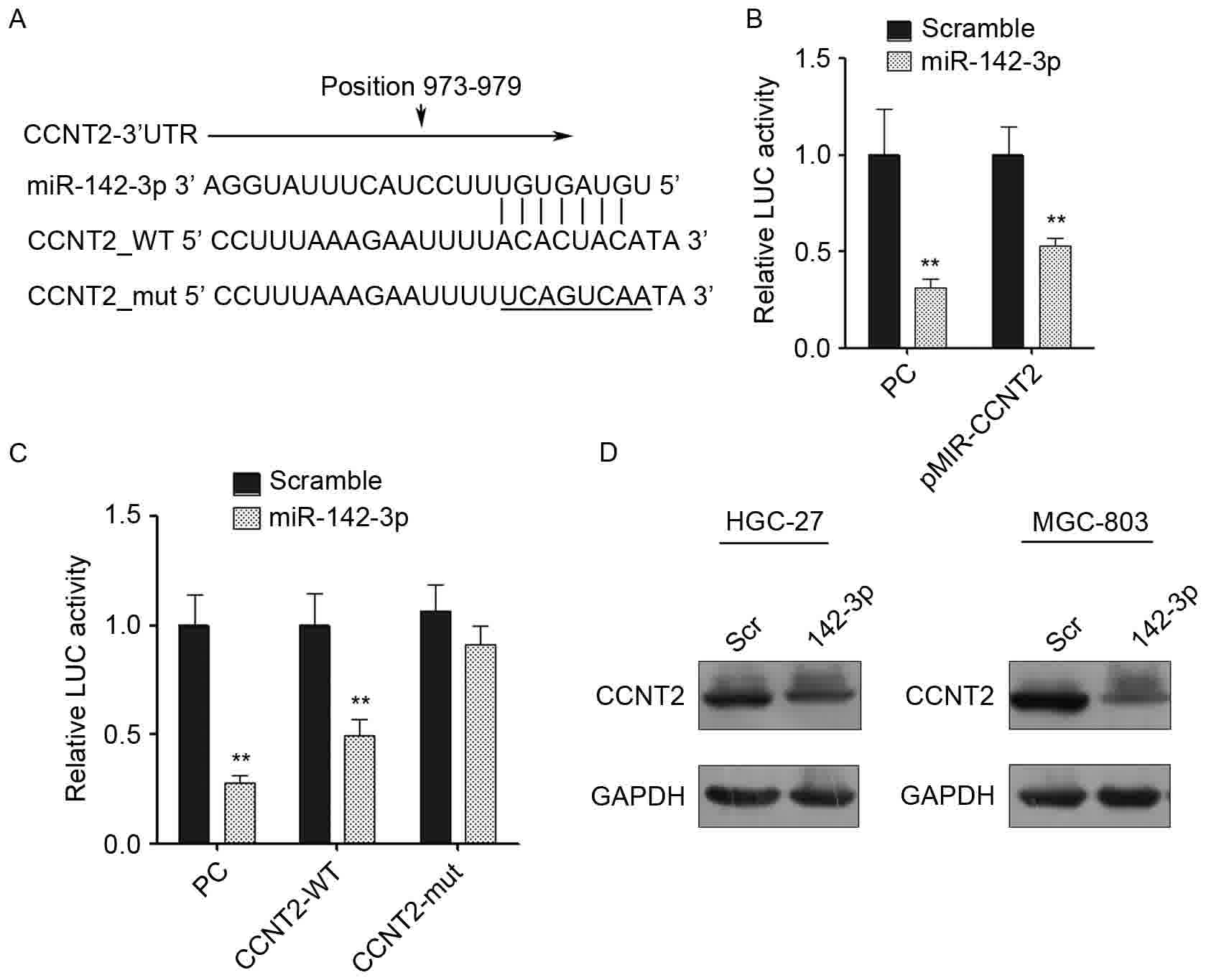
CCNT2 is a direct target of miR-142-3p
in gastric cancer cells
The mechanism by which miR-142-3p regulates the
growth and migration of gastric cancer cells was further examined.
