Introduction
Hepatocellular carcinoma (HCC) is among the most
common types of malignant tumor, and ranked fifth globally in terms
of incidence. It is also the third most common cause of
cancer-associated mortality worldwide (1). HCC has an incidence of ~466,100/year in
China, according to a 2015 study by The National Central Cancer
Registry of China (2). Surgical
resection is the first-line treatment for patients with early-stage
HCC, which can be curative (3,4). However,
the rate of metastasis and recurrence following surgery remains
high, which is a major obstacle for improving the clinical outcome
of patients with HCC (5). Evidence
suggests that tumor recurrence and metastasis are associated with
cancer stem cells (CSCs) in various types of cancer, including HCC
(6). CSCs are reported to have
inherent tumor initiating potential, serving important roles in
tumor relapse, driving primary tumor growth, and the seeding and
establishment of metastases (7).
Recently, CSCs have been investigated as a therapeutic target.
Clinical observations and experimental data have
indicated heterogeneity among cancer cell lines in terms of
aggressive potential, suggesting that CSCs from different sourced
may have varying migration ability (8–10). A
previous study demonstrated that the percentage of CSCs was
positively correlated with the metastatic potential of the parent
cell line (11). However, it remains
to be determined whether CSCs derived from different cell
lines/tumors possess different metastatic capabilities. Side
population (SP) cells, are a subset of cells with the ability to
efflux Hoechst 33342 in flow cytometry through an ATP-binding
cassette (ABC) membrane transporter, and were first described by
Goodell et al (12) in the
bone marrow. SP cells isolated from various cancer cell lines have
been demonstrated to exhibit stem cell-like properties (13–16). In
the present study, SP cells were employed as a model to study the
molecular differences in the metastatic potential of CSCs derived
from different cell lines.
High-throughput quantitative proteomic technologies
provide a powerful tool for systematically characterizing the
overall proteome alterations underlying physiological or
pathological changes. Isobaric tags for relative and absolute
quantification (iTRAQ) is an ultrasensitive and precise approach
for studying protein quantitative changes in ≤8 samples
simultaneously (17,18). Comparative proteomic approaches
coupled with iTRAQ are widely used to investigate the molecular
mechanisms of tumorigenesis, metastasis and recurrence of HCC
(19–21). iTRAQ-based quantitative study of
protein expression profiles between CSCs and their parental cell
lines have also been reported (22).
However, to the best of our knowledge, the application of iTRAQ
labeling in studying the molecular differences among CSCs from cell
lines with different metastatic potentials has not been previously
reported. In the present study, an iTRAQ based quantitative
proteomic approach was used to systematically compare the overall
proteome profiles among different SP cells to reveal the underlying
molecular mechanisms of HCC cell lines with different metastatic
potentials.
Materials and methods
Cell culture
The human HCC HCCLM3, MHCC97-H and MHCC97-L cell
lines were purchased from The Cell Bank of Type Culture Collection
of Chinese Academy of Science, Shanghai Institute for Biological
Sciences (Shanghai, China). The HCC cell line, Hep3B, was purchased
from the America Type Culture Collection (Manassas, VA, USA).
HCCLM3, MHCC97-H, MHCC97-L cells were cultured in high-glucose DMEM
containing 10% FBS, 100 U⁄ml penicillin and 100 µg⁄ml streptomycin
(all reagents from Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA). Hep3B was cultured in MEM (Gibco; Thermo Fisher
Scientific, Inc.) with 10% FBS. All cells were incubated at 37°C in
a humidified atmosphere containing 5% CO2.
Flow cytometry (FCM) analysis of SP
cells
The 4 cell lines were cultured to 80% confluence and
detached using 0.25% Trypsin-EDTA, then suspended in DMEM
supplemented with 3% FBS, at a density of 1×106
cells/ml. The cells were then incubated with 20 µg/ml Hoechst 33342
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) alone or with 25
µg/ml verapamil (Sigma-Aldrich; Merck KGaA) at 37°C for 90 min.
Verapamil was used as a guiding parameter to determine the boundary
between SP and main population (MP) cells. The samples were
centrifuged at 300 × g for 5 min at 4°C, and then re-suspended in
PBS supplemented with 3% FBS. Propidium iodide (PI; Sigma-Aldrich;
Merck KGaA) was added at 1 µg/ml to exclude analysis of any dead
cells. FCM analysis was performed using a Moflo XDP flow cytometer
(Beckman Coulter, Inc., Brea, CA, USA), as previously described
(23). Each assay was performed in
triplicate.
Sphere formation assay and soft agar
colony formation assay
For the sphere formation, SP and MP cells sorted
from the 4 cell lines were suspended separately in serum-free
DMEM/F12 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 20 ng/ml epidermal growth factor, 10 ng/ml basic
fibroblast growth factor and 10 µl/ml B27 (all from Gibco; Thermo
Fisher Scientific, Inc.). The cells were then plated into 6-well
UltraLow Attachment plates (Corning Incorporated, Corning, NY, USA)
at 2×103 cells/well. After 14 days, the number of
spheres were counted under a confocal microscope (magnification,
×50). For the soft agar colony formation assay, sorted SP and MP
cells were seeded into 6-well plates at 5×103 cells/well
in 0.3% agarose (Promega Corporation, Madison, WI, USA) over a 0.6%
agarose layer. After 14 days, the number of colonies were counted
under confocal microscope (magnification, ×50).
Cell migration assay
The migration assays were performed using Transwell
plates (pore diameter, 8-µm; Corning Incorporated), according to
the manufacturer's instructions. A total of 100 µl SP and MP cells
at 1×106 cells/ml were cultured in the upper chamber in
serum-free DMEM supplemented with 0.5% BSA. A total of 600 µl DMEM
supplemented with 10% FBS was added to the lower chamber. After 48
h, non-migrating cells on the upper surface of the filters were
removed with cotton swabs, and cells on the lower membrane of the
filters were fixed with 4% paraformaldehyde, and stained with 0.1%
hematoxylin (Beyotime Institute of Biotechnology, Haimen, China)
for 20 min at room temperature. The stained cells, which had passed
through the filter to the lower surface, were counted under a
confocal microscope in 5 randomly selected fields of view
(magnification, ×200).
