Introduction
Lung cancer is one of the most common types of
malignant tumor and threatens the health and survival of human
beings (1). The majority of cases of
lung cancer are non-small cell lung cancer (NSCLC; 80%), which is
associated with ~400,000 mortalities annually, a generally poor
prognosis and a difficulty of diagnosing in the early stages
(2–4).
The development of lung cancer is often due to environmental or
lifestyle factors including smoking, environmental pollution and
occupational exposure to carcinogens. These factors may induce
mutations in susceptible genes and epigenetic changes, ultimately
resulting in the lung cancer (5).
Among genes that are commonly mutated in NSCLC,
including TP53, epidermal growth factor receptor (EGFR) and KRAS,
the detection of EGFR mutations is the most common (6). EGFR, a transmembrane tyrosine kinase
receptor, includes an intracellular tyrosine kinase domain, and an
extracellular domain with a high affinity for epidermal growth
factor. EGFR regulates a number of biological activities, including
cell proliferation, differentiation, migration and the promotion of
cell survival (7). Therefore, when
EGFR is abnormally expressed, cells may rapidly proliferate and
differentiate, potentially leading to the formation of a tumor. A
previous study reported that 48–80% of cases of NSCLC exhibit
abnormal EGFR expression (8).
At present, small-molecule therapeutic agents,
including gefitinib, erlotinib, cetuximab and bevacizumab, have
been applied as a molecularly-targeted treatment against tumors.
These agents have been demonstrated to be suitable for the
treatment of patients with NSCLC in previous studies (3,9,10). Other previous studies concluded that
patients with NSCLC with EGFR mutations receiving EGFR-TKI
treatment experienced an increased median progression-free survival
time compared with patients treated with conventional chemotherapy,
with a difference of ~8 months (10,11). Thus,
it is important to develop a low-cost, accurate and efficient
method for the clinical detection of EGFR mutations.
Various detection methods are currently used in the
clinic, including capillary electrophoresis (12), denaturing high-performance liquid
chromatography (13), high-resolution
melting analysis (14), quantitative
polymerase chain reaction (PCR) analysis (15) and PCR single-strand conformation
polymorphism (16). However, these
methods may be expensive and time-consuming, require large samples,
and produce hazardous substances during detection. Consequently,
there are no methods that are completely suited to replace the
direct sequencing method. As the current gold standard for clinical
detection, the direct sequencing method has several limitations due
to technical and methodological issues; however, it is accurate,
reliable and capable of detecting almost every mutation of EGFR.
The allele refractory mutation system (ARMS), a highly selective
and sensitive approach that can detect 29 types of EGFR mutation in
a reduced number of samples, has been applied in clinical and
laboratory settings. In the present study, ARMS was compared with
direct sequencing to identify which is the most appropriate method
for detection in the clinic.
Materials and methods
Sample collection
The present study was approved by the First
Affiliated Hospital of Wenzhou Medical University (Wenzhou, China).
Written informed consent was obtained from all patients. Samples
were collected from 1,596 patients with NSCLC who had been
diagnosed in The First Affiliated Hospital of Wenzhou Medical
University between July 2011 and June 2016. Exons 18–21 of the EGFR
genes were analyzed using the ARMS and direct sequencing methods.
All samples were successfully analyzed using the ARMS method,
whereas ~1,140 were successfully analyzed using the direct
sequencing method (direct sequencing failed for 91 samples due to
technical issues, and 365 samples were of insufficient quantity for
direct sequencing). There were 1,329 adenocarcinoma samples, 217
squamous cell carcinoma samples and 19 adenosquamous cell carcinoma
samples; the pathology type of 31 samples was unknown. There were
922 male and 674 female patients, with an age range of 18–89 and a
mean age of 64 years.
DNA extraction
The samples were collected using 3 different
methods: Biopsy, surgical resection and hydrothoracic or ascitic
fluid extraction. The samples were paraffin-embedded and DNA was
extracted and purified using the AmoyDx® FFPE DNA
Extraction kit (cat. no. ADx-FF01; Amoy Diagnostics Co., Ltd.,
Xiamen, China) according to the manufacturer's instructions.
EGFR gene analysis with ARMS
Following the determination of the DNA
concentration, by ultraviolet spectrophotometry, EGFR gene
mutations were analyzed using the AmoyDx® EGFR 29
Mutations Detection kit (cat. no. ADx-EG0X; Amoy Diagnostics Co.,
Ltd.) according to the manufacturer's instructions on a
LightCycler480 II (Roche Diagnostics, Basel, Switzerland).
Fluorescence calibration was performed on a LightCycler480 I (Roche
Diagnostics).
The results of the mutation assay were analyzed
based on different mutant Cq values (Table I).
 | Table I.Definition of Cq
thresholds for strong positive, weak positive and negative EGFR
mutation detection status. |
Table I.
Definition of Cq
thresholds for strong positive, weak positive and negative EGFR
mutation detection status.
| Strong
positive | Strong
positive | 19Del | L858R | T790M | Insertions | G791X | S786I | L861Q |
|---|
| Strong
positive | Mutant
Cq value |
Cq<26 |
Cq<26 |
Cq<26 |
Cq<26 |
Cq<26 |
Cq<26 |
Cq<26 |
| Weak positive | Mutant
Cq value |
26≤Cq<29 |
26≤Cq<29 |
26≤Cq<29 |
26≤Cq<29 |
26≤Cq<29 |
26≤Cq<29 |
26≤Cq<29 |
|
| ΔCq
threshold value | 12 | 11 | 7 | 9 | 7 | 8 | 8 |
| Negative | Mutant
Cq value |
Cq≥29 |
Cq≥29 |
Cq≥29 |
Cq≥29 |
Cq≥29 |
Cq≥29 |
Cq≥29 |
Strong positive
If the sample Cq value was <26, then
the sample was classified as strong positive.
