Introduction
Cervical cancer is one of the most common malignant
tumours in the female reproductive system, with ~500,000 new cases
each year worldwide (1), accounting
for 5% of all new cancer cases. Overall, 78% of cases are reported
in developing countries (2).
Radiotherapy is one of the most well-used tumour
treatments (3). As reported by the
World Health Organization in 1992, 60–70% of patients with
malignant tumours undergo radiotherapy (4). Approximately 45% of all tumours can be
cured using a variety of treatments, 22% of which can be cured by
surgery, 18% by radiotherapy and 5% by chemotherapy (5). Thus, radiotherapy is valuable in tumour
treatment. With the widespread use of precise radiotherapies,
including conformal radiotherapy, intensity-modulated radiotherapy,
image-guarded radiation therapy, and the application of protons and
heavy ions in the clinic, the role of radiotherapy has become
increasingly important in tumour therapy (6). However, in clinical practice, the
majority of radiation practitioners have found that not every
patient with a tumour responds to the treatment (7,8). Certain
patients only exhibit good responses at the beginning of treatment.
Radiation can control the tumour, provide detailed imaging scans
and clinical efficacy (7,8). However, local recurrence or distant
metastasis can occur shortly after treatment (9). A second course of irradiation does not
usually produce good results (9). To
solve this problem, the changes that occur in tumour genomes
following radiotherapy require further understanding.
In traditional research, individual cells of the
same phenotype have been commonly viewed as identical functional
units of a tissue (10). Analyses by
conventional detection methods are always based on the overall
average reaction of cells (10).
Sequencing of DNA or RNA from single cells indicates that
heterogeneous cells enable the system-level functions of a tissue
(11). Single-cell sequencing is a
type of high-throughput sequencing technology at the single-cell
level. Compared with conventional throughput sequencing, it is a
powerful approach for studying the genetic heterogeneity of
individual cells with the same phenotype (11).
Human papillomavirus (HPV) infection is a major
cause of cervical cancer (12,13). In
China, HPV infection is also prevalent (14–16).
Recently, using high throughput sequencing, Hu et al
(17) reported frequent HPV
integration sites in genes such as POU class 5 homeobox 1B
(POU5F1B) (9.7%) and fragile histidine triad (8.7%).
However, to the best of our knowledge, there are no reports
concerning HPV infection prior to and following radiotherapy.
Navin et al (18) applied single-nucleus sequencing to
investigate the tumour population structure and evolution in two
human breast cancer cases. Each analysis of 100 single cells from
the two cases revealed three distinct clonal subpopulations in one
heterogeneous tumour, whereas another tumour consisted of a group
of genetically identical cells (12).
This data indicated that tumours grow by punctuated clonal
expansions, with few persistent intermediates. Xu et al
(19) performed single-cell exome
sequencing of renal cell carcinoma, revealing that the tumour did
not contain any significant clonal subpopulations, and
demonstrating that mutations occurred at different frequencies and
different mutation spectrums. The study demonstrated that renal
cell carcinoma maybe more heterogeneous than was believed, which
would require the development of more effective cellular targeted
therapies (13). This approach is
also conducive for researching the mechanism of tumour development
and metastasis. Felthaus et al (20) analysed oral squamous cell carcinoma
cell lines and revealed that the resistance of this cancer to
conventional chemotherapy or radiotherapy may be caused by cancer
stem cells.
In view of the power of single-cell sequencing
technology, the present study analysed genomic alterations,
particularly in terms of HPV infection, prior to and following
radiotherapy. Furthermore, using this technology, the effect of
radiotherapy could be assessed in patients with cervical cancer and
guide subsequent treatment in the future.
Materials and methods
Sample collection and preparation of
cell suspensions
Fresh tumour and blood samples were obtained from a
46-year-old female patient with the exogenous type of cervical
carcinogenesis at Beijing Obstetrics and Gynaecology Hospital
(Beijing, China) in April 2015. The diagnosis of cervical
carcinogenesis has been described in detail previously (17). The pathological type of cervical
cancer was squamous cell carcinoma and the tumour was classified as
stage IIA2, according to the 2009 International Federation of
Gynaecology and Obstetrics staging system (21). The size of the primary tumour was 5
cm. The HPV type was detected as HPV 16 using flow-through
hybridization. The level of squamous cell carcinoma antigen was
4.74 µg/l. The patient received 10 Gy in 5 fractions of 2 Gy,
following which the tumor tissue was excised and 12 cells were
isolated for gene sequencing. Then, the patient continued to
receive 36 Gy in 18 fractions of 2 Gy (10 Gy). Following radiation
therapy, the level of squamous cell carcinoma antigen was 4.62
µg/l. No improvements were noted in the patient's condition. Tumour
tissues were obtained prior to and following radiotherapy. The
tumour tissues were pathologically confirmed as malignant cervical
carcinogenesis with >90% tumour cells. The present study was
performed with the approval of the Beijing Obstetrics and
Gynaecology Hospital. Signed written consent was obtained from the
patient prior to recruitment to the study.
Collection of single cells and
preparation of cell lysates
Single cells from the tumour samples were prepared
as described previously (19) A
manually controlled pipetting system was used to isolate single
cells under an inverted light microscope (Nikon Instruments Co.,
Ltd.). Each cell was transferred into a precooled polymerase chain
reaction (PCR) tube containing a cell lysis solution (Qiagen GmbH,
Hilden, Germany) (The samples were incubated in a thermocycler for
10 min at 65°C. A physiological saline blank was included as a
negative control. Every step during the experiments was performed
strictly according to the aforementioned protocol. With sufficient
dispersion and cascade-dilution of the cells, single cells were
randomly isolated from tumour tissues into PCR-ready tubes using an
inverted microscope and a mouth-controlled, fine hand-drawn
microcapillary pipetting system made in-house. Single-cell
isolation was visually confirmed by microscopy and documented as
micrographs. The cells were washed three times using the elution
buffer (Qiagen GmbH).