Protein preparation and iTRAQ
labeling
A total of 500 µl lysis buffer [8 M urea
(Sigma-Aldrich, Merck KgaA), 50 mM NH4HCO3,
50 mM iodoacetamide (IAA) and 1X protease inhibitor cocktail (Roche
Diagnostics, Basel, Switzerland)] was added to the 4 types of SP
and MP cells, followed by sonication for 8 min with a period of 1
sec sonication and 5 sec pause at 190 W on ice. Following
centrifugation at 17,000 × g for 10 min at 4°C, the supernatant was
collected. The protein concentration of the supernatant was
determined by BCA assay (TransGen Biotech, Co., Ltd., Beijing,
China), according manufacturer's protocol. Then, 100 µg protein was
adjusted to a final volume of 100 µl in 100 mM triethylammonium
bicarbonate (TEAB). A total of 8 µl DTT (1M) was added to the
protein samples and incubated at 55°C for 1 h, followed by the
addition of 10 µl 500 mM IAA, and another incubation for 30 min in
the dark at room temperature to alkylate the proteins. The proteins
were re-dissolved in 100 µl TEAB (100 mM), and then digested by
sequence-grade modified trypsin (Promega Corporation) at 37°C for
16 h, and the resultant peptide mixture was labeled using chemicals
from the iTRAQ reagent kit (cat. no. 4381663, Sigma-Aldrich Merck
KGaA), according to the manufacturer's protocol. The peptides from
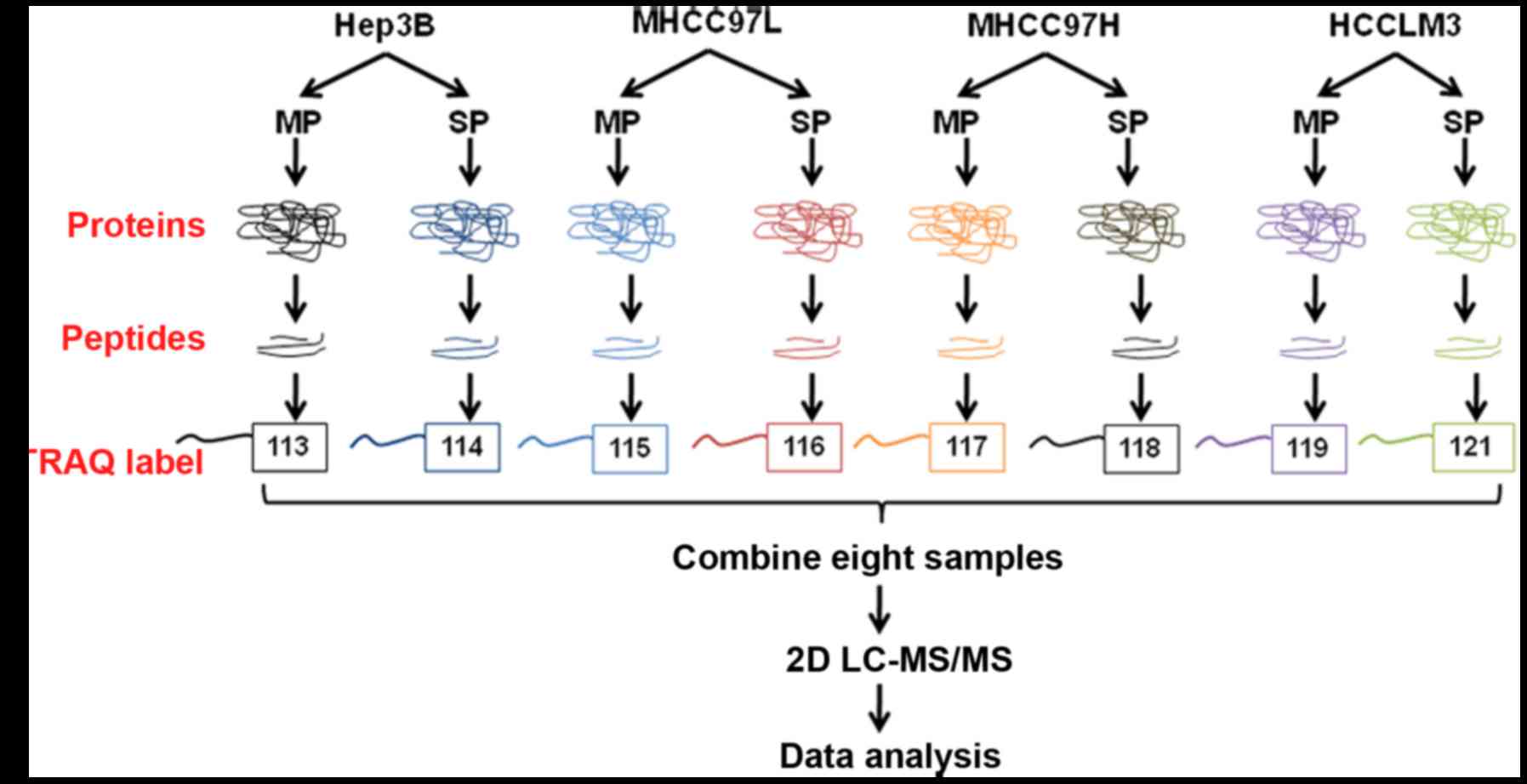
Hep3B, MHCC97-L, MHCC97-H and HCCLM3 MP cells were labeled with
113, 115, 117 and 119 isobaric tags, respectively; and the peptides
from the SP cells of the 4 HCC cell lines were labeled with 114,
116, 118 and 121 isobaric tags, respectively. Equal amounts of
labeled samples were mixed together, and desalted in Sep-Pak Vac
C18 cartridges (Waters Technologies Corporation, Milford, MA, USA)
and dried in a vacuum centrifuge.
High pH reverse phase separation and
low pH two dimensional-liquid chromatography-tandem mass
spectrometry (2D-LC-MS/MS) analysis
The peptide mixture was re-dissolved in solution A
(10% acetonitrile (ACN) in water, pH 10.0), and fractionated by
high pH separation using a 1260 Infinity LC system (Agilent
Technologies, Inc., Santa Clara, CA, USA) connected to a reverse
phase column (Durashell C18, 5 µm, 4.6×250 mm;
Phenomenex®, Torrance, CA, USA). High pH separation was
performed with a linear gradient of 0–80% solution B (95% ACN in
water, pH 10.0) for 80 min at a flow rate of 700 µl/min. Following
separation, the column was re-equilibrated with solution A for 15
min. A total of 10 fractions were collected and dried in a vacuum
concentrator. The fractions were analyzed using an EASY-nLC1000
system (Thermo Fisher Scientific, Inc.) connected to a Q Exactive
Quadrupole-Orbitrap mass spectrometer (Thermo Fisher Scientific,
Inc.) equipped with an online nano-electrospray ion source. Each
fraction was re-suspended in 80 µl solution C (0.1 % formic acid in
water). The samples were then separated by nano-LC and analyzed
using the online electrospray tandem mass spectrometry as follows.
A total of 10 µl peptide sample was loaded onto a trap column
(Acclaim PepMap C18, 100 µm × 2 cm; Thermo Scientific, Inc.).
Subsequently, it was separated in an analytical column (Acclaim
PepMap C18, 75 µm × 15 cm) with a linear gradient of 2–80% solution
D (0.1 % formic acid in ACN) over 2 h with a flow rate of 300
nl/min at 40°C. Finally, the column was re-equilibrated with
solution C for 15 min. The solutions A, B, C and D were all
produced in-house. The Q-Exactive mass spectrometer was operated in
the data dependent mode to switch mechanically between MS and MS/MS
acquisitions. MS spectra (m/z 300–1,500) were acquired with a mass
resolution of 70,000 HZ, followed by 20 sequential high energy
collisional dissociation MS/MS scans with a resolution of 17,500
HZ. One microscan was recorded using a dynamic exclusion of 20 sec
for all cases.
Database searching and criteria
Protein identification was performed using Proteome
Discoverer (version 1.4; Thermo Fisher Scientific, Inc.) against a
database provided by The Universal Protein Resource (www.uniprot.org/uniprot; released, 2014-04-10).
The enzyme specificity of trypsin was used and ≤2 missed cleavages
were allowed for protease digestion. Proteome Discoverer was
searched with a parent ion tolerance of 10 parts per million and a
fragment ion mass tolerance of 0.02 Da. Carboxymethyl of cysteine
was specified as a fixed modification. iTRAQ modification of
peptide N-terminus, oxidation of methionine, deamination of
asparagine and glutamine, and iTRAQ 8-plex labeling of lysine and
tyrosine residues were set as variable modifications. Scaffold
software (version 4.4.5; Proteome Software Inc., Portland, OR, USA)
was used to estimate the false discovery rate (FDR) and validate
the MS/MS-based peptide and protein identifications. The proteins
were assembled using the parsimony method and accepted with a
peptide FDR <1% and a protein probability >90%. Proteins
containing similar peptides, which could not be distinguished based
on MS/MS analysis alone, were grouped to satisfy the principles of
parsimony. All results were then exported into Excel 2010
(Microsoft Corporation, Redmond, WA, USA) for data interpretation.
To identify the differentially expressed proteins (DEPs), the
relative protein expression values were compared among the 4 SP
cell groups. The proteins were considered to be differentially
expressed if the iTRAQ ratios were >1.5 or <0.67 when
LM3-SP/MP cells were compared with Hep3B-SP/MP cells. The DEPs of
SP and MP were analyzed using a Venn diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/),
and trends in the expression values of the DEPs of SP cells
specially were analyzed using Excel 2010 (Microsoft Corporation,
Redmond, WA, USA). The protein expression levels that were
upregulated or downregulated with increasing metastatic potential
in the 4 HCC SP cell groups are summarized in Table I.
 | Table I.Differentially expressed proteins
among SP cells with increasing metastatic potentials. |
Table I.
