Introduction
Preoperative chemoradiotherapy (CRT) followed by
total mesorectal excision (TME) is the standard of treatment for
patients with locally advanced rectal cancer (LARC) (1–3).
Preoperative CRT allows for tumor downstaging and pathologic
complete response (pathCR), which is correlated with 5-year
recurrence free survival (4).
Therefore, identifying patients who may or may not achieve a pathCR
would allow for treatment with alternative approaches in the
preoperative setting (5,6).
Many researchers investigate specific
immunohistochemical biomarkers and genotype biomarkers as potential
predictors of pathCR (7,8). However, genotyping tumors may be
challenging due to the invasiveness of sample collection and
limited availability of tissue biopsies. Instead, circulating tumor
cells (CTCs) are considered an alternative source of tumor cells
present in the peripheral blood of cancer patients. The ability to
isolate and profile CTCs presents an attractive option for
genotyping tumors noninvasively.
In the present study, we produced an integrated
workflow to investigate whether aneuploidy of chromosome 8 and
mutations of CTCs can be specific predictors of pathCR to
preoperative CRT in patients with rectal cancer. Future studies
will be performed to improve the workflow and validate the
conclusions in prospective and larger cohorts with long-term
follow-up survival data.
Materials and methods
Patients
We recruited 33 patients with LARC (cT3-T4 and/or
cN+, The 7th edition of AJCC) treated with neoadjuvant CRT at our
institution between September 2014 and March 2015. All patients
signed informed consent and the Fudan University Shanghai Cancer
Center Ethics Review Board approved the present study.
Patients were treated with CRT, with radiotherapy
dose of 50 Gy and simultaneous fluorouracil-based chemotherapy. All
patients received 50 Gy/25Fx in the study. Surgery was generally
performed 6 to 8 weeks after completion of CRT and included low
anterior resection, or abdominoperineal resection using TME
principles. Following surgical resection, the pathological stage of
the tumor was determined according to TNM classification. Standard
pathological tumor staging of the resected specimens was performed
after resection in accordance with the guidelines of the College of
American Pathologists, with histopathological diagnosis performed
by dedicated gastrointestinal cancer pathologists. The gross tumor
volume was embedded and serially sectioned for subsequent
hematoxylin and eosin staining and microscopic evaluation.
Blood specimen collection and
processing
Blood samples from the 33 patients with rectal
cancer were obtained prior to CRT. Rectal adenocarcinoma was
confirmed pathologically for all patients by biopsy. For all 33
patients, 8.5 ml blood samples were obtained for detection of CTCs.
All blood samples were collected into evacuated ACD anticoagulant
tubes.
Subtraction enrichment of CTCs and
identification of aneuploid CTCs
The enrichment and identification of CTCs was
performed according to CTCseq™ kit instruction (Shanghai
Majorbio Pharmaceutical Technology Co., Ltd., Shanghai, China).
CTCs were confirmed to be negative for CD45 and either positive for
PanCK staining or chromosome 8 aneuploidy. CTCs on slides were
counted and imaged, and individual tumor cell coordinates were
recorded to facilitate subsequent target cell identification. These
slides were kept frozen at −20°C.
Laser capture microdissection
(LCM)
For LCM, samples were loaded onto the stage of a
Zeiss PALM MicroBeam (Carl Zeiss AG, Oberkochen, Germany) under a
40× objective. Following microdissection with a 355-nm laser beam,
target cells were collected into AdhesiveCap 200 opaque tubes (Carl
Zeiss AG). CTCs from one patient were collected in one tube; the
number of CTCs in one tube varied from one to thirty. Lysis buffer
(5 µl; Yikon Genomics, Shanghai, China) was then added to each
sample and stored at −20°C.