Multiple displacement amplification
(MDA)
Whole-genome amplification (WGA) was performed using
a REPLI-g Mini kit (Qiagen GmbH) according to the manufacturer's
protocol. All samples were amplified by MDA, according to the
aforementioned protocol. A total reaction volume of 50 µl was used
at 30°C for 16 h and then terminated at 65°C for 10 min. Amplified
DNA products were then stored at −20°C.
Whole-genome sequencing (WGS)
Paired-end library preparation was conducted using
Illumina protocols (22). Genomic DNA
(400 ng) was fragmented to an insert size of ~400 bp with a Covaris
device (M220 Focused-ultrasonicator; Covaris, Inc., Woburn, MA,
USA), and size selection was performed using 2% agarose gel
excision. Deep sequencing was performed using Illumina X10
instruments (Illumina, Inc., San Diego, CA, USA). Each sample
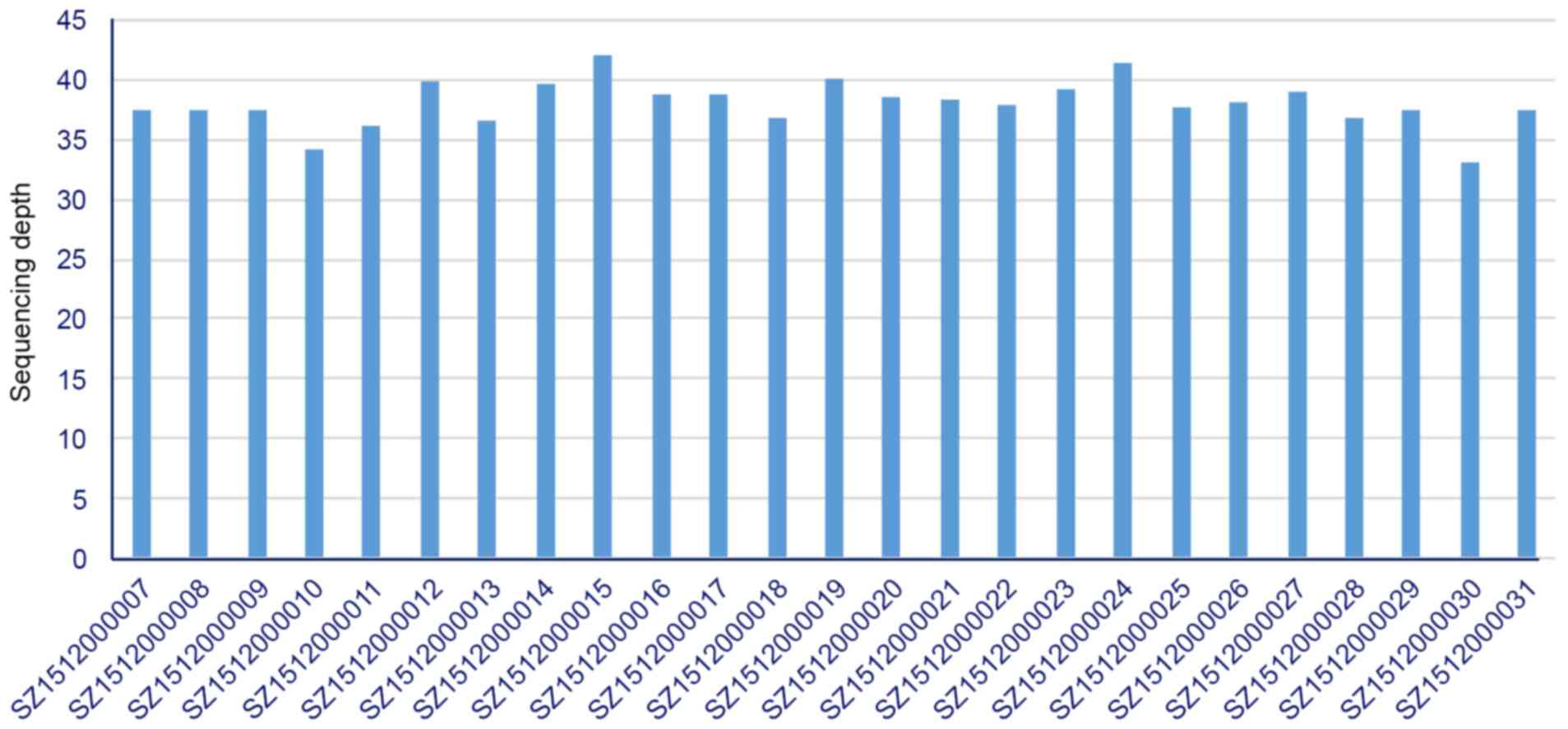
achieved ~38 times genomic coverage.
Somatic mutation detection
Following the removal of adapters and low-quality
reads, all sequencing reads were mapped to the human genome (hg19
build) (23) using Burrows-Wheeler
Aligner (v0.5.9) (24) with default
parameters. The sequence alignment map output was converted to a
sorted binary alignment map file using SAM tools (v1.3) (25). Picard (v1.70) (Broad Institute,
Cambridge, MA, USA) was used to remove PCR duplicates. Somatic
single nucleotide variants (SNVs) were detected by VarScan (v2.3.9)
(26). A candidate somatic mutation
was called if the following criteria were met: i) The somatic
P-values of the variants were <0.05; ii) mutant allele
frequencies in tumour cells were >15%; iii) mutant allele
frequencies in the normal control were <0.5%; iv) reads with a
mutant allele were >4; v) the forward reference count (i.e., the
number of forward reads that match the reference base at the
locus), the reverse reference count (i.e., the number of reverse
reads that match the reference base at the locus), the forward
non-reference count (i.e., the number of forward reads that do not
match the reference base at the locus) and the reverse
non-reference count (i.e., the number of reverse reads that do not
match the reference base at the locus) in the tumour must be ≥1;
and vi) only mutations detected in four samples were retained. To
eliminate common germline variants, SNVs observed in dbSNP137 or
the 1,000 Genomes Project, March 2012 data release project were
excluded (27). Annotation was
performed using snpEff (v4.2) (28).