A number of potential targets of miR-142-3p were predicted using
PicTar and TargetScan.
In particular, CCNT2 was identified as a potential
candidate target. The 3′-UTR of CCNT2 contains a sequence that
matches with the ‘seed’ sequence of miR-142-3p (Fig. 4A). CCNT2 is a member of highly
conserved cyclin family, which regulates cyclin-dependent kinase
(CDK) kinase activity, and it is expressed periodically during cell
cycle. CCNT2 and its kinase partner CDK9 were identified to be
components of the transcription elongation factor complex, p-TEFb,
which was reported to facilitate transcription through
phosphorylating the C-terminus of the large subunit of RNA
polymerase II (26–28).
To validate whether miR-124-3p is able to regulate
CCNT2, the 3′-UTR of CCNT2 (containing the miR-124-3p binding site)
was cloned into a luciferase reporter construct (pMIR-reporter).
The complete complementary sequence of miR-142-3p was also cloned
into the reporter and used as a positive control. The luciferase
activity of wild-type CCNT2 3′-UTR was downregulated by 50% when
miR-142-3p mimic was transfected compared with the scrambled
control (Fig. 4B). To test whether
the effect was dependent on the predicted miRNA binding site, the
mutated CCNT2 3′-UTR reporter was constructed, where the predicted
miRNA binding site was altered. The transfection of the mutated
CCNT2 3′-UTR reporter was able to eliminate the reduction in
luciferase activity, which suggests that the inhibitory effect of
miR-142-3p on the CCNT2 3′-UTR was dependent on the miRNA binding
site (Fig. 4C). To further validate
that CCNT2 is a real target of miR-142-3p, CCNT2 protein expression
was quantified by western blotting. A marked decrease in the level
of CCNT2 expression was observed in HGC-27 and MGC-803 cells that
were transfected with the miR-142-3p mimic compared with the cells
that were transfected with the scrambled control, which supported
the hypothesis that CCNT2 is a direct target of miR-142-3p
(Fig. 4D).
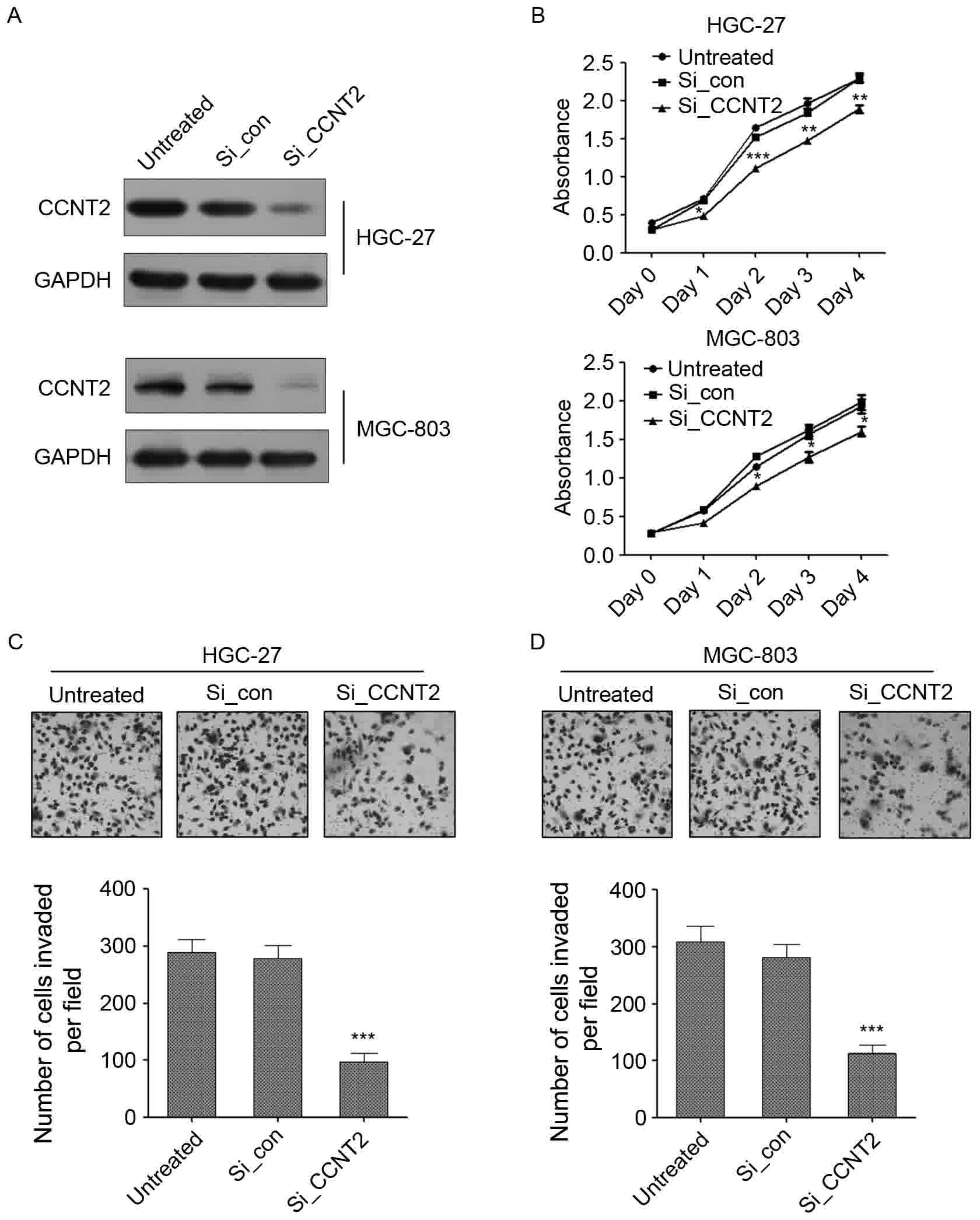
Knock down of CCNT2 suppress the
growth and invasion of gastric cancer cells
The p-TEFb complex, which contains CCNT2 and its
kinase partner CDK9, has been demonstrated to negatively regulate
human immunodeficiency virus type 1 Tat expression (29). CCNT2 was also reported to be able to
inhibit monocytic differentiation by increasing proliferation