Weak positive
If the sample Cq value ranged between 26
and 29, the sample was provisionally classified as weak positive
and the ΔCq of the reaction tube was calculated to
confirm the result. If the ΔCq value was less than the
corresponding threshold value of ΔCq, the sample was
confirmed as weak positive. If the ΔCq value was greater
than the cut-off ΔCq value, the sample was classified as
negative or below the detection limit of the kit. ΔCq
was calculated as follows: ΔCq = mutant Cq
value - external control Cq value.
Negative
If the sample amplification signal Cq
value was ≥ the critical negative value presented in the ‘Negative’
row in Table I, then the sample was
classified as negative or below the detection limit of the kit.
DNA direct sequencing
A total of 1,140 samples-examined using direct
sequencing method were examined using the direct sequencing method
for comparison with the ARMS method. The primers were designed by
Shanghai Shenggong Biology Engineering Technology Service, Ltd.
(Shanghai, China; Table II). A
BigDye™ Terminator v1.1 Cycle Sequencing kit was applied in the
detection, the total reaction system contained 10 ng of purified
PCR product, 8 µl of BigDye (2.5X; Beijing Think-Far Technology
Co., Ltd., Beijing, China), 3.2 pmol of primers and 10 µl aseptic
deionized water. The PCR reaction conditions included an initial
denaturation at 95°C for 1 min, followed by 25 cycles at 94°C for
10 sec and 50°C for 5 sec, then 60°C for 4 min. Subsequently, the
products were purified using ExoSap-IT reagent (Thermo Fisher
Scientific, Inc., Waltham, MA, USA), and the reaction was
terminated using BigDye Terminator v1.1 (cat. no. 4337449; Beijing
Think-Far Technology Co., Ltd.) according to the manufacturer's
instructions. All sequence data was analyzed using Sequencher
4.6® (Gene Codes Corporation, Ann Arbor, MI, USA) and
all positive results were detected a second time for
confirmation.
 | Table II.Primer sequences for direct
sequencing by polymerase chain reaction. |
Table II.
Primer sequences for direct
sequencing by polymerase chain reaction.
| Target | Sequence |
|---|
| Exon 18 |
|
| Sense
(5′-3′) |
TGTCCTGGCACCCAAGCCCA |
|
| TGCCGTGGCT |
|
Antisense (5′-3′) |
GTGGGGAGCCCAGAGTCCTT |
|
| GCAAGCTGTATA |
| Exon 19 |
|
| Sense
(5′-3′) |
CAGTGTCCCTCACCTTCGGG |
|
| GTGCATCGCT |
|
Antisense (5′-3′) |
AACCTCAGGCCCACCTTTT |
|
| CTCATGTCTGG |
| Exon 20 |
|
| Sense
(5′-3′) |
GCCCTGCGTAAACGTCCCTG |
|
| TGCTAGGTCT |
|
Antisense (5′-3′) |
CACATGCGGTCTGCGCTCCT |
|
| GGGATAGCAA |
| Exon 21 |
|
| Sense
(5′-3′) |
CCATTCTTTGGATCAGTAGT |
|
| CACTAACGT |
|
Antisense (5′-3′) |
CCAGGCTGCCTTCCCACTAG |
|
| CTGTATTG |
Data analysis
Following data collection and tissue classification,
a corresponding line chart was constructed according to the year of
tissue collection (Fig. 1; Cohen's κ
coefficient was calculated using SPSS (version 16.0 for Windows;
SPSS, Inc., Chicago, IL, USA) to examine the consistency of the
detection results between ARMS and direct sequencing. A κ-value
between 0.40 and 0.75 was considered to represent a good
consistency level, and a κ-value >0.75 was considered to
represent a high consistency level. Subsequently, the influences of
different sample types on consistency and the association between
EGFR gene mutation with age and sex were examined using the
χ2 test in SPSS (version 16.0 for Windows; SPSS, Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference. GraphPad Prism software
(version 5.0; GraphPad Software, Inc., La Jolla, CA, USA) was used
to produce the figures.