Differentially expressed proteins
among SP cells with increasing metastatic potentials.
| Accession
number | Protein name | iTRAQ ratio (97-L
SP/Hep3B SP) | iTRAQ ratio (97-H
SP/Hep3B SP) | iTRAQ ratio (LM3
SP/Hep3B SP) |
|---|
| NPM_HUMAN | Nucleophosmin | 0.76 | 0.74 | 0.66 |
| NCBP1_HUMAN | Nuclear cap-binding
protein subunit 1 | 0.83 | 0.77 | 0.63 |
| TIGAR_HUMAN |
Fructose-2,6-bisphosphatase TIGAR | 0.83 | 0.82 | 0.66 |
| PRDX4_HUMAN |
Peroxiredoxin-4 | 0.53 | 0.53 | 0.53 |
| ADIRF_HUMAN | Adipogenesis
regulatory factor | 0.65 | 0.56 | 0.56 |
| CIP2A_HUMAN | Protein CIP2A | 0.84 | 0.79 | 0.63 |
| ECI1_HUMAN | Enoyl-CoA
disomerase 1, mitochondrial | 0.75 | 0.72 | 0.66 |
| ITB1_HUMAN | Integrin β-1 | 0.80 | 0.79 | 0.62 |
| PCM1_HUMAN | Pericentriolar
material 1 protein | 0.81 | 0.65 | 0.51 |
| HS74L_HUMAN | Heat shock 70 kDa
protein 4L | 0.87 | 0.77 | 0.67 |
| KV203_HUMAN | Igkchain V–II
region MIL | 0.62 | 0.55 | 0.50 |
| DIC_HUMAN | Mitochondrial
dicarboxylate carrier | 0.98 | 0.83 | 0.62 |
| PAK2_HUMAN | Cluster of
Serine/threonine-protein kinase PAK 2 | 0.72 | 0.70 | 0.58 |
| RBM39_HUMAN | RNA-binding protein
39 | 0.84 | 0.79 | 0.55 |
| DDX17_HUMAN | Probable
ATP-dependent RNA helicase DDX17 | 0.68 | 0.65 | 0.64 |
| XRCC5_HUMAN | X-ray repair
cross-complementing protein 5 | 0.73 | 0.72 | 0.66 |
| TXTP_HUMAN | Tricarboxylate
transport protein, mitochondrial | 0.58 | 0.55 | 0.52 |
| SPCS2_HUMAN | Signal peptidase
complex subunit 2 | 0.71 | 0.66 | 0.56 |
| RM38_HUMAN | 39S ribosomal
protein L38, mitochondrial | 0.65 | 0.59 | 0.52 |
| PPID_HUMAN | Peptidyl-prolyl
cis-trans isomerase D | 1.01 | 1.10 | 1.53 |
| SNX3_HUMAN | Sorting
nexin-3 | 1.16 | 1.26 | 1.53 |
| MRP1_HUMAN | Multidrug
resistance-associated protein 1 | 1.26 | 1.80 | 2.14 |
| GET4_HUMAN | Golgi to ER traffic
protein 4 homolog | 3.97 | 4.07 | 4.62 |
| GNL3_HUMAN | Guanine
nucleotide-binding protein-like 3 | 1.09 | 1.43 | 1.63 |
| DNJB1_HUMAN | DnaJ homolog
subfamily B member 1 | 1.34 | 1.35 | 1.63 |
| CHRD1_HUMAN | Cysteine and
histidine-rich domain-containing protein 1 | 1.07 | 1.22 | 1.52 |
| ALDR_HUMAN | Aldose
reductase | 1.44 | 1.51 | 1.55 |
| PSD12_HUMAN | 26S proteasome
non-ATPase regulatory subunit 12 | 1.39 | 1.41 | 1.55 |
| TKT_HUMAN | Transketolase | 1.40 | 1.42 | 1.59 |
| CERU_HUMAN | Ceruloplasmin | 1.08 | 1.33 | 1.67 |
Functional analysis of the
differentially expressed proteins
The Gene Ontology (GO) annotation and pathway
enrichment analysis of the differentially expressed proteins and
clustered proteins was performed using the Database for Annotation,
Visualization and Integrated Discovery (https://david.ncifcrf.gov/). GO annotation included
biological processes, cellular components and molecular functions.
The biological function and signaling pathway annotations of the
differentially expressed proteins were analyzed by Ingenuity
Pathway Analysis (IPA) software (version 7.5, Qiagen GmbH, Hilden
Germany). The GO annotations were ranked in terms of the level of
enrichment of the differentially expressed proteins.
Statistical analysis
All data are expressed as the mean ± standard
deviation. Statistical analysis was performed using one-way
analysis of variance followed by Tukey's test. P<0.05 was
considered to indicate a statistically significant difference.
Statistical analysis was performed using SPSS software (version 17;
SPSS, Inc., Chicago, IL, USA).
Results
Identification and characterization of
SP cells
97-L, 97-H and LM3 cell lines possess identical
genetic backgrounds and progressively increased metastatic
potential (24). The Hep3B cell line
has low metastatic potential. These cell lines have been used
widely in the study of the HCC metastasis (24,25). In
the present study, the 4 HCC cell lines were analyzed by dual
wavelength fluorescence-activated cell sorting following incubation
with Hoechst 33342. Representative results are presented in
Fig. 1A, the proportion of SP cells
in Hep3B, 97-L, 97-H, and LM3 were 0.79, 2.27, 4.21 and 6.03%,
respectively. The proportion of SP cells increased with the
metastatic potentials of their parent cell lines, which is
consistent with previous reports.
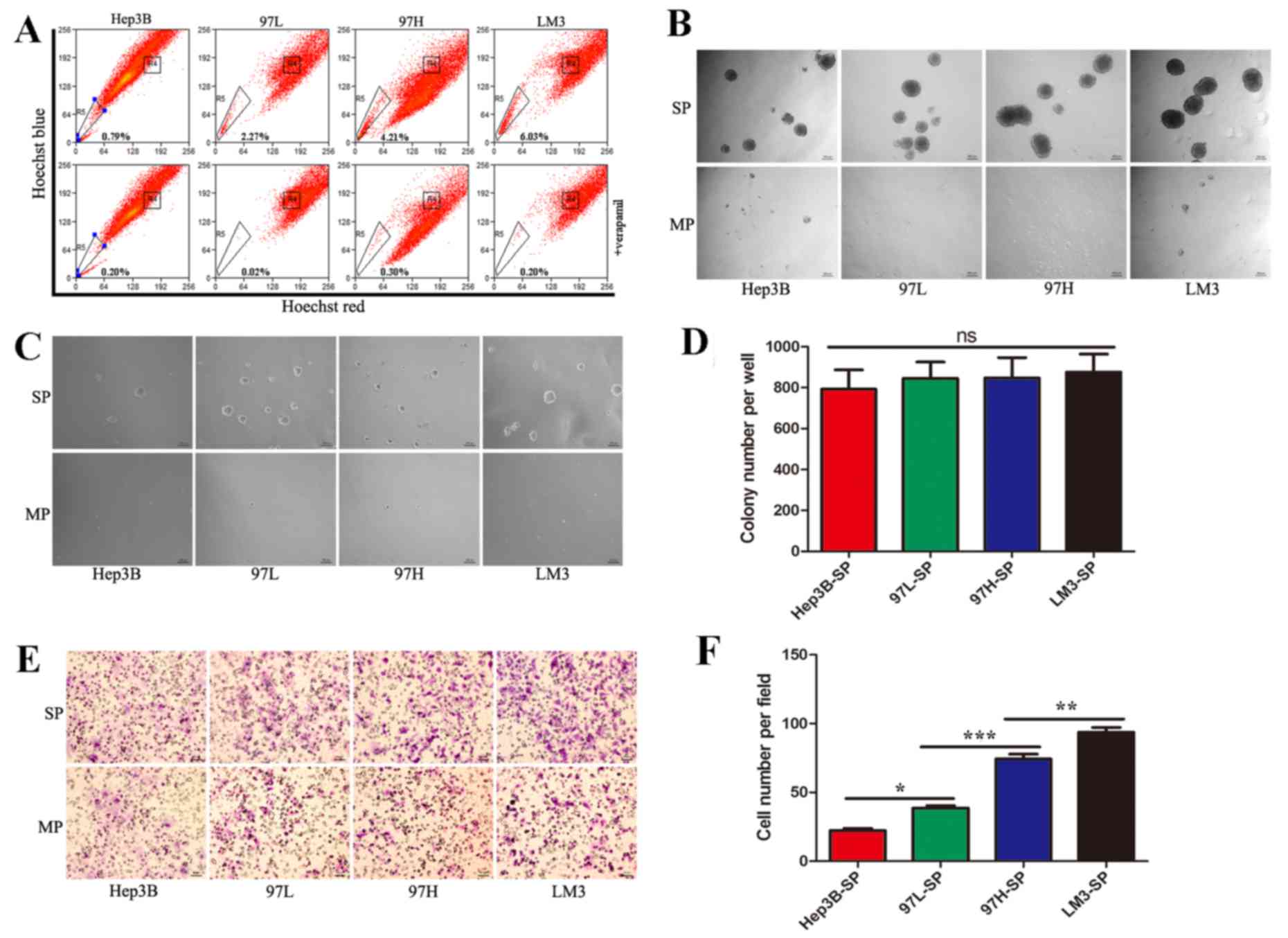 | Figure 1.Identification of side population
cells and the characteristics of each subpopulation. (A)
Identification of SP cells by flow cytometry in the HCC cell lines,
Hep3B, MHCC97L, MHCC97H and HCCLM3. (B) SP and MP cells were
cultured in serum-free medium for 2 weeks (×50, magnification) to
analyze sphere formation. (C) Representative images of the soft
agar colony formation assays, in which SP and MP cells were seeded
into a 6-well plate at 5×103 cells/well in 0.3% agarose
over a 0.6% agarose bottom layer for 2 weeks (×50, magnification).