(22). However, the role of CCNT2 in
regulating gastric cancer carcinogenesis was largely unknown. To
investigate the potential role of CCNT2 in gastric cancer, CCNT2
expression was knocked down using siRNA in HGC-27 and MGC-803
gastric cancer cell lines. A decreased expression of CCNT2 was
observed in CCNT2 siRNA-transfected gastric cancer cells compared
with cells that were transfected with siRNA control and untreated
cells (Fig. 5A). The knockdown of
CCNT2 was able to significantly attenuate the proliferation of GC
cells in the two gastric cancer cell lines (Fig. 5B). By contrast, transection of the
control siRNA exerted no effects on cell proliferation compared
with untreated cells (Fig. 5B). The
effect of CCNT2 on cell invasion was also examined. CCNT2 knockdown
was able to significantly inhibit the number of invaded cells
compared with the siRNA negative control and untreated cells in
HGC-27 and MGC-803 cell lines (Fig. 5C
and D). These results demonstrated that CCNT2 might act as an
oncogene in gastric cancer and the role of miR-142-3p in regulating
gastric cancer cell proliferation, migration and invasion was
through targeting CCNT2.
Discussion
In the present study, miR-142-3p expression was
examined in patients with gastric cancer, and the potential role
and mechanism of miR-142-3p in regulating GC carcinogenesis was
investigated. The expression profiles of miR-142-3p indicated that
miR-142-3p was significantly downregulated in GC tissues compared
with adjacent non-neoplastic tissues. Furthermore, a lower level of
miR-142-3p expression was associated with higher tumor stages. The
overexpression of miR-142-3p was able to the proliferation,
migration and invasion of gastric cancer cells. These results
suggested that miR-142-3p may act as a tumor suppressor in gastric
cancer, and therefore might be a potential diagnostic marker or
therapeutic target for gastric cancer.
miR-142-3p has been reported to be downregulated in
diverse types of cancer and to contribute to carcinogenesis. For
example, downregulation of miR-142-3p was observed in a large
number of follicular thyroid adenomas and carcinomas, and
miR-142-3p was also reported to function as a tumor suppressor in
follicular thyroid tumorigenesis (16). miR-142-3p has also been reported to be
able to inhibit the proliferation and invasion of cervical cancer
cells by targeting frizzled-7 (30).