GR Ch37.75 was used for transcript identification and to determine
amino acid changes. All putative somatic mutations in coding
regions were validated visually. The mean number and the standard
deviation of somatic mutations were calculated. When it was found
that the number of somatic mutations in one sample was less than
the total of the mean number minus the standard deviation, the
sample with the fewest number of somatic mutations was removed and
this sample was considered as the normal cells. In this way, when
the sample with 25 somatic mutations was removed, the number of all
samples remained was greater than the total of the mean number
minus the standard deviation.
Mapping and analysis of HPV
integration sites
The 400-bp paired-end fragment libraries (150 bp
read length) were mapped to the human reference genome (hg19) and
HPV genome (HPV6: FR751337.1, HPV82: AF293961.1, HPV69: AB027020.1,
HPV68: FR7 51039.1, HPV66: EF177191.1, HPV59: E U918767.1, HPV58:
HQ537777.1, HPV56: EF177181.1, HPV52: HQ537751.1, HPV45:
EF202167.1, HPV39: M62849.1, HPV35: HQ537730.1, HPV33: HQ537688.1,
HPV31: HQ537687.1, HPV18: AY262282.1, HPV16: NC_001526.2, and
HPV11: HE574705.1; www.ncbi.nlm.nih.gov). If a paired-end read was
uniquely mapped to hg19 at one end and to HPV at the other end,
integration was reported. All mapping locations were subjected to a
filtering process to remove possible PCR duplicates. Specifically,
if there were two or more paired reads that mapped to
near-identical locations (±2 bp), only one of the reads was
considered. The fusion point was determined by analysing cases in
which one region of the read aligned to HPV and the other aligned
to hg19. These locations were crosschecked with the clusters of
paired-end reads for consistency. Furthermore, to determine the
exact fusion point between hg19 and HPV, all hg19-HPV mapped reads
were extracted, and aligned each read entirely on hg19 and HBV
using Blat (http://genome.ucsc.edu/cgi-bin/hgBlat, -min -Score 25,
-minIdentity 85).
Statistical analysis
Paired Student's t-test was used to compare the
number of somatic mutations in samples prior to and following
radiotherapy. The Wilcoxon rank-sum test was used to compare the
HPV integration events between tumour and normal cells or tumor
cells prior to and following radiotherapy. P<0.05 was considered
to indicate a statistically significant difference. All tests were
performed using R software (v2.14; https://www.R-project.org).
Results
High-throughput isolation and
amplification of single cells from fresh tumour tissues
Fresh tumour tissues prior to and following
radiotherapy were obtained from a 46-year-old woman with cervical
carcinoma classified as stage IIA2. Blood was also collected from
this patient, which was used as a matched normal control. To obtain
detailed cellular genetic information on this tumour, single cell
sequencing in individual cells from the tumour samples was
performed as described previously (19). WGA was performed based on MDA of the
DNA from each single cell of the tumour tissues. In total, 13 cells
were obtained from tumour tissues prior to radiotherapy
(SZ1512000007-SZ1512000019), and 12 cells were obtained from tumour
tissues following radiotherapy (SZ1512000020-SZ1512000031). These
25 single tumour cells met the previously described criteria
(19) and were selected for
subsequent analysis. Massively parallel single-cell WGS was
performed on these samples using paired-end 150-bp reads. The blood
sample also underwent conventional WGS. A mean of 114 Gb of
high-quality mappable WGS data were aligned to the human reference
genome, with 38.0-fold coverage in the tumour prior to
radiotherapy, 37.8-fold coverage in the tumour following
radiotherapy and 38.6-fold coverage in the blood (Fig. 1).
To detect somatic mutations in each cell, Varscan
(v2.3.9) was used to assess tumour-normal pairs, with each tumour
cell as tumour and the blood sample as normal. To avoid
variation-calling biases in single cell sequencing, only mutations
that were detected in at least four samples were retained.
Following the removal of somatic mutations that occurred at
dbSNP135 and in the 1,000 Genomes Project March 2012 data release
project, 149 somatic protein-altering mutations were identified in
the 25 samples. Samples harboured mean of 47 somatic mutations.