(D) Quantification of the colony formation assay demonstrated there
was no significant difference in self-renewal ability among SP
cells derived from the different cell lines. (E) Representative
images of migration assay. (F) Quantification of the migration
assay revealed that the metastatic abilities of SP cells from the
different cell lines increased progressively from Hep3B-SP to
97L-SP, to 97H-SP, to LM3-SP. SP, side population; HCC,
hepatocellular carcinoma; MP, main population; 97L, MHCC97-L; 97H,
MHCC97-H; LM3, HCCLM3. *P<0.05, **P<0.01 and
***P<0.001. |
To assess the CSC characteristics of the sorted SP
and MP cells, they were subjected to a sphere formation assay. On
day 3, sorted SP cells began to form spheres, whereas MP cells
formed no floating spheres and eventually died. Upon spheres
dispersal into single cells, secondary and tertiary spheres also
formed. SP cells of the 4 cell lines demonstrated a similar ability
to form spheres (Fig. 1B).
In order to compare self-renewing capacity, the
sorted SP and MP cells were subjected to soft agar colony formation
assays. After 2 weeks, there was no significant difference in terms
of colony number among SP cells derived from the different cell
lines (Fig. 1C and D).
To assess metastatic ability, the sorted SP and MP
cells were subjected to a Transwell migration assay. SP cells
exhibited a high metastatic ability compared with MP cells.
Furthermore, the metastatic ability increased progressively from
Hep3B-SP to 97L-SP, to 97H-SP, to LM3-SP cells (Fig. 1E and F).
Proteomic analysis of the SP cells and
relative quantification of proteome of the side population
cells
A quantitative MS-based discovery strategy was
applied to study the overall proteome of SP cells. Total proteins
extracted from the cells were analyzed using iTRAQ and 2D-LC-MS/MS,
and the workflow is illustrated in Fig.
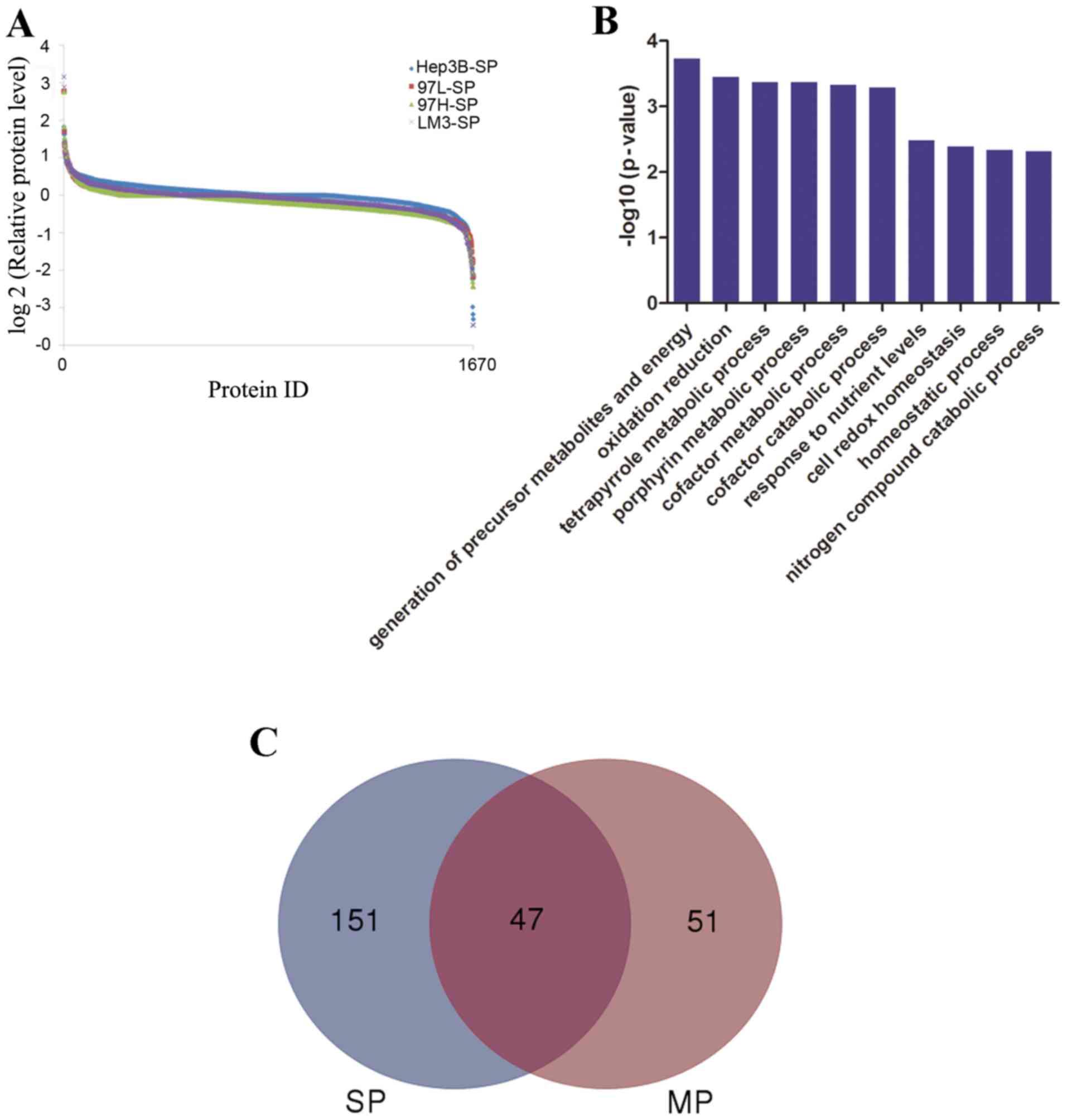
2. Scaffold software was used to identify a total of 1,670
proteins (Fig. 3A), then GO
annotation was applied. As summarized in Fig. 3B, the top 3 molecular functions
associated with the identified proteins were ‘translation’, ‘RNA
processing’ and ‘protein localization’.
Expression trends of proteins and GO
analysis
To systematically analyze the molecular differences
between SP cell groups, the relative protein expression values were
compared. A mean fold change of >1.5 or <0.67 in relative
protein expression between LM3 and Hep3B cells was indicative of
differential expression. A total of 198 differentially expressed
proteins (DEPs; 57 upregulated and 141 downregulated) were
identified when LM3-SP cells were compared with Hep3B-SP cells; 98
DEPs (48 upregulated and 50 downregulated) were identified when
LM3-MP cells were compared with Hep3B-MP cells. The DEPs of SP (198
DEPs) and MP (98 DEPs) were analyzed using a Venn diagram. There
were 47 joint DEPs appeared in both SP and MP cells. Furthermore,
there were 51 DEPs observed in MP cells and 151 DEPs were observed
in the SP cells (Fig. 3C), suggesting
that there may be different molecular mechanisms between SP and MP
cells in HCC.