Results
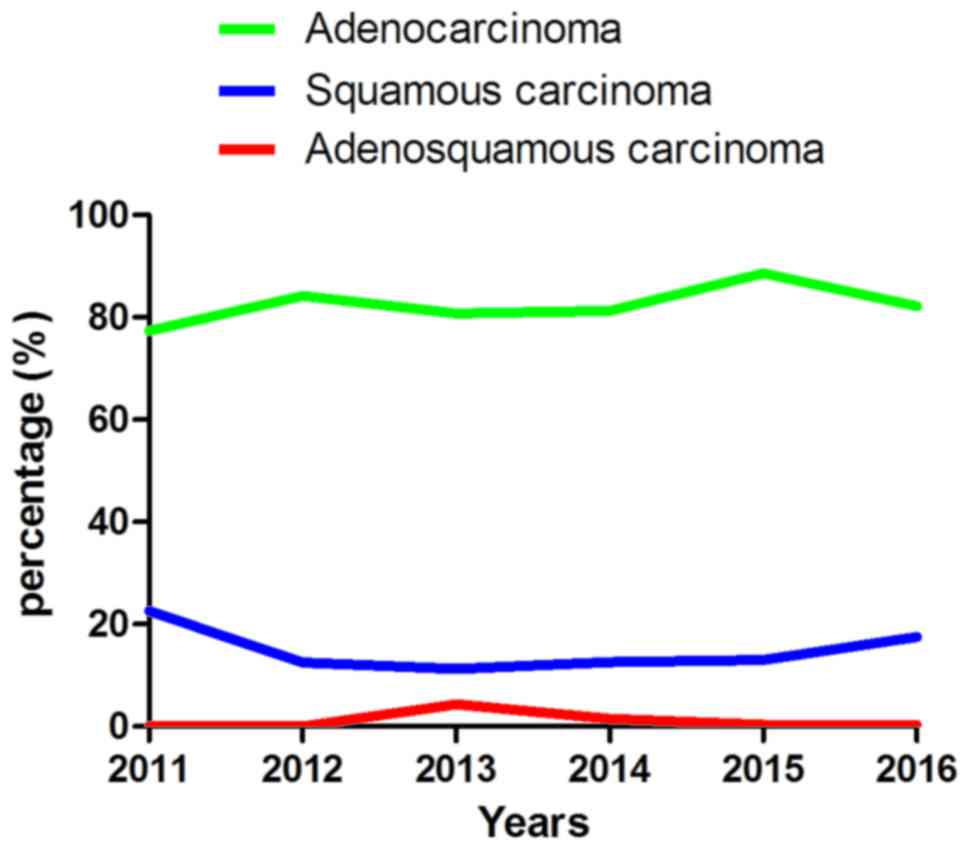
Analysis of the patients diagnosed
with NSCLC
The patients diagnosed in The First Affiliated
Hospital of Wenzhou Medical University were classified into three
types according to their pathology, including adenocarcinoma,
squamous cell carcinoma and adenosquamous cell carcinoma (Fig. 1; Table
III). Adenocarcinoma was the most common and squamous cell
carcinoma second most common, followed by adenosquamous cell
carcinoma. It was also identified that the total number of NSCLC
cases exhibited an increasing incidence, with fewer patients
diagnosed in 2011 than 2016. Data was only collected until June
2016 and the data for 2016 in Fig. 1
was extrapolated from previous years; the tendency towards an
increasing incidence rate may be more apparent from the actual
data.
 | Table III.Summary of the pathology type of all
patients with non-small cell lung cancer diagnosed at The First
Affiliated Hospital of Wenzhou Medical University over 6 years. |
Table III.
Summary of the pathology type of all
patients with non-small cell lung cancer diagnosed at The First
Affiliated Hospital of Wenzhou Medical University over 6 years.
|
| Years, n (%) |
|
|---|
|
|
|
|
|---|
| Pathology type | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | Total, n (%) |
|---|
| Adenocarcinoma | 24 (77.4) | 128 (84.21) | 164 (80.8) | 322 (81.3) | 433 (88.6) | 258 (82.2) | 1,329 (83.2) |
| Squamous cell
carcinoma | 7 (22.6) | 19 (12.5) | 23 (11.3) | 50 (12.6) | 65 (13.0) | 55 (17.5) | 219 (13.7) |
| Adenosquamous cell
carcinoma | 0 (0) | 1 (0.07) | 9 (4.4) | 6 (1.5) | 2 (0.4) | 1 (0.3) | 19 (1.2) |
| Other types | 0 (0) | 4 (2.63) | 7 (3.4) | 18 (4.5) | 0 (0.00) | 0 (0) | 29 (1.7) |
| Total | 31 | 152 | 203 | 396 | 500 | 314 | 1,596 |
Comparison of the detection outcome
between ARMS and direct sequencing
From 1,596 patients with NSCLC, 1,596 cases were
analyzed using ARMS, but direct sequencing was successful for 1,140
samples (in the failed cases, exons 19–21 were complete in 22
samples, partially complete in 48 cases, and insufficient in 2
cases; Table IV). Among the 1,140
cases analyzed with both methods, 1,017 samples had consistent
detection outcomes between the two methods; of the inconsistent
samples, 77 cases were detected as wild-type individuals using the
direct sequencing method and subsequently detected as mutants using
ARMS, whereas 46 samples were identified as wild-type by ARMS and
mutants by direct sequencing. Thus, the consistency was adequate,
with a concordance rate of 89.21% (P=0.007, κ=0.775; Table II). Furthermore, the positive rate of
ARMS (40.96%) was marginally increased compared with that obtained
with direct sequencing (38.25%).
 | Table IV.Consistency of ARMS and direct
sequencing methods for different sample types. |
Table IV.
Consistency of ARMS and direct
sequencing methods for different sample types.
|
| Sample type |
|---|
|
|
|
|---|
| Outcome | Biopsy | Surgical | Cytological | Total |
|---|
| Consistent | 659 | 271 | 77 | 1,007 |
| Inconsistent | 88 | 24 | 11 | 122 |
| ARMS(−)
sequencing(+) | 33 | 9 | 4 | 45 |
| ARMS(+)
sequencing(−) | 55 | 15 | 7 | 77 |
| Direct sequencing
failure | 34 | 25 | 9 | 68 |
EGFR mutation rate and the association
between mutations and population characteristics
EGFR mutation in all disease
subtypes
A total of 12 types of mutation were detected in the
present study, including 3 (G719A, G719S and G719C) in exon 18, 4
[exon 19 deletion (19Del), L747-T751Del, L747-A750>P and
E746-S752>V] in exon 19, 3 (T790M, S768I and an unknown mutation
accounting for 14 cases) in exon 20, and 2 (L858R and L861Q) in
exon 21. A combination of mutations was observed in a number of
cases (Table V); the mutations in 25
cases were caused by previously unidentified DNA alterations
(Table VI). Among the 1,596 cases,
mutations in EGFR were identified in 727 cases; thus, the total
mutation rate was 45.55% (727/1,596). The mutations 19Del and L858R
were the most common, accounting for 319 and 320 cases
respectively, with mutation rates of 43.88% (319/727) and 44.02%
(320/727) respectively. Lung adenocarcinoma and squamous cell
carcinoma had mutation rates of 51.77 (688/1,329) and 8.68%
(19/219) respectively. As lung adenocarcinoma and squamous cell
carcinoma were the dominant types of NSCLC in the data, further
analysis considered these subtypes separately.