Single-cell whole-genome amplification
(WGA) with multiple annealing and looping-based amplification
cycles (MALBAC)
WGA of lysed cells was performed using the MALBAC
method (9) following the standard
protocol provided by the commercial MALBAC amplification kit (Yikon
Genomics). In summary, cells were centrifuged and collected at
8,000 rcf for 5 min, transferred to a new 200 µl PCR tube,
following incubation at 50°C for 2 h and 80°C for 10 min. Then, 30
µl of freshly prepared preamplification mix was added to each tube
and was incubated at 94°C for 3 min. DNA was amplified using 8
cycles of 40 sec at 20°C, 40 sec at 30°C, 30 sec at 40°C, 30 sec at
50°C, 30 sec at 60°C, 4 min at 70°C, 20 sec at 95°C and 10 sec at
58°C; samples were then placed on ice immediately. Amplification
reaction mix (30 µl) was added to each tube and incubated at 94°C
for 30 sec followed by 17 cycles of 20 sec at 94°C, 30 sec at 58°C
and 3 min at 72°C.
WGA quality control on Agilent
2100
MALBAC products were purified with 1.4× Ampure XP
beads (Beckman Coulter, Inc., Brea, CA, USA) and assessed by 2100
BioAnalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA).
Each MALBAC product was diluted to 1 ng/µl. The sample volume for
the analysis was 1 µl. A BioAnalyzer High Sensitivity DNA kit
(Agilent Technologies, Inc.) was used to visualize the size range
of MALBAC products. Samples were injected into the separation
channel and their components were electrophoretically
separated.
Target gene sequencing
A total of 36 genes (ACVR2A, SMAD4, TP53, NRAS, P65
(NFKB3), ATM, PRKCB, AR, SUMO1, PIK3CA, APC, MLH1, BRAF, HDAC1,
ABL1, IGF2, TCF7L2, MLH3, MFS2, KRAS, STAT1, MYC, ERBB3, MSH2,
ERBB2, TGFBR1, AMER1, PTEN, JUN, MSH3, TRIM63, CDK1, SOX9, PMS2,
ARID1A and MSH6), corresponding to 105 kb, which were frequently
mutated in colorectal cancer according to previous studies
(10,11), were shown to be enriched. MygeneSeq
technology (Morgene) was used to identify enriched genes following
the manufacturer's instructions, with a 100 ng input of MALBAC
products used. In summary, barcodes were mapped to multiplex PCR
products and enriched targets were identified using the MygeneSeq
panel. Then, a standard shotgun library was made from amplicons
using NEXTflex Rapid DNA-Seq kit Bundle with DNA Barcodes 1–24
(Bioo Scientific, Austin, TX, USA). Final concentrations of DNA
samples were measured prior to sequencing using the Qubit v.2.0
fluorometer (Thermo Fisher Scientific, Inc., Waltham, MA, USA). A
total of 36 genes were sequenced using 150 cycle paired-end
flowcell lanes on the HiSeq4000 system (Illumina, Inc., San Diego,
CA, USA). All sequence data were processed using CASAVA 1.8.1
pipeline (Illumina, Inc.) converting BCL basecall files to fastq
files.
Software analysis
MALBAC primers and Illumina adaptors were trimmed by
cutadapt 1.12 (12). Trimmed
sequencing reads were aligned to the human genome (hg19) using the
Burrows-Wheeler Aligner (BWA-0.7.12-r1039) (13). The mapping files in SAM format were
converted to BAM format and sorted by SAMtools-1.2 (14). The Genome Analysis Toolkit (GATK
v3.3–0-g37228af; http://www.broadinstitute.org/gatk/) was used to
locally realign the sorted BAM files (15). Then, sequencing data was re-calibrated
and insertion/deletion (indel)-realigned using GATK (http://www.broadinstitute.org/gatk/) before
variant detection. Base-level sequencing coverage was enumerated by
the Depth Of Coverage module from bedtools.
Next, we respectively identified single nucleotide
variants (SNVs) and indels from the BAM files generated with all
samples using GATK UnifiedGenotyper (parameters: -mbq 20). BAM
files of all samples were processed together to generate a single
VCF4 file. We used the GATK VariantFiltration to filter using the
parameters MQ<40, HaplotypeScore>200.0, FS>60.0 (SNP),
FS>200.0 (Indel), then the VCF file was annotated using SnpSift
and snpEff and ANNOVAR. A minimum coverage depth of 10 and >2
reads with variant alleles was used for further filtering of SNVs.
