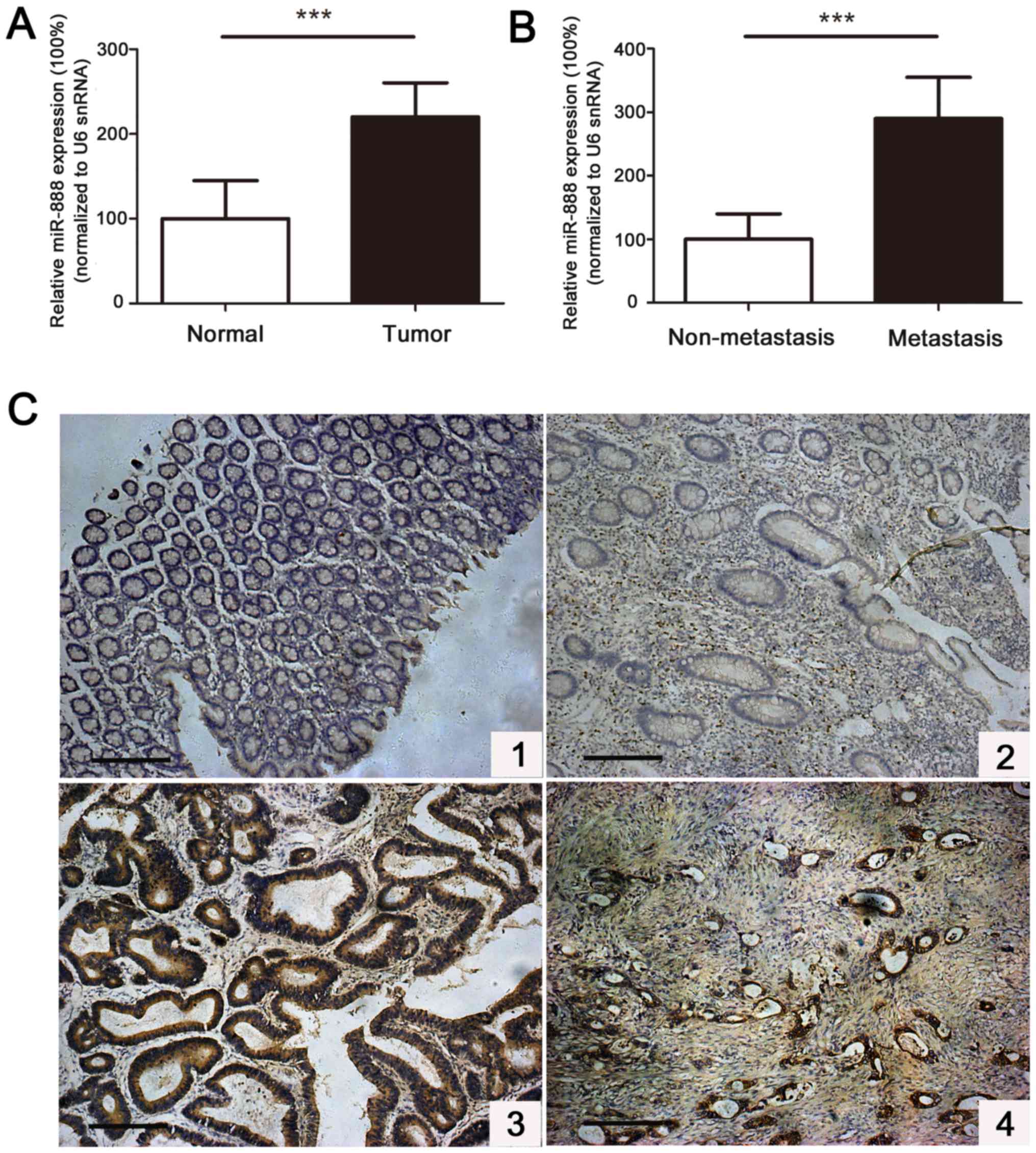
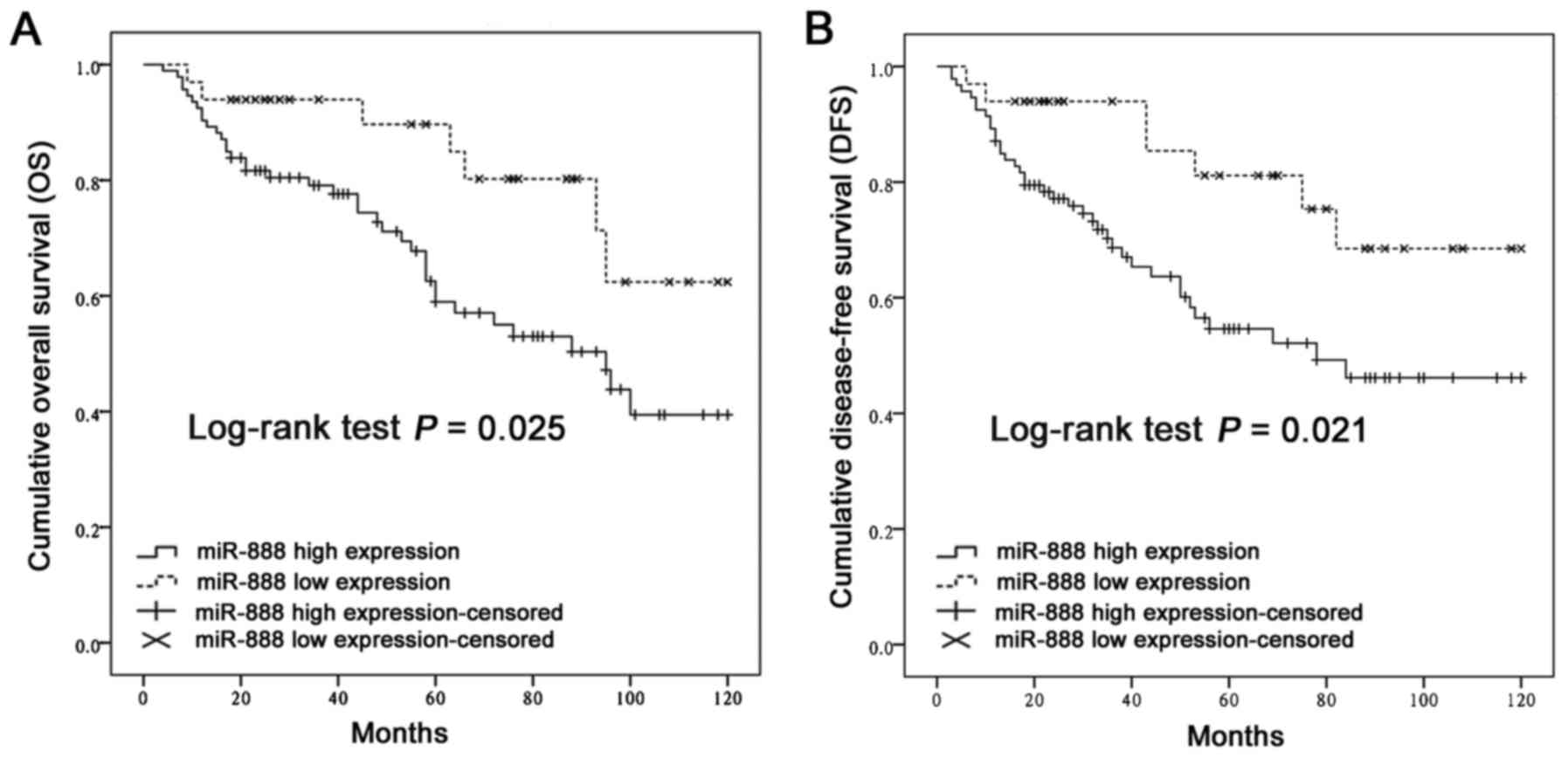
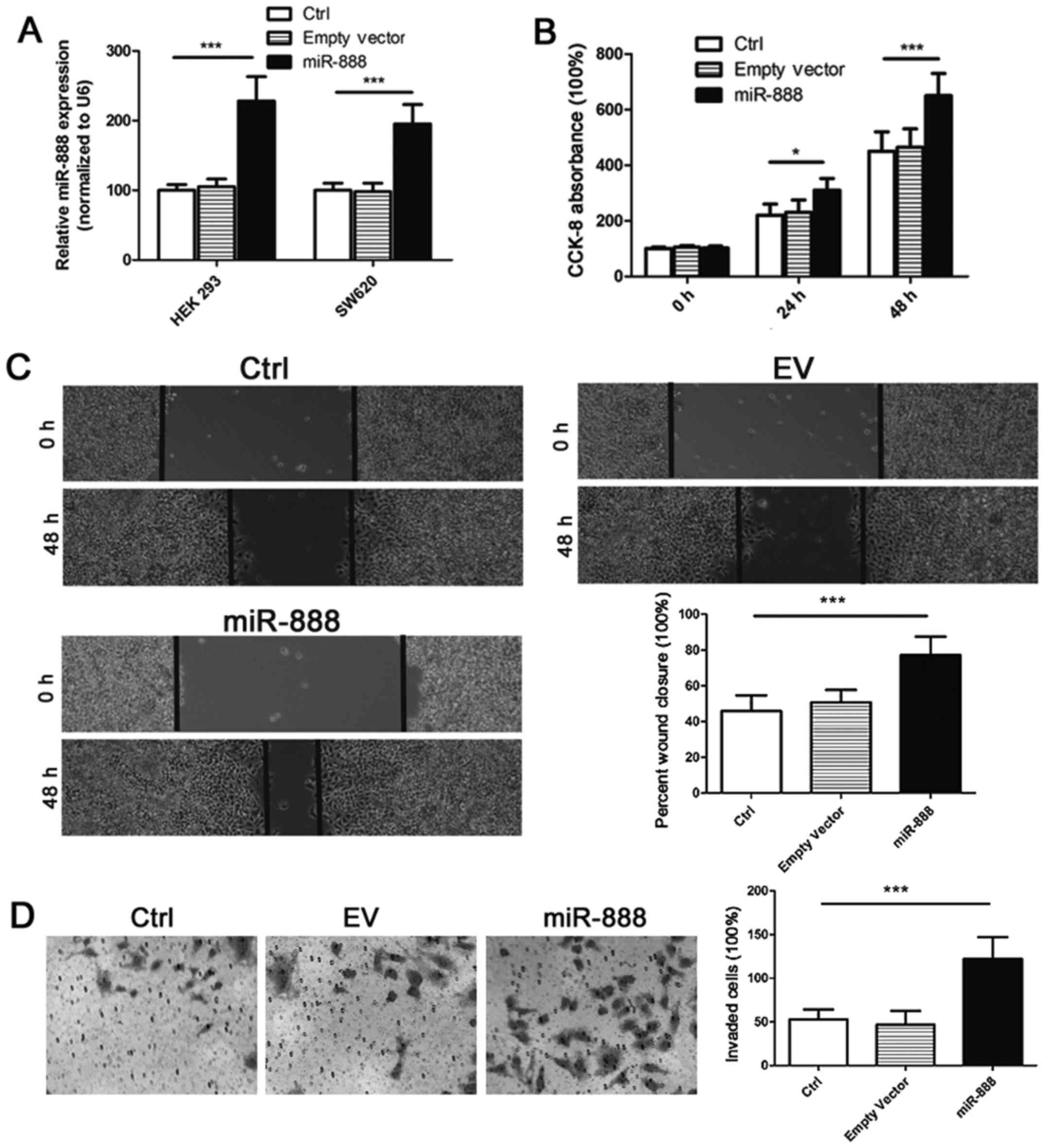
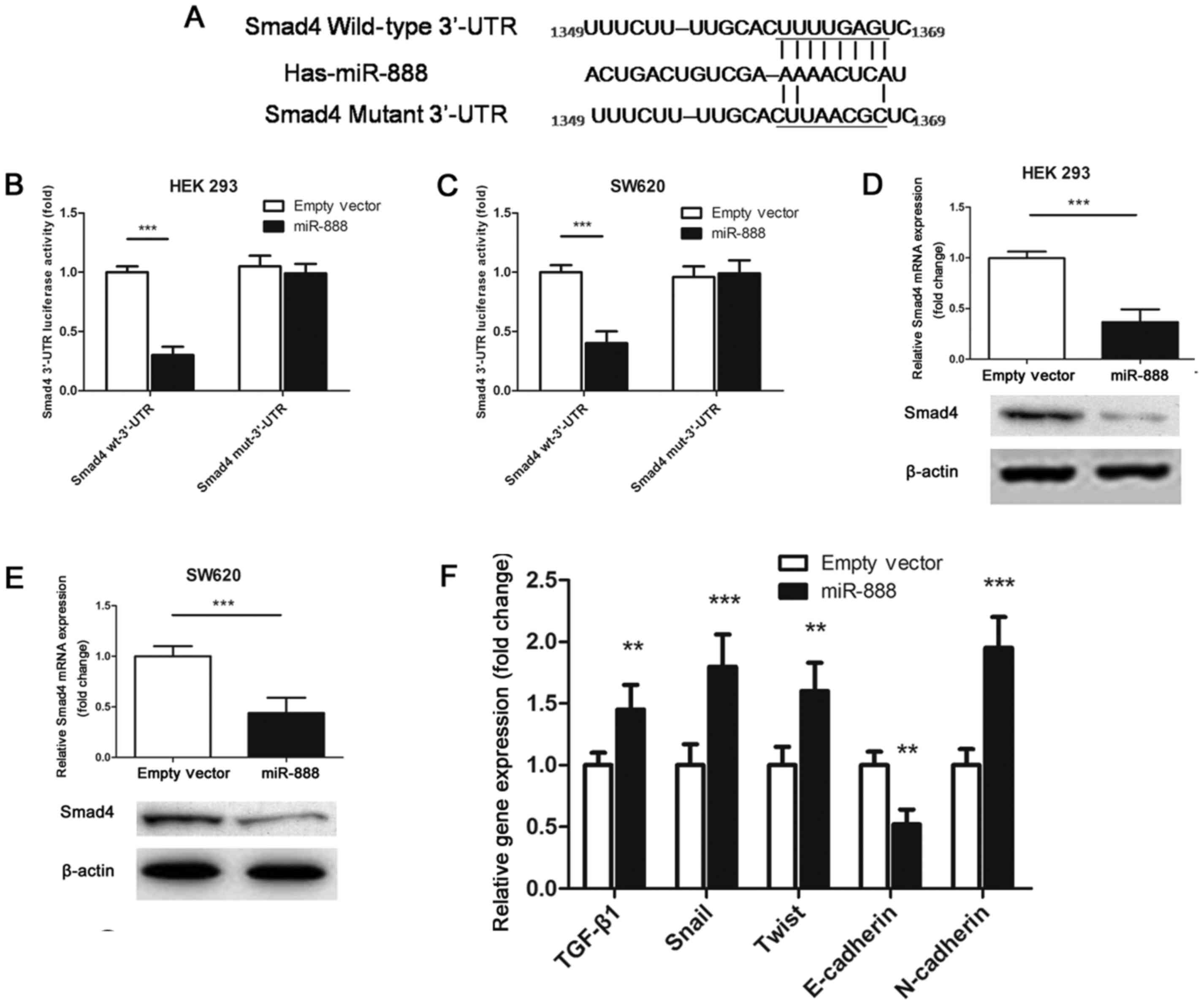
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Verma AM, Patel M, Aslam MI, Jameson J,
Pringle JH, Wurm P and Singh B: Circulating plasma microRNAs as a
screening method for detection of colorectal adenomas. Lancet. 385
Suppl 1:S1002015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lan YT, Yang SH, Chang SC, Liang WY, Li
AF, Wang HS, Jiang JK, Chen WS, Lin TC and Lin JK: Analysis of the
seventh edition of American Joint Committee on colon cancer
staging. Int J Colorectal Dis. 27:657–663. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Van Cutsem E, Cervantes A, Nordlinger B