 | Table V.Frequency of simultaneous
mutations. |
Table V.
Frequency of simultaneous
mutations.
| Mutation type | Cases, n |
|---|
| 19Del + L858R | 13 |
| 19Del + T790M | 8 |
| L858R + T790M | 8 |
| 20Ins + L858R | 1 |
| G719X + L861Q | 4 |
| G719 + S768I | 2 |
 | Table VI.Rare mutations detected in 25
patients using the direct sequencing method. |
Table VI.
Rare mutations detected in 25
patients using the direct sequencing method.
| Mutation type | Cases |
|---|
| 2572 C>A | 1 |
| 2361 G>A | 16 |
| GCGTGGACA | 1 |
| 2571 G>A | 1 |
| T751-I759>N | 1 |
| 2240-2252Del | 1 |
| 2237-2250>T | 1 |
| 2240-2257Del | 1 |
| 2253-2276Del | 1 |
| 2259 G>A | 1 |
EGFR mutation in lung
adenocarcinoma
As presented in Table
VII, lung adenocarcinoma accounted for 1,329 cases, of which
688 cases, including 283 male and 405 female patients, exhibited
EGFR mutations. The difference in the mutation rate between male
and female patients with adenocarcinoma was significant
(P<0.001); the mutation rate in female patients (65.53%,
405/618) was distinctly increased compared with that in males
(39.80%, 283/711). However, there was no difference in the mutation
rate between patients ≥60 and <60 (P=0.145).
 | Table VII.Characteristics and EGFR gene
mutation rate of patients with lung adenocarcinoma. |
Table VII.
Characteristics and EGFR gene
mutation rate of patients with lung adenocarcinoma.
|
|
EGFR gene
mutation type, n |
|
|
|
|---|
|
|
|
|
|
|
|---|
| Characteristic | 19Del | L858R | T79M | 19Del + L858R | 19Del + T79M | L858R + T79M | 20Ins | 20Ins + L858R | G719× | L861Q | G719× + L861Q | G719× + S768I | Detected, n | Total, n | Mutation rate,
% |
|---|
| Total | 312 | 310 | 1 | 12 | 9 | 7 | 10 | 1 | 9 | 11 | 4 | 2 | 688 | 1,329 | 51.77 |
| Sex |
|
Male | 126 | 127 | 1 | 8 | 6 | 2 | 4 | 0 | 5 | 4 | 2 | 1 | 283 | 711 | 39.8 |
|
Female | 186 | 183 | 0 | 4 | 3 | 5 | 6 | 1 | 4 | 7 | 2 | 1 | 405 | 618 | 65.53a |
| Age, years |
|
<60 | 115 | 105 | 1 | 3 | 6 | 2 | 7 | 0 | 4 | 1 | 0 | 1 | 245 | 449 | 54.57 |
|
≥60 | 197 | 205 | 0 | 9 | 3 | 5 | 3 | 1 | 5 | 10 | 4 | 1 | 443 | 870 | 50.92b |
EGFR mutation in squamous cell
carcinoma
Similar results were also observed in squamous cell
carcinoma (Table VIII); among 219
squamous cell carcinoma cases, including 187 male and 32 female
patients, there was a significant difference in the mutation rate
between male (5.35%, 10/187) and female (28.13%, 9/32) patients
(P<0.001), whereas there was no difference between patients ≥60
and <60 (P=1.00).
 | Table VIII.Characteristics and EGFR gene
mutation rate of patients with squamous cell carcinoma. |
Table VIII.
Characteristics and EGFR gene
mutation rate of patients with squamous cell carcinoma.
|
| EGFR gene mutation,
n |
|
|
|---|
|
|
|
|
|
|---|
| Characteristic | 19Del | L858R | 19Del + L858R | 19Del - T790M | 20Ins | Total | Detected
number | Mutation rate,
% |
|---|
| Sex |
|
Male | 5 | 5 | 0 | 0 | 0 | 10 | 187 | 5.35a |
|
Female | 2 | 5 | 0 | 0 | 2 | 9 | 32 | 28.13 |
| Age, years |
|
<60 | 2 | 2 | 0 | 0 | 0 | 4 | 59 | 6.78b |
|
≥60 | 5 | 8 | 0 | 0 | 2 | 15 | 160 | 9.38 |
| Total | 7 | 10 | 0 | 0 | 2 | 19 | 219 | 8.68 |
EGFR mutation in adenosquamous cell
carcinoma
From a total of 19 adenosquamous cell carcinoma
cases, 9 exhibited EGFR mutations, including 4 cases with 19Del, 4
cases with L858R and 1 case with both 19Del and L858R.