Only SNVs covered in all samples remained following filtering. SNVs
in clustered regions with neighboring SNVs within 10 bp were
filtered from the data to remove false positives (16). Finally, we filtered benign SNPs from
1000g2015aug_eas and exac03nontcga, and selected the remaining
variants to separate SNVs into VCF4 files.
Statistical analyses
Statistical analyses were performed using SPSS
software (v.16.0, SPSS Inc., Chicago, IL, USA). Age, number of CTCs
and aneuploidy were expressed as continuous variables. Categorical
variables included gender, stage and presence of aneuploidy and
mutations. Univariate logistic regression was used to analyze the
association between clinical characteristics and aneuploidy of
chromosome 8 with pathCR. The incidence of pathCR between
nonsynonymous mutations and wild type genes was compared using
Pearson's chi-square test.
Results
Patient and tumor characteristics
Patient and tumor characteristics are presented in
Table I. All patients completed
neoadjuvant treatment. Overall, 6 (18.2%) patients had a pathCR, 21
patients got pathPR and 6 patients got pathSD. CTCs were observed
in all 33 patients with a median number of 7 cells per 8.5 ml
(range 1–30). Additionally, 12 patients had 1–3 CTCs and 21
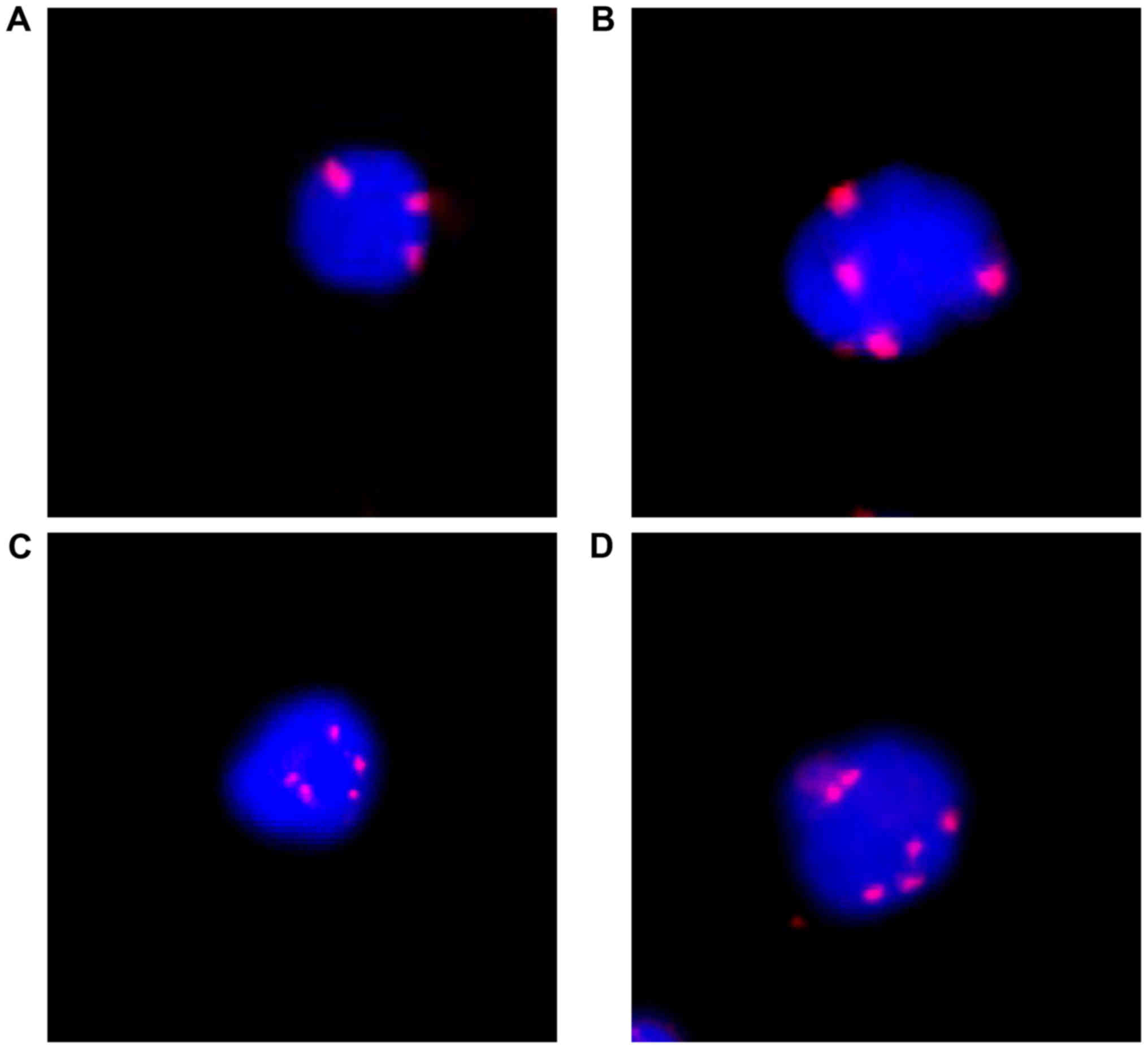
patients had >3 CTCs. Triploidy, tetraploidy, pentaploidy or
>5 copies of chromosome 8 were observed in CTCs from rectal
cancer patients (Fig. 1). There was
no apparent green staining of PanCK in Fig. 1 for two reasons. First, the expression
of PanCK is not necessary to identify CTCs. In our study, CTC was
defined as CEP8+/DAPI+/CD45-. The high sensitivity of this method