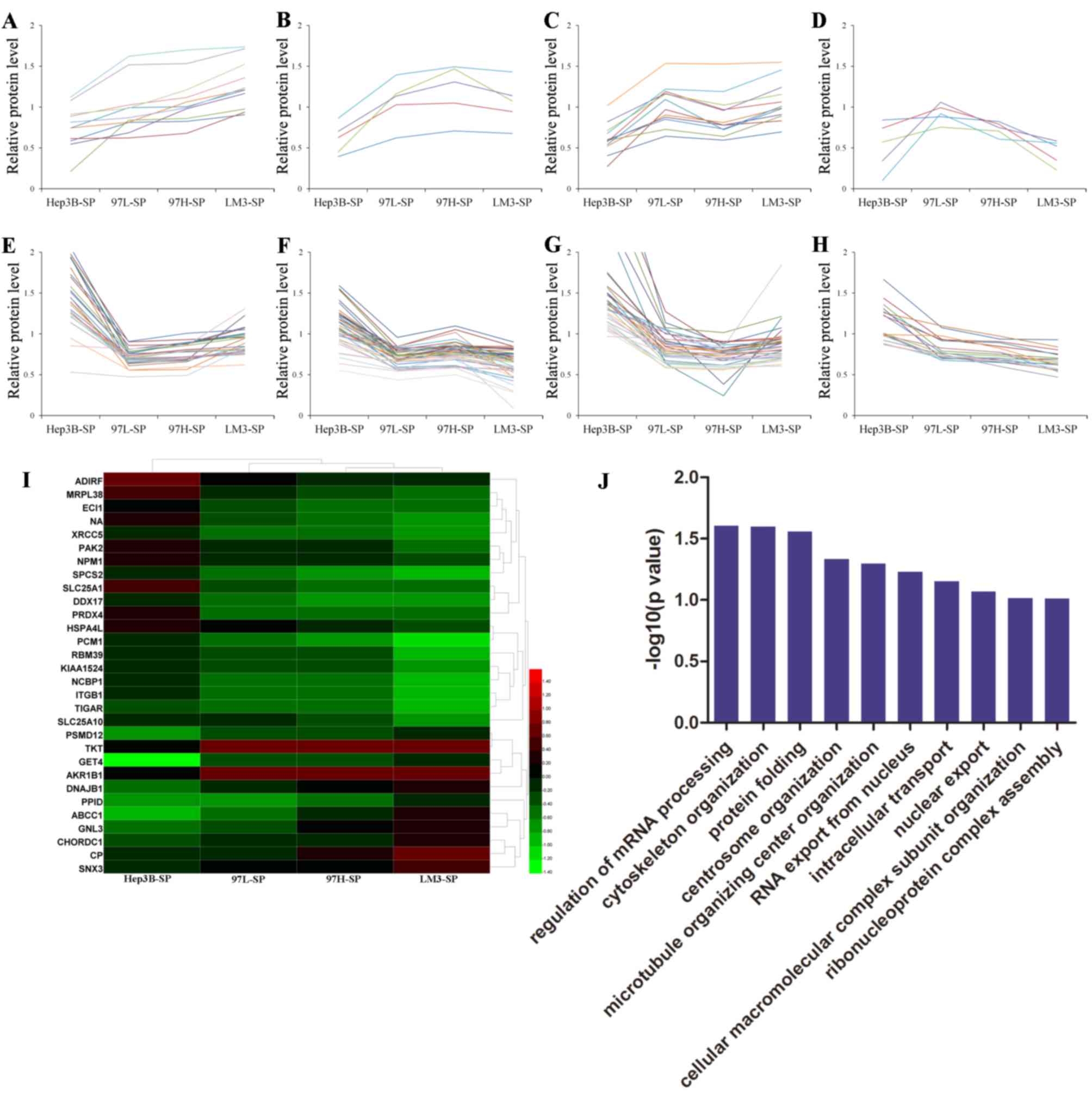
Furthermore, the relative protein levels of 151 DEPs
observed in the SP cells were analyzed using Excel 2010. There were
8 trends identified according to the the relative protein level of
Hep3B-SP, 97L-SP, 97H-SP and LM3-SP. A number of DEPs increased
progressively in expression from Hep3B-SP to LM3-SP (Fig. 4A) and a number of DEPs decreased
progressively from Hep3B-SP to LM3-SP (Fig. 4H). The expression of other DEPs
altered non-linearly between groups (Fig.
4B-G). A total of 11 and 19 differentially expressed proteins
(DEPs) were upregulated and downregulated, respectively, with
increasing metastatic potentials (Table
I). These 30 DEPs formed clearly distinct clusters, as
illustrated in the heatmap in Fig.
4I, and were mainly involved in ‘regulation of mRNA processing’
and ‘cytoskeleton organization’ biological processes according to
GO analysis (Fig. 4J). Furthermore,
the majority of these proteins were involved in the process of
‘self-renewal’, ‘chemoresistance’ and ‘metastasis’ in various types
of cancer, and they may promote HCC migration in a synergistic
manner. Transketolase (TKT) has been demonstrated to promote cell
proliferation and metastasis in various types of cancer and is
associated with poor survival of patients with HCC (26–28).
Targeting TKT has been demonstrated to lead to increased oxidative
stress, causing increased sensitivity of cancer cells to
therapeutic treatments, including Sorafenib (29). ATP binding cassette subfamily C member
1 (30), integrin subunit β1 (ITGB1)
(31,32) and aldo-keto reductase family 1 member
B (33,34) have been reported to be involved in
chemoresistance/radioresistance and cancer metastasis.
P21-activated kinase 2 (35), G
protein nucleolar 3 (36),
nucleophosmin (37) and
pericentriolar material 1 (38) have
been associated with metastatic characteristics in various types of
cancer. These results indicate that different SP cells may have
different molecular mechanisms for metastatic initiation,
regulation, and evasion of conventional HCC chemotherapies.
Pathways and networks identified by
IPA analysis
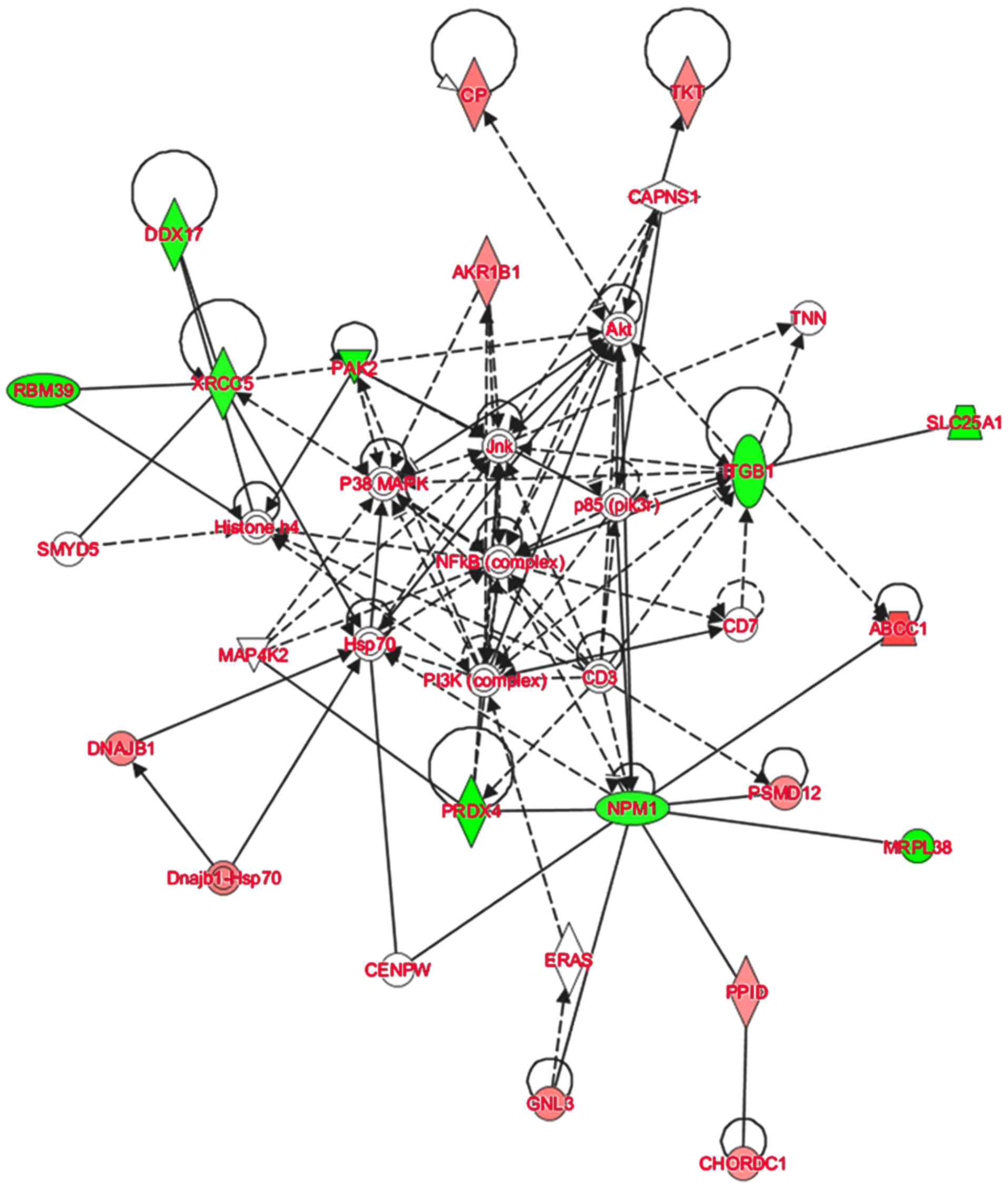
To further elucidate the possible molecular
mechanisms of tumorigenesis and metastasis of SP cell groups, IPA
software was used to analyze the signaling pathways in which the
DEPs were involved. The IPA analysis demonstrated that the majority
of the 30 DEPs altered linearly between SP cell groups were
involved in the regulation of AKT and nuclear factor-κB (NF-κB)
signaling (Fig. 5). AKT signaling is
associated with the regulation of cell proliferation,
differentiation, apoptosis and glucose transport (39). The AKT signaling pathway is also
associated with cell invasion and migration (40). Cluster of differentiation (CD)133 has
been demonstrated to promote gallbladder carcinoma cell migration
by activating AKT phosphorylation (41). The AKT signaling pathway has been
associated with CSC-like properties and epithelial-mesenchymal
transition (EMT) features of A549/CDDP cells (42). In the network assembled in the present
study, ITGB1 was indicated to affect AKT indirectly, and TKT and CP
are downstream of AKT signaling. Furthermore, NF-κB signaling is
central to the network. It has been reported that the NF-κB
signaling contributes to the invasive and metastatic capabilities
of CSCs through modulation of the extracellular environment or
through cell-intrinsic changes, including EMT (43,44).
Therefore, the results of the present study suggest that in HCC,
AKT and NF-κB signaling may contribute to the regulation of HCC
metastasis through regulating the DEPs expressed in CSC-like SP
cells. These results suggest that the quantitative proteomics
approach used was suitable for studying the overall molecular
profile changes of different SP cells in HCC, and provide insight
into the potential molecular mechanisms of this effect.
Discussion
Although surgical resection has been the first-line
treatment for HCC in recent decades, the prognosis of HCC patients
remains poor (45). Post-surgical
metastasis and recurrence are the main causes of the high mortality
rate and poor prognosis of HCC (46).
To achieve a longer survival time, the identification of novel
therapeutic targets associated with HCC invasion and metastasis is
required. CSCs are thought to cause cancer development, and to be
the primary cause of tumor relapse and metastasis (47). Thus, CSC-targeted therapeutic
approaches may have great potential in HCC.
CSCs have been identified in several types of
malignant tumor, including breast (48), brain (49), prostate (50), lung (51), gastric (52), colon (53) and pancreatic (54) cancers, as well as melanoma (55). Recently, research has revealed the
existence of CSCs in HCC (56). With
the exception of SP identification, several surface markers are
used including CD133 (57), CD90
(58), CD44 (59), epithelial cell adhesion molecule
(60), oval cell marker 6 (61), and aldehyde dehydrogenase (62) are employed. Although cells expressing
different combinations of these markers possess CSC properties,
heterogeneity in metastatic potential is observed. It was reported
that CD90+CD44+ cells demonstrated a more aggressive phenotype