and Arnold D: ESMO Guidelines Working Group: Metastatic colorectal
cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment
and follow-up. Ann Oncol. 25 Suppl 3:iii1–9. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
De Rosa M, Rega D, Costabile V, Duraturo
F, Niglio A, Izzo P, Pace U and Delrio P: The biological complexity
of colorectal cancer: Insights into biomarkers for early detection
and personalized care. Therap Adv Gastroenterol. 9:861–886. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Aghagolzadeh P and Radpour R: New trends
in molecular and cellular biomarker discovery for colorectal
cancer. World J Gastroenterol. 22:5678–5693. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guan X: Cancer metastases: Challenges and
opportunities. Acta Pharm Sin B. 5:402–418. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Svoronos AA, Engelman DM and Slack FJ:
OncomiR or tumor suppressor? The duplicity of microRNAs in cancer.
Cancer Res. 76:3666–3670. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shah MY, Ferrajoli A, Sood AK,
Lopez-Berestein G and Calin GA: microRNA therapeutics in cancer-an
emerging concept. EBioMedicine. 12:34–42. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Behbahani GD, Ghahhari NM, Javidi MA,
Molan AF, Feizi N and Babashah S: MicroRNA-mediated
post-transcriptional regulation of epithelial to mesenchymal
transition in cancer. Pathol Oncol Res. 23:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mohammadi A, Mansoori B and Baradaran B:
The role of microRNAs in colorectal cancer. Biomed Pharmacother.