was due to the high sensitivity of CEP8. The Panck marker was only
an observational biomarker in our study. Second, one possible
explanation for this observation was that those PanCK negative CTCs
were undergoing epithelial-mesenchymal transition (EMT). In the
CTCs, triploidy, tetraploidy and ≥5 copies of chromosome 8 were
observed in 25, 18 and 27 patients, respectively (Table I).
 | Table I.Patient and tumor characteristics
(n=33). |
Table I.
Patient and tumor characteristics
(n=33).
| Characteristics | No. of patients
(%) |
|---|
| Sex |
|
| Male | 21 (63.6) |
|
Female | 12 (36.4) |
| Age (years) |
|
| Mean | 54.5 |
|
Median | 57 |
|
Range | 30–73 |
| Clinical stage |
|
| II | 3 (9.1) |
| III | 30 (90.1) |
| No. of CTCs |
|
| 1–3 | 12 (36.4) |
|
>3 | 21 (63.6) |
| Triploidy |
|
| Yes | 25 (75.8) |
| No | 8 (24.2) |
| Tetraploidy |
|
|
Yes | 18 (54.5) |
| No | 15 (45.5) |
| ≥5 copies |
|
|
Yes | 27 (81.8) |
| No | 6 (18.2) |
Association between pathCR and
chromosome 8 aneuploidy in CTCs
Aneuploidy of chromosome 8 in CTCs from 33 patients
was examined. As shown in Fig. 1,
triploidy, tetraploidy, pentaploidy or >5 copies of chromosome 8
were observed in in CTCs from rectal cancer patients. Furthermore,
univariate logistic regression indicated that ≥5 copies of
chromosome 8 was associated with pathCR (P=0.042; Table II). Of the patients whose CTCs had
<5 copies of chromosome 8 (n=6), 3 exhibited pathCR (3/6, 50%),
and of the 27 patients whose CTCs had ≥5 copies of chromosome 8, 3
exhibited pathCR (3/27, 11.1%; Chi-square test, P=0.0255).
 | Table II.Univariate logistic regression
predicting pathCR. |
Table II.