Analysis of all somatic mutations showed a preference for
C>T/G>A, the apolipoprotein B mRNA editing enzyme catalytic
subunit 3Bmutation signature (29),
and was similar to the previously reported mutation mechanism
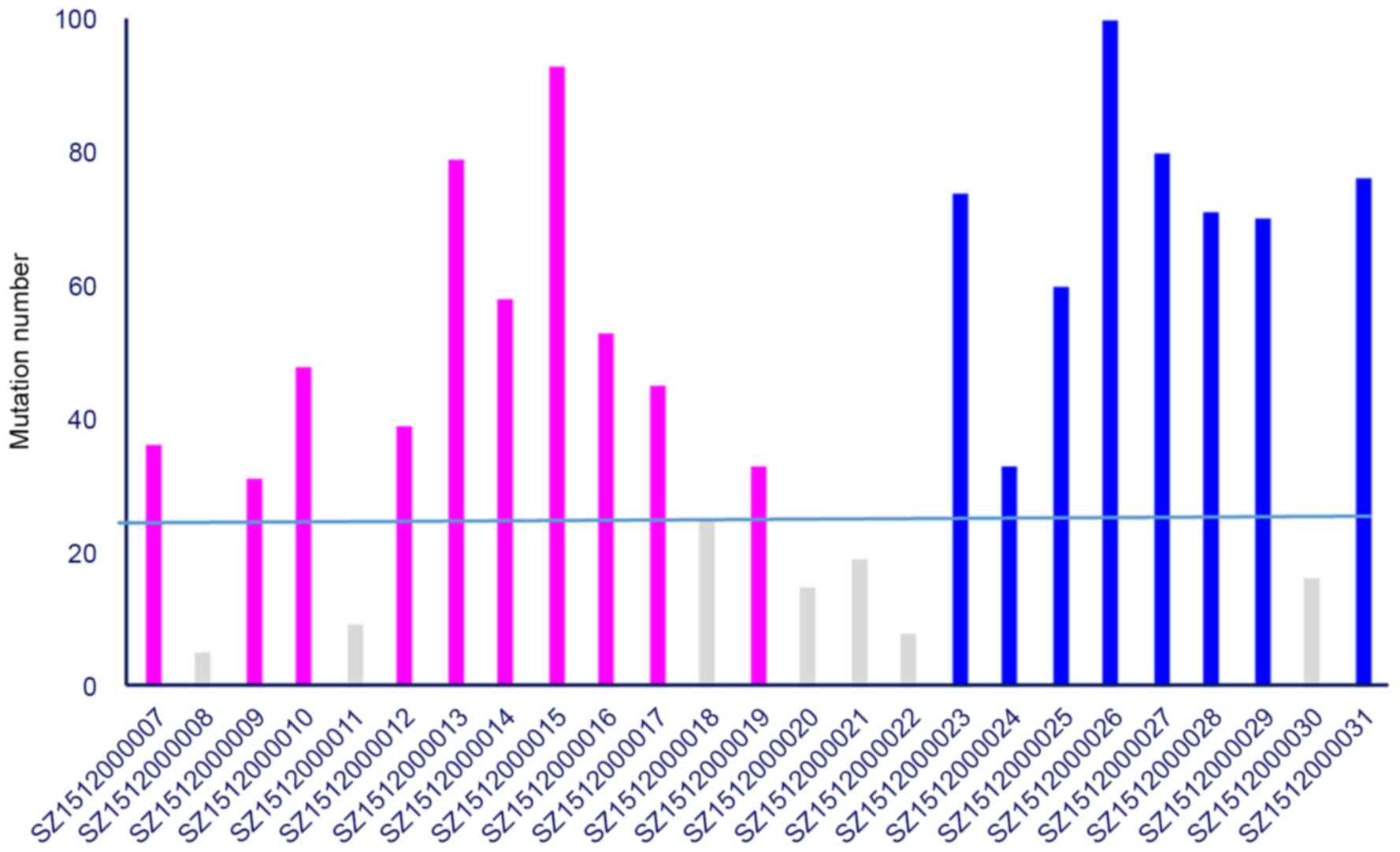
pattern observed in cervical cancer (30). A total of 7 samples with <25
mutations were recognised as normal cells owing to the small amount
of variance between sample and adjacent tissues (3 samples were
obtained from tumours prior to radiotherapy and 4 samples were from
tumours following radiotherapy). This is an explainable phenomenon,
as tumour tissues are mixed tissues, which may be mixed with normal
cells. In pre-radiotherapy samples, mean of 51.5 mutations were
detected, whereas 70.5 mutations were detected in samples following
radiotherapy. Significantly more somatic mutations were identified
in samples following radiotherapy (P=0.030; single-end Student's
t-test; Fig. 2), suggesting that
radiotherapy may cause genetic instability (31).
Principal component analysis and
hierarchical cluster tree analysis
To investigate the intercellular heterogeneity
structure in tumours prior to and following radiotherapy,
population genetic analyses was applied to the comprehensive 25 WGS
data sets. A total of 149 non-synonymous mutations were subjected
to principal component analysis (PCA) to characterise the genetic
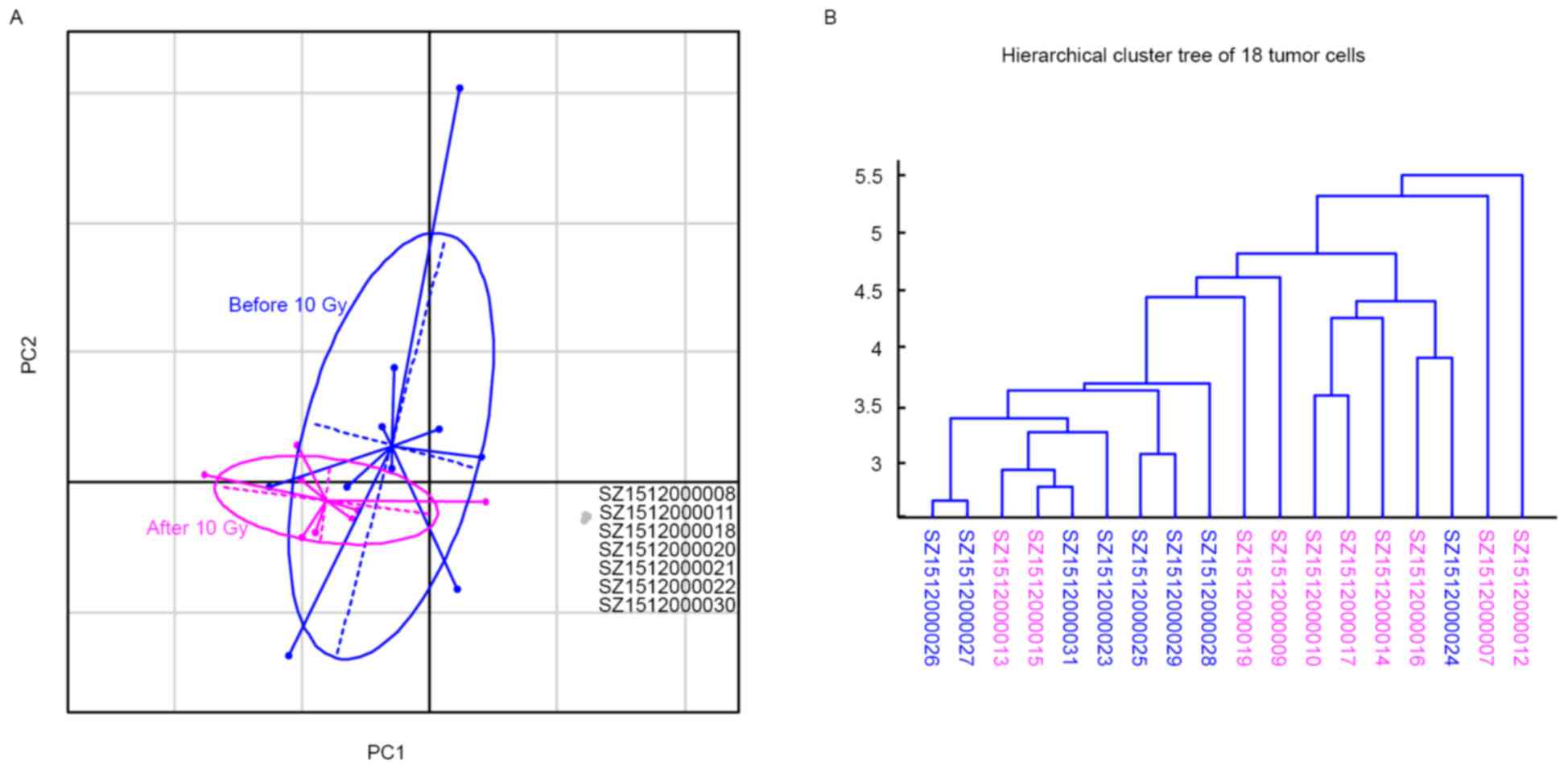
heterogeneity of this tumour. It was found that two first principal
components (PC1 and PC2), which represented 26% of the whole
inertia (Table I), clearly divided
isolated single cells into two subsets: Normal-cell and tumour-cell
subsets (Fig. 3A). The normal-cell
subset consisted of the 7 samples recognised as normal cells with
<25 somatic mutations. The tumour-cell subsets were also
clustered into two subgroups that could almost distinguish samples
prior to and following radiotherapy (Fig.