compared with CD90+CD44-cells, and formed metastatic lesions in the
lungs of immunodeficient mice (58).
CD133+CD44+ HCC cells isolated from HCC cell lines have been
demonstrated to exhibit an improved ability to initiate tumor
formation and lung metastasis compared with CD133+CD44-cells
(63).
In the present study, SP cells were isolated from 4
HCC cell lines and their abilities to self-renew and migrate were
compared. The results demonstrate that SP cells from different HCC
lines possess similar self-renewing abilities and differing
metastatic potentials, which is consistent with the results of
previous studies (64,65). Furthermore, through a comprehensive
analysis of DEPs, significant differences among the 4 SP cell
groups were demonstrated at the proteome level. The expression
levels of 11 and 19 proteins were upregulated and downregulated,
respectively, with increased metastatic potential. The majority of
these proteins were demonstrated to be involved in ‘cell
proliferation’, ‘migration’ and ‘invasion of cancer’, and they may
promote HCC metastasis in a synergistic manner. This hypothesis was
supported by the results of IPA analysis, which indicated
alteration of the AKT and NF-κB signaling pathways, which have been
demonstrated to be associated with the regulation of migration and
invasion in a number of types of cancer (66–68).
Numerous markers have been identified which are
indicative of CSCs in the context of HCC. However, such cells
exhibit significant heterogeneity between laboratories (69). The proportion of cells expressing
specific markers also varies between HCC cell lines. For example,
CD133+ cells cannot be detected in numerous HCC cell lines and
clinical HCC samples (11). SP cells
are also heterogeneous; however, they are mainly identified by the
expression of ABC transporters, including ABCB1 and ABCG2 (70). SP identification provides an
alternative method of identification when definite markers of CSCs
are uncharacterized. There are limitations to all CSC isolation
strategies, therefore, a combination of different isolation methods
may be required.
To the best of our knowledge, this is the first
report to investigate the overall proteome variations among SP
cells from HCC cell lines with different metastatic potentials. The
present study provides novel information regarding of the
metastatic potential of CSCs, which will facilitate further
investigation. Further research, such as a genomic-transcriptomic
combination study, is required to fully elucidate the underlying
molecular mechanisms of CSC metastatic potential.
Acknowledgments
Not applicable.
Funding
The present study was supported by the specialized
Science and Technology Key Project of Fujian Province (grant no.
2013YZ0002-3), the Science and Technology Infrastructure
Construction Program of Fujian Province (grant no. 2014Y2005), the
National Natural Science Foundation of China (grant nos. 31201008
and 31400634), the Natural Science Foundation of Fujian Province
(grant no. 2015J05174), the Scientific research project of Health
and Family Planning Commission of Fujian Province (grant no.
2015-1-94), the Scientific Foundation of Fuzhou Health Department
(grant no. 2014-S-139-3, 2015-S-wq12) and the Scientific Foundation
of Fuzhou City (grant no. 2013-S-125-4).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available at [http://www.iprox.org/index] with the iProx ID:
IPX00078600.
Authors' contributions
HL, YW, XL and JL participated in the conception and
design of the study. HL, XX and YS performed the proteomic studies
and LC-MS/MS analysis. HL and YW performed cell culture and cell
functional assays. QL and SC performed FCM analysis. HL, XX, GC and
DW performed data analysis. HL, YW, XX and DW participated in data
interpretation. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
CSCs
|
cancer stem cells
|
|
SP
|
side population
|
|
MP
|
main population
|
|
ABC
|
ATP-binding cassette
|
|
MS
|
mass spectrometry
|
|
iTRAQ
|
isobaric tags for relative and
absolute quantitation
|
|
2D LC-MS/MS
|
two-dimensional liquid
chromatography-tandem mass spectrometry
|
|
TEAB
|
triethylammonium bicarbonate
|
|
DTT
|
D-dithiothreitol
|
|
IAA
|
iodoacetamide
|
|
FDR
|
false discovery rate
|
|
GO
|
Gene Ontology
|
|
IPA
|
ingenuity pathway analysis
|
|
DEPs
|
differentially expressed proteins
|
|
EMT
|
epithelial-mesenchymal transition
|
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shah SA, Cleary SP, Wei AC, Yang I, Taylor
BR, Hemming AW, Langer B, Grant DR, Greig PD and Gallinger:
Recurrence after liver resection for hepatocellular carcinoma: Risk
factors, treatment, and outcomes. Surgery. 141:330–339. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bruix J and Sherman M: American
Association for the Study of Liver Diseases: Management of
hepatocellular carcinoma: An update. Hepatology. 53:1020–1022.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tang Z: Hepatocellular carcinoma-cause,
treatment and metastasis. World J Gastroenterol. 7:445–454. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Guo Z, Li LQ, Jiang JH, Ou C, Zeng LX and
Xiang BD: Cancer stem cell markers correlate with early recurrence
and survival in hepatocellular carcinoma. World J Gastroenterol.