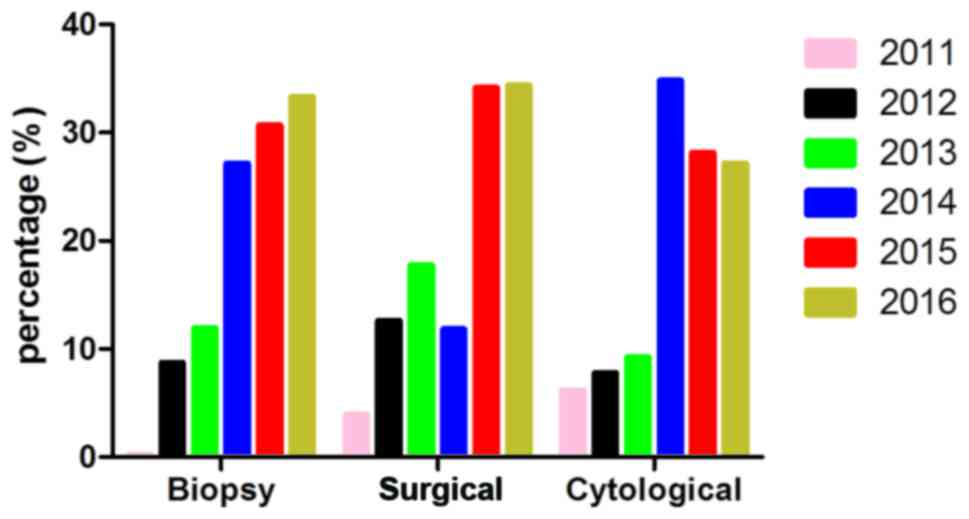
Associations between sample type and
detection methods
The sample types in all cases were classified into 5
types according to their origin and extraction method (Fig. 2). Paraffin-embedded biopsy sections
accounted for 939 cases; paraffin-embedded samples from surgical
resection accounted for 372 cases; paraffin-embedded hydrothorax or
ascitic fluid samples accounted for 270 cases; fresh tissue samples
accounted for 9 cases; and 6 samples were of unknown origin.
Samples in 2014, 2015, 2016 year presented an obvious increasing
tendency (samples in 2016 only collected up to June), in which
paraffin-embedded biopsy sections were the majority in every year,
making up ~2/3 of all samples (939/1596). No differences were
observed in the consistency between sample types (P=0.052).
EGFR mutation characteristics of
retrospective cases
A total of 29 cases were re-examined. Among these
cases, 1 case revealed a different pathological type, from
adenocarcinoma to adenosquamous cell carcinoma. A total of 9 cases
exhibited mutation type variation; 1 case changed from all exons
intact to 19Del, whereas 1 case changed from 19Del to wild-type,
and 7 cases exhibited multiple mutations whereas they were
originally detected as single-mutation or wild-type samples
(Table IX). The 29 samples were
classified depending on the consistency and whether the sample type
in the first instance was consistent with the second. The results
were statistically insignificant (Table
X), suggesting that different gene detection results did not
result from the different sample types.
 | Table IX.Results of the re-inspection of 9
cases. |
Table IX.
Results of the re-inspection of 9
cases.
| Detection
instance | Epidermal growth
factor receptor mutation type |
|---|
| First | 19Del | WT | WT | L858R | 19Del |
| Second | WT | 19Del | 19Del + T790M | L858R + T790M | 19Del + T790M |
| Cases, n | 1 | 1 | 2 | 3 | 2 |
 | Table X.Influence of different sample type on
the consistency of the result over multiple assessments. |
Table X.
Influence of different sample type on
the consistency of the result over multiple assessments.
|
| Consequence, n |
|
|---|
|
|
|
|
|---|
| Sample types | Same outcome | Different
outcome | Total |
|---|
| Total | 20 | 9 | 29 |
| Same type | 10 | 5 | 15 |
| Different type | 10 | 4 | 14 |
Discussion
Reflecting the heterogeneity of NSCLC, molecularly
targeted therapy requires that different patients should receive
different treatment strategies. Patients with EGFR mutations
respond well to tyrosine kinase inhibitors, whereas patients with
wild-type EGFR genes respond poorly (17–19). Chu
et al (20) demonstrated that
the rate of EGFR-TKI effectiveness against tumors with EGFR
mutations (80%) was increased compared with tumors with EGFR
wild-type (12.5%). Gefitinib may also be a more optimal treatment
for tumors with common EGFR mutations than for tumors with rare
EGFR mutations based on progression-free and overall survival (OS)
(21). In addition, the improvement
in OS is more evident for first-line afatinib than conventional
chemotherapy for tumors with EGFR exon 19 mutations, whereas this
does not occur for tumors with exon 21 mutations (17). Thus, an efficient, economic and
convenient detection method for EGFR mutations may assist in the
development of personalized treatment regimes.
In the present study, two methods were applied to
detect EGFR gene mutations. Direct sequencing was perceived as a
standard reference method to compare with ARMS; however, direct
sequencing is hyposensitive and time consuming (~2 days for one
detection), and these shortcomings prevent its widespread
application in clinical and laboratory settings. On the contrary,
ARMS is a time-saving method with high sensitivity (30% vs. 1%)
(22).
The number of patients diagnosed with NSCLC at The
First Affiliated Hospital of Wenzhou Medical University increased
each year between 2011 and 2016. This may have been caused by a
deteriorating living environment, including haze, sand storms and
occupational exposure, and unhealthy lifestyle factors, including
smoking. The increasing diagnosis rates may also result from
improved detection techniques. Among the 1,596 cases, the total
EGFR mutation frequency was 45.55% (727 cases), a frequency
increased compared with that observed by Ueno et al
(23) (33%), possibly as a result of
the different sex distribution of the patients or the misdiagnosis
of squamous cell carcinoma.