3A). Next, hierarchical cluster tree analysis of the 18 tumour
cells was performed to infer their population substructure. Samples
from the tumour prior to radiotherapy were clustered together, as
were the samples from the tumour following radiotherapy (Fig. 3B). This observation indicated the
clear genetic variance between tumour cells prior to and following
radiotherapy.
 | Table I.Percentage of each PC as a proportion
of the whole inertia. |
Table I.
Percentage of each PC as a proportion
of the whole inertia.
| PC | Percentage |
|---|
| PC1 | 16.40 |
| PC2 | 9.30 |
| PC3 | 8.50 |
| PC4 | 7.60 |
| PC5 | 7.20 |
Notably, 8 tumour cells prior to radiotherapy and 1
tumour cell following radiotherapy were clustered together, while
only 2 tumour cells prior to radiotherapy and 7 tumour cells
following radiotherapy were clustered together (Fig. 3B). This result indicated that at least
2 subpopulations existed in the tumour prior to radiotherapy (the
subpopulation of 8 tumour cells prior to radiotherapy were defined
as subpopulation 1, and the subpopulation of only 2 tumour cells
prior to radiotherapy were defined as subpopulation 2).
Subpopulation 1 was the main population in the tumour prior to
radiotherapy. Following radiotherapy, the majority of the cells in
subpopulation 1 were killed, and the majority of the cells in
subpopulation 2 had survived. Thus, following radiotherapy,
although the 2subpopulations existed, subpopulation 2 became the
main population.
HPV integration analyses
To identify driver mutations in the tumour genome,
which may be responsible for the radiotherapy resistance, HPV
integration was detected. The patient was infected with HPV16. A
total of 17 HPV-integration genes were identified in the 25 cells
from the cervical carcinoma (Table
II). Among these genes, an integration site in alkylglycerone
phosphate synthase (AGPS) was supported by the highest number of
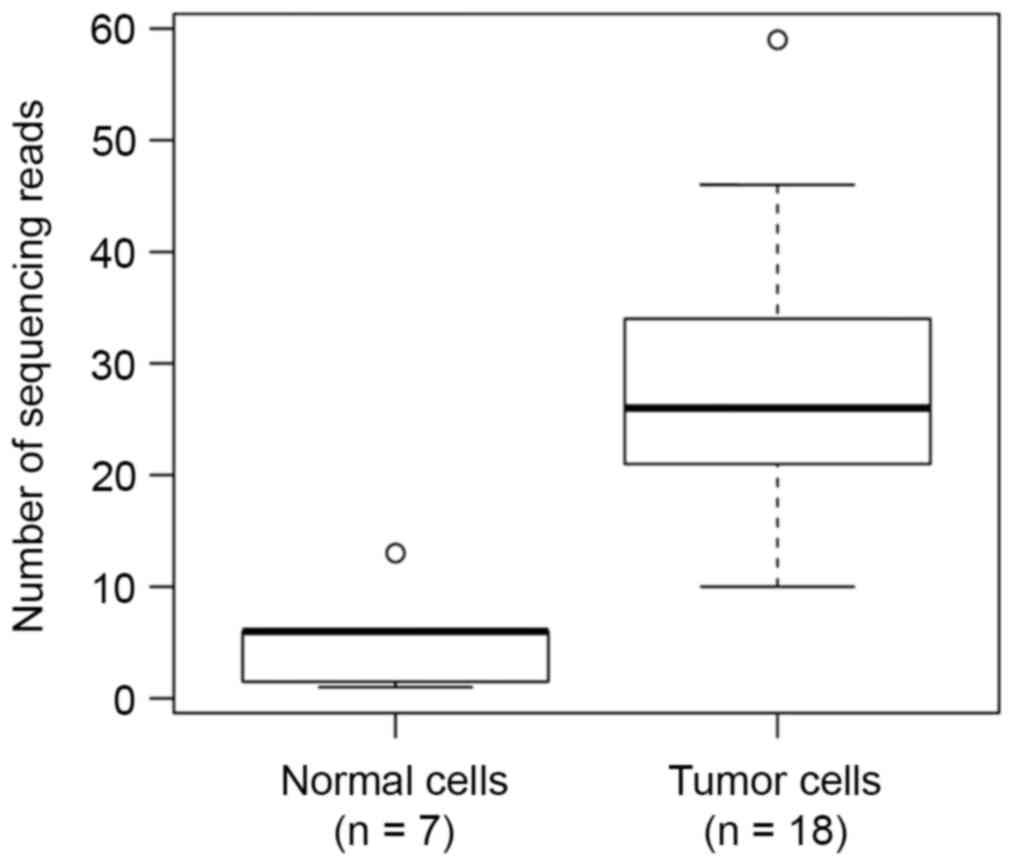
paired-reads (512 reads). Notably, the number of sequencing reads
of HPV-integration events in the 7 samples regarded as normal cells
were significantly less than that in other samples
(P=1.9×10−4; Wilcoxon rank-sum test; 5.0 vs. 27.7;
Fig. 4), further supporting that
these 7 samples were normal cells. This result was consistent with
the results of a previous study (32). No significant difference was found in
the number of sequencing reads of HPV integration events between
samples from the tumour prior to and following radiotherapy (P=1.0,
Wilcoxon rank-sum test).