20:2098–2106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yu Z, Pestell TG, Lisanti MP and Pestell
RG: Cancer stem cells. Int J Biochem Cell Biol. 44:2144–2151. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Giannelli G, Bergamini C, Fransvea E,
Marinosci F, Quaranta V and Antonaci S: Human hepatocellular
carcinoma (HCC) cells require both alpha3beta1 integrin and matrix
metalloproteinases activity for migration and invasion. Lab Invest.
81:613–617. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen W, Chen L, Cai Z, Liang D, Zhao B,
Zeng Y, Liu X and Liu J: Overexpression of annexin A4 indicates
poor prognosis and promotes tumor metastasis of hepatocellular
carcinoma. Tumor Biol. 37:9343–9355. 2016. View Article : Google Scholar
|
|
10
|
Hermann PC, Huber SL, Herrler T, Aicher A,
Ellwart JW, Guba M, Bruns CJ and Heeschen C: Distinct populations
of cancer stem cells determine tumor growth and metastatic activity
in human pancreatic cancer. Cell Stem Cell. 1:313–323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shi GM, Xu Y, Fan J, Zhou J, Yang XR, Qiu
SJ, Liao Y, Wu WZ, Ji Y, Ke AW, et al: Identification of side
population cells in human hepatocellular carcinoma cell lines with
stepwise metastatic potentials. J Cancer Res Clin Oncol.
134:1155–1163. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Goodell MA, Brose K, Paradis G, Conner AS
and Mulligan RC: Isolation and functional properties of murine
hematopoietic stem cells that are replicating in vivo. J Exp Med.
183:1797–1806. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chiba T, Kita K, Zheng YW, Yokosuka O,
Saisho H, Iwama A, Nakauchi H and Taniguchi H: Side population
purified from hepatocellular carcinoma cells harbors cancer stem
cell-like properties. Hepatology. 44:240–251. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang J, Guo LP, Chen LZ, Zeng YX and Lu
SH: Identification of cancer stem cell-like side population cells
in human nasopharyngeal carcinoma cell line. Cancer Res.
67:3716–3724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ho MM, Ng AV, Lam S and Hung JY: Side
population in human lung cancer cell lines and tumors is enriched
with stem-like cancer cells. Cancer Res. 67:4827–4833. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Haraguchi N, Utsunomiya T, Inoue H, Tanaka
F, Mimori K, Barnard GF and Mori M: Characterization of a side
population of cancer cells from human gastrointestinal system. Stem
Cells. 24:506–513. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ross PL, Huang YN, Marchese JN, Williamson
B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et
al: Multiplexed protein quantitation in Saccharomyces cerevisiae
using amine-reactive isobaric tagging reagents. Mol Cell
Proteomics. 3:1154–1169. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Aggarwal K, Choe LH and Lee KH: Shotgun
proteomics using the iTRAQ isobaric tags. Brief Funct Genomic
Proteomic. 5:112–120. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xing X, Huang Y, Wang S, Chi M, Zeng Y,
Chen L, Li L, Zeng J, Lin M, Han X, et al: Comparative analysis of
primary hepatocellular carcinoma with single and multiple lesions
by iTRAQ-based quantitative proteomics. J Roteomics. 128:262–271.
2015. View Article : Google Scholar
|
|
20
|
Wei D, Zeng Y, Xing X, Liu H, Lin M, Liu X
and Liu J: Proteome differences between hepatitis B virus
genotype-B- and genotype-C-induced hepatocellular carcinoma
revealed by iTRAQ-based quantitative proteomics. J Proteome Res.
15:487–498. 2015. View Article : Google Scholar
|
|
21
|
Huang X, Zeng Y, Xing X, Zeng J, Gao Y,
Cai Z, Xu B, Liu X, Huang A and Liu J: Quantitative proteomics
analysis of early recurrence/metastasis of huge hepatocellular
carcinoma following radical resection. Proteome Sci. 12:222014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ko CH, Cheng CF, Lai CP, Tzu TH, Chiu CW,
Lin MW, Wu SY, Sun CY, Tseng HW, Wang CC, et al: Differential
proteomic analysis of cancer stem cell properties in hepatocellular
carcinomas by isobaric tag labeling and mass spectrometry. J
Proteome Res. 12:3573–3585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xu Y, Xie Y, Wang X, Chen X, Liu Q, Ying M
and Zheng Q: Identification of cancer stem cells from
hepatocellular carcinoma cell lines and their related microRNAs.
Oncol Rep. 30:2056–2062. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yu Y, Shen H, Yu H, Zhong F, Zhang Y,
Zhang C, Zhao J, Li H, Chen J, Liu Y and Yang P: Systematic
proteomic analysis of human hepotacellular carcinoma cells reveals
molecular pathways and networks involved in metastasis. Mol
Biosyst. 7:1908–1916. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen N, Sun W, Deng X, Hao Y, Chen X, Xing
B, Jia W, Ma J, Wei H, Zhu Y, et al: Quantitative proteome analysis
of HCC cell lines with different metastatic potentials by SILAC.
Proteomics. 8:5108–5118. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ricciardelli C, Lokman NA, Cheruvu S, Tan
IA, Ween MP, Pyragius CE, Ruszkiewicz A, Hoffmann P and Oehler MK:
Transketolase is upregulated in metastatic peritoneal implants and
promotes ovarian cancer cell proliferation. Clin Exp Metastasis.
32:441–455. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Zhu SC, Li SG, Zhao Y, Xu JR and
Song CY: TKTL1 promotes cell proliferation and metastasis in
esophageal squamous cell carcinoma. Biomed Pharmacother. 74:71–76.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Tan GS, Lim KH, Tan HT, Khoo ML, Tan SH,
Toh HC and Chung Ching Ming M: Novel proteomic biomarker panel for
prediction of aggressive metastatic hepatocellular carcinoma
relapse in surgically resectable patients. J Proteome Res.
13:4833–4846. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xu IM, Lai RK, Lin SH, Tse AP, Chiu DK,
Koh HY, Law CT, Wong CM, Cai Z, Wong CC and Ng IO: Transketolase
counteracts oxidative stress to drive cancer development. Proc Natl
Acad Sci USA. 113:E725–E734. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chow AK, Ng L, Lam CS, Wong SK, Wan TM,
Cheng NS, Yau TC, Poon RT and Pang RW: The Enhanced metastatic
potential of hepatocellular carcinoma (HCC) cells with sorafenib
resistance. PLoS One. 8:e786752013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lu Y, Hu J, Sun W, Li S, Deng S and Li M:
MiR-29c inhibits cell growth, invasion, and migration of pancreatic
cancer by targeting ITGB1. Onco Targets Ther. 9:992015.PubMed/NCBI
|
|
32
|
Broustas CG and Lieberman HB: RAD9
enhances radioresistance of human prostate cancer cells through
regulation of ITGB1 protein levels. Prostate. 74:1359–1370. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nakarai C, Osawa K, Akiyama M, Matsubara
N, Ikeuchi H, Yamano T, Hirota S, Tomita N, Usami M and Kido Y:
Expression of AKR1C3 and CNN3 as markers for detection of lymph
node metastases in colorectal cancer. Clin Exp Med. 15:333–341.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dan S, Shirakawa M, Mukai Y, Yoshida Y,
Yamazaki K, Kawaguchi T, Matsuura M, Nakamura Y and Yamori T:
Identification of candidate predictive markers of anticancer drug
sensitivity using a panel of human cancer cell lines. Cancer Sci.