Among patients with adenocarcinoma, the low
proportion of female patients (male/female, 707/608) may have
resulted in an EGFR mutation frequency which was reduced relative
to other studies, including those by Wang et al (24) (63.1% vs. 51.77% in the present study).
The EGFR genes of female patients with adenocarcinoma were more
susceptible to mutation in a previous study (25), which was consistent with the present
study (female vs. male patients, P<0.01). It has been
hypothesized that women are more sensitive to the carcinogens from
cigarette smoke (26), whereas a
number of studies conclude that there is no association between
smoking status and EGFR gene mutations in lung adenocarcinoma
(27,28). Although a number of previous studies
have reported the inverse outcome, non-smoking patients develop
adenocarcinoma with a higher EGFR gene mutation frequency (25,29–31). We
hypothesize that the target gene for the carcinogens in tobacco is
not EGFR, but an as-yet unidentified gene.
Squamous cell carcinoma, a type of NSCLC with the
highest morbidity after adenocarcinoma, exhibited a relatively high
EGFR mutation frequency in the present study (15.55%). The results
were consistent with a previous study that identified a higher
mutation frequency in female patients compared with male patients
with squamous cell carcinoma (23).
Furthermore, Ueno et al and Chen et al (23,32)
concluded that there was no distinct association between EGFR
mutation frequency and age; the results of the present study were
consistent with this hypothesis. Among the 1,596 patients, 19
patients presented with adenosquamous cell carcinoma, a
pathologically mixed tumor of cells from both adenocarcinoma and
squamous cell carcinoma. Due to its rarity, there are a low number
of reports concerning this phenomenon in terms of its EGFR gene
mutation status (33–35), and it remains unknown whether
EGFR-TKIs are suitable for the treatment of patients with
adenosquamous cell carcinoma with mutant EGFR or normal EGFR
genes.
There was consistency (89.21%) between the standard
reference method of direct sequencing and ARMS among the 1,140
cases detected using both methods. Additionally, the direct
sequencing method has relatively low sensitivity (36,37),
requiring 200 ng of DNA and a mutation in >20% of the sample
(38), whereas ARMS may detect 1%
mutant DNA in a background of 99% normal DNA in a 10 ng DNA sample.
The existing ARMSDx® EGFR 29 Mutations Detection kit was
able to detect 29 types of mutation and amplified only the mutant
sequences, demonstrating that the kit is more sensitive than direct
sequencing, which amplifies the whole sequence of the target gene
(39). Thus, ARMS may be a viable
alternative for patients who have not undergone surgery, as a
smaller sample is required.
We hypothesized that the optimal sample type for the
two methods may differ, as different sample types may suit
different detection methods. Therefore, a χ2 test was
used to detect discrepancies in formalin-fixed paraffin-embedded
tissue samples from biopsy and surgery, which accounted for ~1/2 of
all samples. The outcome revealed a trending, but non-significant
tendency towards a difference in the accuracy of biopsy and surgery
materials between methods (P=0.052). Theoretically, the samples
obtained from surgery include more tissue compared with biopsy
samples. The process of fixing samples with formalin and embedding
in paraffin may cause DNA degradation (40), which may cause a decreased
detectability in samples from biopsy when using a sequencing
method, due to the reduced availability of tissue or DNA. However,
this tendency was not reflected in the present study, possibly due
to the relatively small size of surgically collected samples.
The ARMS method is currently used for routine
clinical detection, treatment guidelines and laboratory research;
it is associated with relatively rapid processing, accuracy,
decreased cost and decreased sample size demand compared with
direct sequencing. However, certain drawbacks of the estimation of
mutation status by ARMS were identified in the present study, as 33
cases that were identified as wild-type using ARMS were identified
as EGFR mutation-positive with direct sequencing, whereas 55 cases
revealed the opposite result, likely reflective of insufficient
sample volume for direct sequencing. It was also verified that the
T790M mutation, which is associated with drug resistance (41,42), was
more likely to be detected in the patients subsequent to treatment,
with 7 of the 9 cases re-tested exhibiting the T790M mutant.
The present study verified the results of previous
studies using large amounts of data, which comparatively expound on
the application of ARMS for EGFR mutant detection. The deficiency
of the present study is the lack of additional research in
treatment and prognosis, which would generate more promising
results.
Acknowledgements
The present study was supported by grants from the
Natural Science Foundation of Zhejiang Province (grant no.
LY16H160047), the Scientific Research Foundation of Wenzhou,
Zhejiang Province, China (grant no. Y20130073), the National
Natural Sciences Foundation of China (grant nos. 81201589 and
81472651) and the Health and Family Planning Commission of Zhejiang
Province (grant no. 2013ZDA014).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistic, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: Epidemiology, risk
factors, treatment and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou C, Wu YL, Chen G, Feng J, Liu XQ,
Wang C, Zhang S, Wang J, Zhou S, Ren S, et al: Erlotinib versus
chemotherapy as first-line treatment for patients with advanced
EGFR mutation-positive non-small-cell lung cancer (OPTIMAL,
CTONG-0802): A multicentre, open-label, randomised, phase 3 study.
Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang Y, Wang WY, Zheng K, Jiang L, Zou Y,
Su XY, Chen J, Zhang WY and Liu WP: EGFR mutations in non-small
cell lung cancer: An audit from West China Hospital. Expert Rev Mol
Diagn. 16:915–919. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Minna JD, Fong K, Zöchbauer-Müller S and
Gazdar AF: Molecular pathogenesis of lung cancer and potential
translational applications. Cancer J. 8(Suppl 1): S41–S46.
2002.PubMed/NCBI
|
|
6
|
Villaflor V, Won B, Nagy R, Banks K,
Lanman RB, Talasaz A and Salgia R: Biopsy-free circulating tumor
DNA assay identifies actionable mutations in lung cancer.
Oncotarget. 7:66880–66890. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Olayioye MA, Neve RM, Lane HA and Hynes
NE: The ErbB signaling network: Receptor heterodimerization in
development and cancer. EMBO J. 19:3159–3167. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Charpidou A, Blatza D, Anagnostou V and
Syrigos KN: Review. EGFR mutations in non-small cell lung
cancer-clinical implications. In vivo. 22:529–536. 2008.
|
|
9
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: Gefitinib or chemotherapy for non-small-cell lung cancer
with mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mitsudomi T, Morita S, Yatabe Y, Negoro S,
Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et
al: Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): An open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han JY, Park K, Kim SW, Lee DH, Kim HY,
Kim HT, Ahn MJ, Yun T, Ahn JS, Suh C, et al: First-SIGNAL:
First-line single-agent iressa versus gemcitabine and cisplatin
trial in never-smokers with adenocarcinoma of the lung. J Clin
Oncol. 30:1122–1128. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang CC, Chao KH, Chen YL, Chang JG and Wu
SM: Capillary electrophoretic genotyping of epidermal growth factor
receptor for pharmacogenomic assay of lung cancer therapy. J
Chromatogra A. 1256:276–279. 2012. View Article : Google Scholar
|
|
13
|
Liu W, Smith DI, Rechtzigel KJ, Thibodeau
SN and James CD: Denaturing high performance liquid chromatography
(DHPLC) used in the detection of germline and somatic mutations.
Nucleic Acids Res. 26:1396–1400. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Do H, Krypuy M, Mitchell PL, Fox SB and
Dobrovic A: High resolution melting analysis for rapid and
sensitive EGFR and KRAS mutation detection in formalin fixed
paraffin embedded biopsies. BMC Cancer. 8:1422008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tuononen K, Maki-Nevala S, Sarhadi VK,
Wirtanen A, Rönty M, Salmenkivi K, Andrews JM, Telaranta-Keerie AI,
Hannula S, Lagström S, et al: Comparison of targeted
next-generation sequencing (NGS) and real-time PCR in the detection
of EGFR, KRAS and BRAF mutations on formalin-fixed,
paraffin-embedded tumor material of non-small cell lung
carcinoma-superiority of NGS. Genes Chromosomes Cancer. 52:503–511.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Marchetti A, Martella C, Felicioni L,
Barassi F, Salvatore S, Chella A, Camplese PP, Iarussi T, Mucilli
F, Mezzetti A, et al: EGFR mutations in non-small-cell lung cancer:
Analysis of a large series of cases and development of a rapid and
sensitive method for diagnostic screening with potential
implications on pharmacologic treatment. J Clin Oncol. 23:857–865.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang JC, Wu YL, Schuler M, Sebastian M,
Popat S, Yamamoto N, Zhou C, Hu CP, O'Byrne K, Feng J, et al:
Afatinib versus cisplatin-based chemotherapy for EGFR
mutation-positive lung adenocarcinoma (LUX-Lung 3 and LUX-Lung 6):
Analysis of overall survival data from two randomised, phase 3
trials. Lancet Oncol. 16:141–151. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rosell R, Carcereny E, Gervais R,
Vergnenegre A, Massuti B, Felip E, Palmero R, Garcia-Gomez R,
Pallares C, Sanchez JM, et al: Erlotinib versus standard
chemotherapy as first-line treatment for European patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(EURTAC): A multicentre, open-label, randomised phase 3 trial.
Lancet Oncol. 13:239–246. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang
Y, Li W, Hou M, Shi JH, Lee KY, et al: Afatinib versus cisplatin
plus gemcitabine for first-line treatment of Asian patients with
advanced non-small-cell lung cancer harbouring EGFR mutations
(LUX-Lung 6): An open-label, randomised phase 3 trial. Lancet
Oncol. 15:213–222. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chu H, Zhong C, Xue G, Liang X, Wang J,
Liu Y, Zhao S, Zhou Q and Bi J: Direct sequencing and amplification
refractory mutation system for epidermal growth factor receptor
mutations in patients with non-small cell lung cancer. Oncol Rep.
30:2311–2315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Watanabe S, Minegishi Y, Yoshizawa H,
Maemondo M, Inoue A, Sugawara S, Isobe H, Harada M, Ishii Y, Gemma
A, et al: Effectiveness of gefitinib against non-small-cell lung
cancer with the uncommon EGFR mutations G719X and L861Q. J Thorac
Oncol. 9:189–194. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Y, Liu B, Li XY, Li JJ, Qin HF, Tang
CH, Guo WF, Hu HX, Li S, Chen CJ, et al: A comparison of ARMS and
direct sequencing for EGFR mutation analysis and tyrosine kinase
inhibitors treatment prediction in body fluid samples of
non-small-cell lung cancer patients. J Exp Clin Cancer Res.
30:1112011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ueno T, Toyooka S, Suda K, Soh J, Yatabe
Y, Miyoshi S, Matsuo K and Mitsudomi T: Impact of age on epidermal
growth factor receptor mutation in lung cancer. Lung cancer.