Furthermore, lower miR-142-3p expression in hepatic cancer was
associated with poorer survival. miR-142-3p was reported to
negatively regulate CD133, which is a hepatic cancer stem cell
marker: (23,31). In non-small cell lung cancer,
miR-142-3p is able to repress transforming growth factor
(TGF)-β-induced growth inhibition through inhibiting TGFβ receptor
1 (20). In human acute lymphoblastic
leukemia, miR-142-3p was reported to inhibit cell proliferation by
targeting the MLL-AF4 oncogene (21).
In colon cancer cells, miR-142-3p was also reported to function as
a tumor suppressor by targeting CD133, ATP-binding cassette
sub-family G member 2 and leucine-rich repeat-containing G-protein
coupled receptor 5 (23).
However, the aberrant expression of miR-142-3p in
gastric cancer and its potential role in gastric carcinogenesis was
largely unknown; there were few reports investigating the aberrant
expression of miR-142-3p in gastric cancer tissues. Inoue et
al (32) examined the expression
level of several miRNAs in 5 patients with gastric cancer and
observed that miR-142-3p was upregulated in tumor tissues compared
with normal tissues. However, the study used a small number of
clinical samples, which would not reflect the real expression and
regulatory role of miR-142-3p in gastric cancer. In the present
study, TaqMan probe-based qPCR was employed to examine the
expression level of miR-142-3p in 100 pairs of gastric cancer
tissues and the corresponding adjacent normal tissues. miR-142-3p
expression was detected to be significantly decreased in gastric
cancer tissues compared with normal tissues. To the best of our
knowledge, this is the first study to report on the aberrant
downregulation of miR-142-3p in a large number of gastric cancer
tissues. The inhibitory role of miR-142-3p on the proliferation,
migration and invasion of GC cells was also examined, which
supports the view that miR-142-3p has a tumor suppressor role in GC
carcinogenesis.
Investigating the tumor suppressor role of
miR-142-3p in GC in vivo will be the future direction of the
present authors. As a lower expression of miR-142-3p was associated
with a higher tumor stage, the potential therapeutic and diagnostic
roles of miR-142-3p are to be examined further in a larger number
of clinical specimens.
miRNAs regulate gene expression at the
transcriptional and post-transcriptional levels by binding to
target mRNAs and initiating the degradation of target mRNAs or
inducing translational repression (33,34). CCNT2
identified as a direct target of miR-142-3p that regulates gastric
cancer carcinogenesis. CCNT2 is a component of the P-TEFb complex,
which is essential for transcription initiation and elongation that
is mediated by RNA polymerase II. P-TEFb complexes have critical
roles in embryonic development and multiple cellular processes
(35,36). The role of CCNT2 in regulating cell
growth and tumorigenesis are not clear, with the exception of one
study which reported that CCNT2 was able to increase the
proliferation of THP-1 cells and inhibit monocytic differentiation
(22). In the present study, the role
of CCNT2 in regulating the proliferation, migration and invasion of
gastric cancer cells was investigated. The knockdown of CCNT2 was
able to inhibit the growth, migration and invasion of gastric
cancer cells, which suggests that CCNT2 has an oncogenic role in
gastric cancer. Therefore, the tumor-inhibitory effect of
miR-142-3p in GC needs to be validated in vivo and the
detailed mechanism of CCNT2 in promoting GC cell proliferation and
invasion also needs to be explored further.
Taken together, miR-142-3p is able to suppress
growth, migration and invasion of gastric cancer cells by
down-regulating CCNT2. The downregulation of miR-142-3p and the
resulting elevated CCNT2 level may contribute to GC carcinogenesis.
miR-142-3p might be a potential novel diagnostic marker or a target
for the treatment of gastric cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from the
National High-Tech Research and Development Program of China (grant
no. 2015AA020408).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
JC and YW conceived the study. YW designed the
experiments and performed the majority of them. ZC contributed to
the performance of experiments. LW and SL performed cell culture
and collected clinical samples. YW wrote the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committees of Department of VIP, National Cancer Center/Cancer
Hospital, Chinese Academy of Medical Sciences (Beijing, China) and
written informed consent was obtained from all patients.