94:1074–1082. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sato M, Matsuda Y, Wakai T, Kubota M,
Osawa M, Fujimaki S, Sanpei A, Takamura M, Yamagiwa S and Aoyagi Y:
p21-activated kinase-2 is a critical mediator of transforming
growth factor-β-induced hepatoma cell migration. J Gastroenterol
Hepatol. 28:1047–1055. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lee M, Williams KA, Hu Y, Andreas J, Patel
SJ, Zhang S and Crawford NP: GNL3 and SKA3 are novel prostate
cancer metastasis susceptibility genes. Clin Exp Metastasis.
32:769–782. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ching RH, Lau EY, Ling PM, Lee JMF, Ma MK,
Cheng BY, Lo RC, Ng IO and Lee TK: Phosphorylation of Nucleophosmin
at Threonine 234/237 is associated with HCC metastasis. Oncotarget.
6:43483–43495. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ueo H, Takano Y, Matsumura T, Kurashige J,
Shinden Y, Eguchi H, Sudo T, Sugimachi K, Saeki H, Oki E, et al:
Identification of genes that predict lymph node metastasis in
colorectal cancer cases. Fukuoka Igaku Zasshi. 104:559–563.
2013.(In Japanese). PubMed/NCBI
|
|
39
|
Song G, Ouyang G and Bao S: The activation
of Akt/PKB signaling pathway and cell survival. J Cell Mol Med.
9:59–71. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Martini M, De Santis MC, Braccini L,
Gulluni F and Hirsch E: PI3K/AKT signaling pathway and cancer: An
updated review. Ann Med. 46:372–383. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li C, Wang C, Xing Y, Zhen J and Ai Z:
CD133 promotes gallbladder carcinoma cell migration through
activating Akt phosphorylation. Oncotarget. 7:17751–17759.
2016.PubMed/NCBI
|
|
42
|
Wang H, Zhang G, Zhang H, Zhang F, Zhou B,
Ning F, Wang HS, Cai SH and Du J: Acquisition of
epithelial-mesenchymal transition phenotype and cancer stem
cell-like properties in cisplatin-resistant lung cancer cells
through AKT/β-catenin/Snail signaling pathway. Eur J Pharmacol.
723:156–166. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rinkenbaugh AL and Baldwin AS: The NF-κB
pathway and cancer stem cells. Cells. 5:pii: E16. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang FS, Fan JG, Zhang Z, Gao B and Wang
HY: The global burden of liver disease: The major impact of China.
Hepatology. 60:2099–2108. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Margonis GA, Sasaki K, Andreatos N,
Nishioka Y, Sugawara T, Amini N, Buettner S, Hashimoto M, Shindoh J
and Pawlik TM: Prognostic impact of complications after resection
of early stage hepatocellular carcinoma. J Surg Oncol. 115:791–804.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chiba T, Iwama A and Yokosuka O: Cancer
stem cells in hepatocellular carcinoma: Therapeutic implications
based on stem cell biology. Hepatol Res. 46:50–57. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
50
|
Collins AT, Berry PA, Hyde C, Stower MJ
and Maitland NJ: Prospective identification of tumorigenic prostate
cancer stem cells. Cancer Res. 65:10946–10951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kim CF, Jackson EL, Woolfenden AE,
Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT and Jacks T:
Identification of bronchioalveolar stem cells in normal lung and
lung cancer. Cell. 121:823–835. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Takaishi S, Okumura T, Tu S, Wang SS,
Shibata W, Vigneshwaran R, Gordon SA, Shimada Y and Wang TC:
Identification of gastric cancer stem cells using the cell surface
marker CD44. Stem Cells. 27:1006–1020. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ricci-Vitiani L, Lombardi DG, Pilozzi E,
Biffoni M, Todaro M, Peschle C and De Maria R: Identification and
expansion of human colon-cancer-initiating cells. Nature.
445:111–115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li C, Heidt DG, Dalerba P, Burant CF,
Zhang L, Adsay V, Wicha M, Clarke MF and Simeone DM: Identification
of pancreatic cancer stem cells. Cancer Res. 67:1030–1037. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Schatton T, Murphy GF, Frank NY, Yamaura
K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM,
Weishaupt C, et al: Identification of cells initiating human
melanomas. Nature. 451:345–349. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Yang ZF, Ngai P, Ho DW, Yu WC, Ng MN, Lau
CK, Li ML, Tam KH, Lam CT, Poon RT and Fan ST: Identification of
local and circulating cancer stem cells in human liver cancer.
Hepatology. 47:919–928. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ma S, Chan KW, Hu L, Lee TK, Wo JY, Ng IO,
Zheng BJ and Guan XY: Identification and characterization of
tumorigenic liver cancer stem/progenitor cells. Gastroenterology.
132:2542–2556. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Yang ZF, Ho DW, Ng MN, Lau CK, Yu WC, Ngai
P, Chu PW, Lam CT, Poon RT and Fan ST: Significance of CD90+ cancer
stem cells in human liver cancer. Cancer Cell. 13:153–166. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhu Z, Hao X, Yan M, Yao M, Ge C, Gu J and
Li J: Cancer stem/progenitor cells are highly enriched in
CD133+CD44+ population in hepatocellular carcinoma. Int J Cancer.
126:2067–2078. 2010.PubMed/NCBI
|
|
60
|
Yamashita T, Ji J, Budhu A, Forgues M,
Yang W, Wang HY, Jia H, Ye Q, Qin LX, Wauthier E, et al:
EpCAM-positive hepatocellular carcinoma cells are tumor-initiating
cells with stem/progenitor cell features. Gastroenterology.
136:1012–1024. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yang W, Wang C, Lin Y, Liu Q, Yu LX, Tang
L, Yan HX, Fu J, Chen Y, Zhang HL, et al: OV6+ tumor-initiating
cells contribute to tumor progression and invasion in human
hepatocellular carcinoma. J Hepatol. 57:613–620. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ma S, Chan KW, Lee TK-W, Tang KH, Wo JY,
Zheng BJ and Guan XY: Aldehyde dehydrogenase discriminates the
CD133 liver cancer stem cell populations. Mol Cancer Res.
6:1146–1153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ma S: Biology and clinical implications of
CD133(+) liver cancer stem cells. Exp Cell Res. 319:126–132. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Yang X, Wang J, Qu S, Zhang H, Ruan B, Gao
Y, Ma B, Wang X, Wu N, Li X, et al: MicroRNA-200a suppresses
metastatic potential of side population cells in human
hepatocellular carcinoma by decreasing ZEB2. Oncotarget.
6:7918–7929. 2015.PubMed/NCBI
|
|
65
|
Zhou L, Yang ZX, Song WJ, Li QJ, Yang F,
Wang DS, Zhang N and Dou KF: MicroRNA-21 regulates the migration
and invasion of a stem-like population in hepatocellular carcinoma.
Int J Oncol. 43:661–669. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Li J, Deng Z, Wang Z, Wang D, Zhang L, Su
Q, Lai Y, Li B, Luo Z, Chen X, et al: Zipper-interacting protein
kinase promotes epithelial-mesenchymal transition, invasion and
metastasis through AKT and NF-kB signaling and is associated with
metastasis and poor prognosis in gastric cancer patients.
Oncotarget. 6:8323–8338. 2015.PubMed/NCBI
|
|
67
|
Tang Y, Lv P, Sun Z, Han L and Zhou W:
14-3-3β promotes migration and invasion of human hepatocellular
carcinoma cells by modulating expression of MMP2 and MMP9 through
PI3K/Akt/NF-κB pathway. PLoS One. 11:e01460702016. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wang YH, Dong YY, Wang WM, Xie XY, Wang
ZM, Chen RX, Chen J, Gao DM, Cui JF and Ren ZG: Vascular
endothelial cells facilitated HCC invasion and metastasis through
the Akt and NF-κB pathways induced by paracrine cytokines. J Exp
Clin Cancer Res. 32:512013. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Chen Y, Yu D, Zhang H, He H, Zhang C, Zhao
W and Shao R: CD133(+)EpCAM(+) phenotype possesses more
characteristics of tumor initiating cells in hepatocellular
carcinoma Huh7 cells. Int J Biol Sci. 8:992–1004. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Moitra K: Overcoming multidrug resistance
in cancer stem cells. Bio Med Res Int. 2015:6357452015.
|