78:207–211. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang R, Zhang Y, Pan Y, Li Y, Hu H, Cai D,
Li H, Ye T, Luo X, Zhang Y, et al: Comprehensive investigation of
oncogenic driver mutations in Chinese non-small cell lung cancer
patients. Oncotarget. 6:34300–34308. 2015.PubMed/NCBI
|
|
25
|
Chen ZY, Zhong WZ, Zhang XC, Li Y, Hu H,
Cai D, Li H, Ye T, Luo X, Zhang Y, et al: EGFR mutation
heterogeneity and the mixed response to EGFR tyrosine kinase
inhibitors of lung adenocarcinomas. Oncologist. 17:978–985. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
International Early Lung Cancer Action
Program I; Henschke CI, Yip R and Miettinen OS: Women's
susceptibility to tobacco carcinogens and survival after diagnosis
of lung cancer. JAMA. 296:180–184. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bain C, Feskanich D, Speizer FE, Thun M,
Hertzmark E, Rosner BA and Colditz GA: Lung cancer rates in men and
women with comparable histories of smoking. J Natl Cancer Inst.
96:826–834. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chan-Yeung M and Dimich-Ward H:
Respiratory health effects of exposure to environmental tobacco
smoke. Respirology. 8:131–139. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Greenhalgh J, Dwan K, Boland A, Bates V,
Vecchio F, Dundar Y, Jain P and Green JA: First-line treatment of
advanced epidermal growth factor receptor (EGFR) mutation positive
non-squamous non-small cell lung cancer. Cochrane Database Syst
Rev. 25:CD0103832016.
|
|
30
|
Jida M, Toyooka S, Mitsudomi T, Takano T,
Matsuo K, Hotta K, Tsukuda K, Kubo T, Yamamoto H, Yamane M, et al:
Usefulness of cumulative smoking dose for identifying the EGFR
mutation and patients with non-small-cell lung cancer for gefitinib
treatment. Cancer Sci. 100:1931–1934. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Midha A, Dearden S and McCormack R: EGFR
mutation incidence in non-small-cell lung cancer of adenocarcinoma
histology: A systematic review and global map by ethnicity
(mutMapII). Am J Cancer Res. 5:2892–2911. 2015.PubMed/NCBI
|
|
32
|
Chen YM, Lai CH, Rau KM, Huang CH, Chang
HC, Chao TY, Tseng CC, Fang WF, Chen YC, Chung YH, et al: Advanced
non-Small cell lung cancer patients at the extremes of age in the
era of epidermal growth factor receptor tyrosine kinase inhibitors.
Lung cancer. 98:99–105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Filosso PL, Ruffini E, Asioli S, Giobbe R,
Macri L, Bruna MC, Sandri A and Oliaro A: Adenosquamous lung
carcinomas: A histologic subtype with poor prognosis. Lung Cancer.
74:25–29. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gawrychowski J, Brulinski K, Malinowski E
and Papla B: Prognosis and survival after radical resection of
primary adenosquamous lung carcinoma. Eur J Cardiothorac Surg.
27:686–692. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Riquet M, Perrotin C, Lang-Lazdunski L,
Hubsch JP, Dujon A, Manac'h D, Le Pimpec Barthes F and Briere J: Do
patients with adenosquamous carcinoma of the lung need a more
aggressive approach? J Thorac Cardiovasc Surg. 122:618–619. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li J, Wang L, Mamon H, Kulke MH, Berbeco R
and Makrigiorgos GM: Replacing PCR with COLD-PCR enriches variant
DNA sequences and redefines the sensitivity of genetic testing.
Nature medicine. 14:579–584. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ellison G, Donald E, McWalter G, Knight L,
Fletcher L, Sherwood J, Cantarini M, Orr M and Speake G: A
comparison of ARMS and DNA sequencing for mutation analysis in
clinical biopsy samples. J Exp Clin Cancer Res. 29:1322010.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li C, Wu J, Wang Z and Feng J: A
comparison of direct sequencing and ARMS assay performance in EGFR
mutation analysis of non-small cell lung cancer patients. Zhongguo
Fei Ai Za Zhi. 17:606–611. 2014.(In Chinese). PubMed/NCBI
|
|
39
|
Jiang J, Wang C, Yu X, Sheng D, Zuo C, Ren
M, Wu Y, Shen J, Jin M and Xu S: PCR-sequencing is a complementary
method to amplification refractory mutation system for EGFR gene
mutation analysis in FFPE samples. Exp Mol Pathol. 99:581–589.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
von Ahlfen S, Missel A, Bendrat K and
Schlumpberger M: Determinants of RNA quality from FFPE samples.
PLoS One. 2:e12612007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hata A, Katakami N, Yoshioka H, Takeshita
J, Tanaka K, Nanjo S, Fujita S, Kaji R, Imai Y, Monden K, et al:
Rebiopsy of non-small cell lung cancer patients with acquired
resistance to epidermal growth factor receptor-tyrosine kinase
inhibitor: Comparison between T790M mutation-positive and
mutation-negative populations. Cancer. 119:4325–4332. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhu VW, Upadhyay D, Schrock AB, Gowen K,
Ali SM and Ou SH: TPD52L1-ROS1, a new ROS1 fusion variant in lung
adenosquamous cell carcinoma identified by comprehensive genomic
profiling. Lung cancer. 97:48–50. 2016. View Article : Google Scholar : PubMed/NCBI
|