 | Table II.Total number of sequencing reads
supporting HPV integration. |
Table II.
Total number of sequencing reads
supporting HPV integration.
| Protein-coding
gene |
Samples
SZ1512000007-SZ1512000031 | Sum |
|---|
| AGPS | 17 | 2 | 59 | 22 | 0 | 46 | 34 | 17 | 28 | 21 | 14 | 5 | 28 | 5 | 13 | 1 | 25 | 20 | 26 | 33 | 27 | 23 | 4 | 6 | 36 | 512 |
| MT-CYB | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| C1orf143 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| CDH8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| AGPAT9 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| DYX1C1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| HNRNPA3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| PTPRT | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| AL445989.1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| MT-ND1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| SLC6A20 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| AKR1B10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 |
| BCL2L14 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 2 |
| TRPC5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| POU5F1B | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 4 | 0 | 0 | 5 |
| USP9X | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 |
| TLE4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| Sum | 17 | 2 | 59 | 23 | 1 | 46 | 34 | 17 | 28 | 21 | 15 | 6 | 30 | 6 | 13 | 1 | 26 | 21 | 26 | 34 | 29 | 24 | 10 | 6 | 38 | 533 |
A total of 4 and 1 sequencing reads of HPV
integration events were identified in POU5F1B of two different
samples from the tumour following radiotherapy, respectively;
POU5F1B has been reported as a frequently integrated gene (17,33). The
supporting reads to the human and HPV genome were remapped using
Blat (UCSC Genome Browser), and it was found that there were no
artefacts. However, no evidence of HPV integration events was found
in POU5F1B in samples from the tumour prior to radiotherapy.
The most likely explanation for this phenomenon is
that the tumour tissue prior to radiotherapy was of polyclonal
origin. The tumour cells with HPV integrations in POU5F1B
represent only a small proportion of these clones. Thus, the
integration events in POU5F1B could not be detected by WGS.
Following radiotherapy, tumour cells with HPV integrations in
POU5F1B had survived, whereas the majority of other cells
were killed by the radiotherapy. Consequently, the proportion of
tumour cells with HPV integrations in POU5F1B increased.
Thus, this integration could be detected by WGS. The genetic
mechanism under radiotherapy resistance is complex. The HPV
integration site in POU5F1B was only one of the genetic
mechanisms that caused radiotherapy resistance. The development of
radiotherapy resistance may be due to somatic mutations, copy
number variants or structure variants. Thus, further study is
required.
AGPS was the most common integration gene in all the
cells. The number of AGPS integrations in tumour cells was
significantly higher than that in normal cells (P=0.00040; Wilcoxon
rank-sum test; 26.7 vs. 4.6). However, there was no significant
difference in the number of AGPS integrations between cells prior
to and following radiotherapy (P=0.89). In fact, no evidence that
AGPS was associated with radiotherapy resistance was found in
previous reports either.
Taken together, these results indicated that tumour
cells prior to and following radiotherapy exhibit distinct genetic
characteristics, and tumour cells with HPV integrations in
POU5F1B are more likely to survive following
radiotherapy.
Discussion
The present study performed single-cell sequencing
on 25 cells from cervical tumour samples prior to and following
radiotherapy. WGS was also performed as a normal control for the 25
tumour cells. To the best of our knowledge, this is the first
detailed genetic landscape of a tumour prior to and following
radiotherapy at the single-cell level.
Population analysis of identified somatic mutations
allowed for tumour cells to be distinguished from normal cells. The
number of somatic mutations in cells from tumours following
radiotherapy was significantly higher than that in cells from
tumours prior to radiotherapy. Through PCA and hierarchical cluster
tree analysis, it was found that cells from the tumour prior to or
following radiotherapy tended to cluster together. Sequencing data
revealed that the HPV type was HPV 16, which was consistent with
the traditional detection method. HPV integration events were
detected in POU5F1B in tumour cells following radiotherapy.
The present study indicated that a proportion of tumour cells could
survive radiotherapy, which informs on the mechanism of
radiotherapy resistance.
Previous studies have suggested that radiotherapy
resistance is the result of the intratumoural heterogeneity and
subpopulation diversity of cancer cells (34,35). The
results of the present study demonstrated the subpopulation
diversity of cancer cells in the cervical tumour prior to
radiotherapy. Certain subpopulations may be sensitive to
radiotherapy and could thus be effectively killed. If all
subpopulations are sensitive to radiotherapy, the patient may
recover following radiotherapy. However, if at least one
subpopulation is resistant to radiotherapy, the radiotherapy may
not be effective for the patient. The patient in the present study
evidently belongs to the latter category.
Radiotherapy resistance could be predicted by
expression analysis based on a defined set of genes (36). In addition to gene expression levels,
somatic mutations are reported to be involved in radiotherapy
resistance (37,38). For example, all patients with
medulloblastomas harbouring tumour protein p53 mutations
experienced early recurrence, and the 5-year survival rate of these
patients was 0% (37). Somatic
mutations may also be involved in radiotherapy resistance in
cervical cancer. For example, patients with KRAS proto-oncogene,
GTPase-mutant cervical cancer who received radiation therapy
experienced poorer outcomes (39).