Consent for publication
Written informed consent was obtained from all
examined patients for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sitarz R, Skierucha M, Mielko J, Offerhaus
GJA, Maciejewski R and Polkowski WP: Gastric cancer: Epidemiology,
prevention, classification, and treatment. Cancer Manag Res.
10:239–248. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fock KM: Review article: The epidemiology
and prevention of gastric cancer. Aliment Pharmacol Ther.
40:250–260. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ang TL and Fock KM: Clinical epidemiology
of gastric cancer. Singapore Med J. 55:621–628. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jang JY and Chun HJ: Efficacy of
Helicobacter pylori eradication for the prevention of metachronous
gastric cancer after endoscopic resection for early gastric cancer.
World J Gastroenterol. 20:2760–2764. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meyer HJ and Wilke H: Treatment strategies
in gastric cancer. Dtsch Arztebl Int. 108:698–705. 2011.PubMed/NCBI
|
|
6
|
Peters MD: Postsurgical chemotherapy vs.
surgery alone for resectable gastric cancer. Am J Nurs. 114:242014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wu WK, Lee CW, Cho CH, Fan D, Wu K, Yu J
and Sung JJ: MicroRNA dysregulation in gastric cancer: A new player
enters the game. Oncogene. 29:5761–5771. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Fang Y, Liu Y and Yang X: MicroRNAs
in ovarian function and disorders. J Ovarian Res. 8:512015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu D, Chen H, Yang X, Chen W, Wang L, Xu
J and Yu L: miR-32 functions as a tumor suppressor and directly
targets SOX9 in human non-small cell lung cancer. Onco Targets
Ther. 8:1773–1783. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner
MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ and Schmittgen
TD: Expression profiling identifies microRNA signature in
pancreatic cancer. Int J Cancer. 120:1046–1054. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Murakami Y, Yasuda T, Saigo K, Urashima T,
Toyoda H, Okanoue T and Shimotohno K: Comprehensive analysis of
microRNA expression patterns in hepatocellular carcinoma and
non-tumorous tissues. Oncogene. 25:2537–2545. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W,
Liu J, Yu J and Chen J: miRNA-96 suppresses KRAS and functions as a
tumor suppressor gene in pancreatic cancer. Cancer Res.
70:6015–6025. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu C, Yu J, Yu S, Lavker RM, Cai L, Liu
W, Yang K, He X and Chen S: MicroRNA-21 acts as an oncomir through
multiple targets in human hepatocellular carcinoma. J Hepatol.
53:98–107. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ventura A and Jacks T: MicroRNAs and
cancer: Short RNAs go a long way. Cell. 136:586–591. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hui A, How C, Ito E and Liu FF: Micro-RNAs
as diagnostic or prognostic markers in human epithelial
malignancies. BMC Cancer. 11:5002011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Colamaio M, Puca F, Ragozzino E, Gemei M,
Decaussin-Petrucci M, Aiello C, Bastos AU, Federico A, Chiappetta
G, Del Vecchio L, et al: miR-142-3p downregulation contributes to
thyroid follicular tumorigenesis by targeting ASH1L and MLL1. J
Clin Endocrinol Metab. 100:E59–E69. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chai S, Tong M, Ng KY, Kwan PS, Chan YP,
Fung TM, Wong N, Xie D, Yuan YF, Guan XY and Ma S: Regulatory role
of miR-142-3p on the functional hepatic cancer stem cell marker
CD133. Oncotarget. 5:5725–5735. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu S, Wei J, Wang F, Kong LY, Ling XY,
Nduom E, Gabrusiewicz K, Doucette T, Yang Y, Yaghi NK, et al:
Effect of miR-142-3p on the M2 macrophage and therapeutic efficacy
against murine glioblastoma. J Natl Cancer Inst.