84:705–713. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hovey AM, Devor EJ, Breheny PJ, Mott SL,
Dai D, Thiel KW and Leslie KK: miR-888: A Novel cancer-testis
antigen that targets the progesterone receptor in endometrial
cancer. Transl Oncol. 8:85–96. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lewis H, Lance R, Troyer D, Beydoun H,
Hadley M, Orians J, Benzine T, Madric K, Semmes OJ, Drake R and
Esquela-Kerscher A: miR-888 is an expressed prostatic
secretions-derived microRNA that promotes prostate cell growth and
migration. Cell Cycle. 13:227–239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang S and Chen L: MiR-888 regulates side
population properties and cancer metastasis in breast cancer cells.
Biochem Biophys Res Commun. 450:1234–1240. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bader AG: miR-888: Hit it when you see it!
Cell Cycle. 13:3512014. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang S, Cai M, Zheng Y, Zhou L, Wang Q
and Chen L: miR-888 in MCF-7 side population sphere cells directly
targets E-cadherin. J Genet Genomics. 41:35–42. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen Z, Liu S, Tian L, Wu M, Ai F, Tang W,
Zhao L, Ding J, Zhang L and Tang A: miR-124 and miR-506 inhibit
colorectal cancer progression by targeting DNMT3B and DNMT1.
Oncotarget. 6:38139–38150. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cao ZG, Li JJ, Yao L, Huang YN, Liu YR, Hu
X, Song CG and Shao ZM: High expression of microRNA-454 is
associated with poor prognosis in triple-negative breast cancer.