HPV integration is a specific type of somatic variant; thus, HP
integration may also result in radiotherapy resistance. However, it
has been reported that certain tumours with HPV integration may
have enhanced radiation sensitivity, such as those of the head and
neck (40), and squamous cell
carcinomas (41). The HPV infection
and the HPV type have also been shown to be associated with
radiotherapy resistance (42).
However, to the best of our knowledge, no reported
study has associated HPV integration sites or genes affected by HPV
with radiotherapy resistance. In the present study, in tumour cells
following radiotherapy, but not prior to radiotherapy, an HPV
integration site in was detected in POU5F1B, which may be a driver
mutation in the tumour genome and thus responsible for the
radiotherapy resistance. Several lines of evidence indicate that
POU5F1B may be responsible for radiotherapy resistance: First,
POU5F1B was reported to be a hotspot for HPV integration in
cervical tumours (17,32); second, amplifications of POU5F1B have
been reported to promote an aggressive phenotype in gastric cancer
(43); and third, POU5F1B is located
at 8q24, a well-known susceptibility locus for various tumours
(44,45). These observations indicate that the
HPV integration site in POU5F1B may responsible for radiotherapy
resistance.
The current study presents an analysis of mutations
prior to and following radiotherapy in a cervical tumour using
single-cell sequencing. The results indicate that tumour cells with
HPV integration in POU5F1B survive radiotherapy, and that
tumour cells prior to and following radiotherapy exhibit distinct
characteristics. The results obtained provide novel insights into
the specific molecular events that drive radiotherapy resistance in
cervical cancer.
Acknowledgements
The present study was supported by a grant from
Beijing Municipal Science and Technology commission (no.
D0906008040491).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ravichandran R: Has the time come for
doing away with Cobalt-60 teletherapy for cancer treatments. J Med
Phys. 34:63–65. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang YJ, Wei L, Li J, Zheng YQ and Li XR:
Status quo and development trend of breast biopsy technology. Gland
Surg. 2:15–24. 2013.PubMed/NCBI
|
|
6
|
Malicki J: Medical physics in
radiotherapy: The importance of preserving clinical
responsibilities and expanding the profession's role in research,
education, and quality control. Rep Pract Oncol Radiother.
20:161–169. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tanderup K, Eifel PJ, Yashar CM, Potter R
and Grigsby PW: Curative radiation therapy for locally advanced
cervical cancer: Brachytherapy is NOT optional. Int J Radiat Oncol
Biol Phys. 88:537–539. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
McCormack M, Kadalayil L, Hackshaw A,
Hall-Craggs MA, Symonds RP, Warwick V, Simonds H, Fernando I,
Hammond M, James L, et al: A phase II study of weekly neoadjuvant
chemotherapy followed by radical chemoradiation for locally
advanced cervical cancer. Br J Cancer. 108:2464–2469. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Amichetti M and Amelio D: A review of the
role of re-irradiation in recurrent high-grade glioma (HGG).
Cancers (Basel). 3:4061–4089. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hao JJ, Lin DC, Dinh HQ, Mayakonda A,
Jiang YY, Chang C, Jiang Y, Lu CC, Shi ZZ, Xu X, et al: Spatial
intratumoral heterogeneity and temporal clonal evolution in
esophageal squamous cell carcinoma. Nat Genet. 48:1500–1507. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu C, Yu J, Yao X, Wu WK, Lu Y, Tang S, Li
X, Bao L, Li X, Hou Y, et al: Discovery of biclonal origin and a
novel oncogene SLC12A5 in colon cancer by single-cell sequencing.
Cell Res. 24:701–712. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pett M and Coleman N: Integration of
high-risk human papillomavirus: A key event in cervical
carcinogenesis? J Pathol. 212:356–367. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chandrani P, Kulkarni V, Iyer P, Upadhyay
P, Chaubal R, Das P, Mulherkar R, Singh R and Dutt A: NGS-based
approach to determine the presence of HPV and their sites of
integration in human cancer genome. Br J Cancer. 112:1958–1965.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao R, Zhang WY, Wu MH, Zhang SW, Pan J,
Zhu L, Zhang YP, Li H, Gu YS and Liu XZ: Human papillomavirus
infection in Beijing, people's republic of China: A
population-based study. Br J Cancer. 101:1635–1640. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dai M, Bao YP, Li N, Clifford GM,
Vaccarella S, Snijders PJ, Huang RD, Sun LX, Meijer CJ, Qiao YL and
Franceschi S: Human papillomavirus infection in shanxi province,
people's republic of China: A population-based study. Br J Cancer.
95:96–101. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li LK, Dai M, Clifford GM, Yao WQ, Arslan
A, Li N, Shi JF, Snijders PJ, Meijer CJ, Qiao YL and Franceschi S:
Human papillomavirus infection in shenyang city, people's republic
of China: A population-based study. Br J Cancer. 95:1593–1597.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hu Z, Zhu D, Wang W, Li W, Jia W, Zeng X,
Ding W, Yu L, Wang X, Wang L, et al: Genome-wide profiling of HPV
integration in cervical cancer identifies clustered genomic hot
spots and a potential microhomology-mediated integration mechanism.
Nat Genet. 47:158–163. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Navin N, Kendall J, Troge J, Andrews P,
Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et
al: Tumour evolution inferred by single-cell sequencing. Nature.
472:90–94. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu X, Hou Y, Yin X, Bao L, Tang A, Song L,
Li F, Tsang S, Wu K, Wu H, et al: Single-cell exome sequencing
reveals single-nucleotide mutation characteristics of a kidney
tumor. Cell. 148:886–895. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Felthaus O, Ettl T, Gosau M, Driemel O,
Brockhoff G, Reck A, Zeitler K, Hautmann M, Reichert TE, Schmalz G
and Morsczeck C: Cancer stem cell-like cells from a single cell of
oral squamous carcinoma cell lines. Biochem Biophys Res Commun.