106:pii:dju1622014. View Article : Google Scholar
|
|
19
|
Xu G, Wang J, Jia Y, Shen F, Han W and
Kang Y: MiR-142-3p functions as a potential tumor suppressor in
human osteosarcoma by targeting HMGA1. Cell Physiol Biochem.
33:1329–1339. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lei Z, Xu G, Wang L, Yang H, Liu X, Zhao J
and Zhang HT: MiR-142-3p represses TGF-β-induced growth inhibition
through repression of TGFβR1 in non-small cell lung cancer. FASEB
J. 28:2696–2704. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dou L, Li J, Zheng D, Li Y, Gao X, Xu C,
Gao L, Wang L and Yu L: MicroRNA-142-3p inhibits cell proliferation
in human acute lymphoblastic leukemia by targeting the MLL-AF4
oncogene. Mol Biol Rep. 40:6811–6819. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang XS, Gong JN, Yu J, Wang F, Zhang XH,
Yin XL, Tan ZQ, Luo ZM, Yang GH, Shen C and Zhang JW: MicroRNA-29a
and microRNA-142-3p are regulators of myeloid differentiation and
acute myeloid leukemia. Blood. 119:4992–5004. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Shen WW, Zeng Z, Zhu WX and Fu GH:
MiR-142-3p functions as a tumor suppressor by targeting CD133,
ABCG2, and Lgr5 in colon cancer cells. J Mol Med (Berl).
91:989–1000. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Carlson RW, Allred DC, Anderson BO,
Burstein HJ, Carter WB, Edge SB, Erban JK, Farrar WB, Goldstein LJ,
Gradishar WJ, et al: Breast cancer. Clinical practice guidelines in
oncology. J Natl Compr Canc Netw. 7:122–192. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
de Falco G and Giordano A: CDK9 (PITALRE):
A multifunctional multifunctional cdc2-related kinase. J Cell
Physiol. 177:501–506. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
De Luca A, Esposito V, Baldi A, Claudio
PP, Fu Y, Caputi M, Pisano MM, Baldi F and Giordano A: CDC2-related
kinase PITALRE phosphorylates pRb exclusively on serine and is
widely expressed in human tissues. J Cell Physiol. 172:265–273.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Simone C, Bagella L, Bellan C and Giordano
A: Physical interaction between pRb and cdk9/cyclinT2 complex.
Oncogene. 21:4158–4165. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Napolitano G, Licciardo P, Gallo P,
Majello B, Giordano A and Lania L: The CDK9-associated cyclins T1
and T2 exert opposite effects on HIV-1 Tat activity. AIDS.
13:1453–1459. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deng B, Zhang Y, Zhang S, Wen F, Miao Y
and Guo K: MicroRNA-142-3p inhibits cell proliferation and invasion
of cervical cancer cells by targeting FZD7. Tumour Biol.
36:8065–8073. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu L, Cai C, Wang X, Liu M, Li X and Tang
H: MicroRNA-142-3p, a new regulator of RAC1, suppresses the
migration and invasion of hepatocellular carcinoma cells. FEBS
Lett. 585:1322–1330. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Inoue T, Iinuma H, Ogawa E, Inaba T and
Fukushima R: Clinicopathological and prognostic significance of
microRNA-107 and its relationship to DICER1 mRNA expression in
gastric cancer. Oncol Rep. 27:1759–1764. 2012.PubMed/NCBI
|
|
33
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Krützfeldt J: Strategies to use microRNAs
as therapeutic targets. Best Pract Res Clin Endocrinol Metab.
30:551–561. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wei P, Gender ME, Fang SM, Fisher WH and
Jones KA: A novel CDK9-associated C-type cyclin interacts directly
with HIV-1 Tat and mediates its high-affinity, loop-specific
binding to TAR RNA. Cell. 92:451–462. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kohoutek J, Li Q, Blazek D, Luo Z, Jiang H
and Peterlin BM: Cyclin T2 is essential for mouse embryogenesis.
Mol Cell Biol. 29:3280–3285. 2009. View Article : Google Scholar : PubMed/NCBI
|