Oncotarget. 7:64900–64909. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shen J, Song G, An M, Li X, Wu N, Ruan K,
Hu J and Hu R: The use of hollow mesoporous silica nanospheres to
encapsulate bortezomib and improve efficacy for non-small cell lung
cancer therapy. Biomaterials. 35:316–326. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Duff EK and Clarke AR: Smad4 (DPC4)-a
potent tumour suppressor? Br J Cancer. 78:1615–1619. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Demagny H and De Robertis EM: Smad4/DPC4:
A barrier against tumor progression driven by RTK/Ras/Erk and
Wnt/GSK3 signaling. Mol Cell Oncol. 3:e9891332016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weng M, Wu D, Yang C, Peng H, Wang G, Wang
T and Li X: Noncoding RNAs in the development, diagnosis, and
prognosis of colorectal cancer. Transl Res. 181:108–120. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sinicrope FA, Okamoto K, Kasi PM and
Kawakami H: Molecular biomarkers in the personalized treatment of
colorectal cancer. Clin Gastroenterol Hepatol. 14:651–658. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marrero JA and Lok AS: Newer markers for
hepatocellular carcinoma. Gastroenterology. 127 5 Suppl
1:S113–S119. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Heneghan HM, Miller N and Kerin MJ: MiRNAs
as biomarkers and therapeutic targets in cancer. Curr Opin
Pharmacol. 10:543–550. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang J, Du Y, Liu X, Cho WC and Yang Y:
MicroRNAs as regulator of signaling networks in metastatic colon
cancer. Biomed Res Int. 2015:8236202015.PubMed/NCBI
|
|
28
|
Zhang Q, Yu N and Lee C: Vicious cycle of
TGF-β signaling in tumor progression and metastasis. Am J Clin Exp
Urol. 2:149–155. 2014.PubMed/NCBI
|
|
29
|
Drabsch Y and ten Dijke P: TGF-β
signalling and its role in cancer progression and metastasis.
Cancer Metastasis Rev. 31:553–568. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Matsuzaki K, Seki T and Okazaki K: TGF-β
signal shifting between tumor suppression and fibro-carcinogenesis
in human chronic liver diseases. J Gastroenterol. 49:971–981. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Inamoto S, Itatani Y, Yamamoto T,
Minamiguchi S, Hirai H, Iwamoto M, Hasegawa S, Taketo MM, Sakai Y
and Kawada K: Loss of SMAD4 promotes colorectal cancer progression
by accumulation of myeloid-derived suppressor cells through the
CCL15-CCR1 Chemokine Axis. Clin Cancer Res. 22:492–501. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fleming NI, Jorissen RN, Mouradov D,
Christie M, Sakthianandeswaren A, Palmieri M, Day F, Li S, Tsui C,
Lipton L, et al: SMAD2, SMAD3 and SMAD4 mutations in colorectal
cancer. Cancer Res. 73:725–735. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Singh S, Narang AS and Mahato RI:
Subcellular fate and off-target effects of siRNA, shRNA and miRNA.
Pharm Res. 28:2996–3015. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jackson AL and Linsley PS: Recognizing and
avoiding siRNA off-target effects for target identification and
therapeutic application. Nat Rev Drug Discov. 9:57–67. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Serrano-Gomez SJ, Maziveyi M and Alahari
SK: Regulation of epithelial-mesenchymal transition through
epigenetic and post-translational modifications. Mol Cancer.
15:182016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Steinestel K, Eder S, Schrader AJ and
Steinestel J: Clinical significance of epithelial-mesenchymal
transition. Clin Transl Med. 3:172014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Saitoh M: Epithelial-mesenchymal
transition is regulated at post-transcriptional levels by
transforming growth factor-β signaling during tumor progression.
Cancer Sci. 106:481–488. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Katsuno Y, Lamouille S and Derynck R:
TGF-β signaling and epithelial-mesenchymal transition in cancer
progression. Curr Opin Oncol. 25:76–84. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shah PP and Kakar SS: Pituitary tumor
transforming gene induces epithelial to mesenchymal transition by
regulation of Twist, Snail, Slug, and E-cadherin. Cancer Lett.
311:66–76. 2011. View Article : Google Scholar : PubMed/NCBI
|