407:28–33. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Koskas M and Rouzier R: Comparative
performance of the 2009 International Federation of Gynecology and
Obstetrics' staging system for uterine corpus cancer. Obstet
Gynecol. 117:1225–1226; author reply 1226. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sung WK, Zheng H, Li S, Chen R, Liu X, Li
Y, Lee NP, Lee WH, Ariyaratne PN, Tennakoon C, et al: Genome-wide
survey of recurrent HBV integration in hepatocellular carcinoma.
Nat Genet. 44:765–769. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kent WJ, Sugnet CW, Furey TS, Roskin KM,
Pringle TH, Zahler AM and Haussler D: The human genome browser at
UCSC. Genome Res. 12:996–1006. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li H: A statistical framework for SNP
calling, mutation discovery, association mapping and population
genetical parameter estimation from sequencing data.
Bioinformatics. 27:2987–2993. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koboldt DC, Zhang Q, Larson DE, Shen D,
McLellan MD, Lin L, Miller CA, Mardis ER, Ding L and Wilson RK:
VarScan 2: Somatic mutation and copy number alteration discovery in
cancer by exome sequencing. Genome Res. 22:568–576. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
1000 Genomes Project Consortium, . Auton
A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini
JL, McCarthy S, McVean GA and Abecasis GR: A global reference for
human genetic variation. Nature. 526:68–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cingolani P, Platts A, le Wang L, Coon M,
Nguyen T, Wang L, Land SJ, Lu X and Ruden DM: A program for
annotating and predicting the effects of single nucleotide
polymorphisms, SnpEff: SNPs in the genome of Drosophila
melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6:80–92.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kuong KJ and Loeb LA: APOBEC3B mutagenesis
in cancer. Nat Genet. 45:964–965. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ojesina AI, Lichtenstein L, Freeman SS,
Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio
L, Cibulskis K, Bertelsen B, et al: Landscape of genomic
alterations in cervical carcinomas. Nature. 506:371–375. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sabatier L, Lebeau J and Dutrillaux B:
Radiation-induced carcinogenesis: Individual sensitivity and
genomic instability. Radiat Environ Biophys. 34:229–232. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang R, Shen C, Zhao L, Wang J, McCrae M,
Chen X and Lu F: Dysregulation of host cellular genes targeted by
human papillomavirus (HPV) integration contributes to HPV-related
cervical carcinogenesis. Int J Cancer. 138:1163–1174. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Annunziata C, Buonaguro L, Buonaguro FM
and Tornesello ML: Characterization of the human papillomavirus
(HPV) integration sites into genital cancers. Pathol Oncol Res.
18:803–808. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Marusyk A and Polyak K: Tumor
heterogeneity: Causes and consequences. Biochim Biophys Acta.
1805:105–117. 2010.PubMed/NCBI
|
|
35
|
Xia Q, Guo Y, Zhang Z, Li D, Xuan Z, Li Z,
Dai F, Li Y, Cheng D, Li R, et al: Complete resequencing of 40
genomes reveals domestication events and genes in silkworm
(Bombyx). Science. 326:433–436. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Eschrich S, Zhang H, Zhao H, Boulware D,
Lee JH, Bloom G and Torres-Roca JF: Systems biology modeling of the
radiation sensitivity network: A biomarker discovery platform. Int
J Radiat Oncol Biol Phys. 75:497–505. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tabori U, Baskin B, Shago M, Alon N,
Taylor MD, Ray PN, Bouffet E, Malkin D and Hawkins C: Universal
poor survival in children with medulloblastoma harboring somatic
TP53 mutations. J Clin Oncol. 28:1345–1350. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Krakstad C and Chekenya M: Survival
signalling and apoptosis resistance in glioblastomas: Opportunities
for targeted therapeutics. Mol Cancer. 9:1352010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sreelekha TT, Nair MK, Jayaprakash PG and
Pillai MR: Immunophenotype of mutant ras p21 and early response to
radiotherapy in cancer of the uterine cervix. J Exp Clin Cancer
Res. 18:337–341. 1999.PubMed/NCBI
|
|
40
|
Kimple RJ, Smith MA, Blitzer GC, Torres
AD, Martin JA, Yang RZ, Peet CR, Lorenz LD, Nickel KP, Klingelhutz
AJ, et al: Enhanced radiation sensitivity in HPV-positive head and
neck cancer. Cancer Res. 73:4791–4800. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vozenin MC, Lord HK, Hartl D and Deutsch
E: Unravelling the biology of human papillomavirus (HPV) related
tumours to enhance their radiosensitivity. Cancer Treat Rev.
36:629–636. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Badaracco G, Savarese A, Micheli A, Rizzo
C, Paolini F, Carosi M, Cutillo G, Vizza E, Arcangeli G and Venuti
A: Persistence of HPV after radio-chemotherapy in locally advanced
cervical cancer. Oncol Rep. 23:1093–1099. 2010.PubMed/NCBI
|
|
43
|
Hayashi H, Arao T, Togashi Y, Kato H,
Fujita Y, De Velasco MA, Kimura H, Matsumoto K, Tanaka K, Okamoto
I, et al: The OCT4 pseudogene POU5F1B is amplified and promotes an
aggressive phenotype in gastric cancer. Oncogene. 34:199–208. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pomerantz MM, Ahmadiyeh N, Jia L, Herman
P, Verzi MP, Doddapaneni H, Beckwith CA, Chan JA, Hills A, Davis M,
et al: The 8q24 cancer risk variant rs6983267 shows long-range
interaction with MYC in colorectal cancer. Nat Genet. 41:882–884.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yeager M, Orr N, Hayes RB, Jacobs KB,
Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee
N, et al: Genome-wide association study of prostate cancer
identifies a second risk locus at 8q24. Nat Genet. 39:645–649.
2007. View
Article : Google Scholar : PubMed/NCBI
|