Univariate logistic regression
predicting pathCR.
| Variables | P-value | Odds ratio |
|---|
| Age | 0.16 | 1.56 |
| Sex | 0.44 | 2.63 |
| Stage | 0.34 | 2.45 |
| No. of CTCs | 0.50 | 0.94 |
| No. of
triploidy | 0.56 | 0.90 |
| % triploidy | 0.10 | 10.99 |
| No. of
tetraploidy | 0.30 | 0.57 |
| % tetraploidy | 0.30 | 0.03 |
| No. of ≥5
copies | 0.73 | 0.97 |
| % ≥5 copies | 0.30 | 0.22 |
| Presence of
triploidy | 0.64 | 1.75 |
| Presence of
tetraploidy | 0.26 | 0.34 |
| Presence of ≥5
copies | 0.04 | 0.13 |
Detection of mutational status in
pathCR and non-pathCR groups
Among the 33 patients with mutations assessed, 6
(18.2%) exhibited pathCR. One patient was excluded for its low
coverage depth (data not shown), the average depth of the other 32
sequencing data was ~4,000× (data not shown). The average coverage
breadth (sites with ≥10× coverage) varied largely from 45.5–96.18%.
In order to find mutational predictors of response to preoperative
CRT, all 6 patients in pathCR group were analyzed, with coverage
breadth (sites with ≥10× coverage) from 50.20–73.68%, and 10
patients were selected from the non-pathCR group, with coverage
breadth of ~80%. Of the 16 patients, we assessed the mutation
status of target regions that all the samples covered and selected
positions with at least one SNV. We identified 22 genes that had
mutations with a significant difference in frequency between the
groups. These genes included TRIM63, ARID1A, HDAC1, MSH2, ACVR2A,
STAT1, SUMO1, TGFBR2, APC, BRAF, ABL1, TCF7L2, IGF2, RELA, ATM,
ERBB3, PRKCB, TP53, ERBB2, SOX9, AMER1 and AR. There were 64
nonsynonymous SNVs and 28 synonymous SNVs (data not shown).
Subsequently, a chi-square test was performed to
examine the significant difference in pathCR group (n=6) and
non-pathCR group (n=10). Through comparing the nonsynonymous
mutational status of the pathCR group and non-pathCR group, 9
statistically significant nonsynonymous mutations were identified
(Chi-square test; Table IIIA).
 | Table III.Mutations with significant difference
in frequency between pathCR and non-pathCR patients. |
Table III.
Mutations with significant difference
in frequency between pathCR and non-pathCR patients.
| A, Mutations
between pathCR (n=6) and non-pathCR patients (n=10) |
|---|
| Gene | Amino acid
change | Overall (%)
n=16 | pathCR (%) n=6 | Non-pathCR (%)
n=10 | Excluded | P-value |
|---|
| ARID1A | p.L2117M | 3 (18.8) | 3 (50) | 0 (0) | – | 0.0131 |
| HDAC1 | p.F341V | 4 (25) | 4 (66.7) | 0 (0) | – | 0.0029 |
| APC | p.K1310M | 4 (25) | 4 (66.7) | 0 (0) | – | 0.0029 |
| BRAF | p.P75S | 4 (25) | 4 (66.7) | 0 (0) | – | 0.0029 |
| ERBB3 | p.Y86H | 4 (25) | 4 (66.7) | 0 (0) | – | 0.0029 |
| TP53 | p.T155I | 5 (31.3) | 5 (83.3) | 0 (0) | – | 0.0005 |
| AMER1 | p.A504V | 5 (31.3) | 5 (83.3) | 0 (0) | – | 0.0005 |
| AMER1 | p.V490D | 3 (18.8) | 3 (50) | 0 (0) | – | 0.0131 |
| AR | p.A700V | 5 (31.3) | 5 (83.3) | 0 (0) | – | 0.0005 |
|
| B, Mutations
between pathCR (n=6) and non-pathCR patients (total number=16-26,
some patients were excluded) |
|
| Gene | Amino acid
change | Overall
(%) | pathCR (%)
n=6 | Non-pathCR
(%) |
Excluded | P-value |
|
| ARID1A | p.L2117M | 5 (15.6) | 3 (50) | 2 (7.7) | 0 | 0.0101 |
| HDAC1 | p.F341V | 8 (30.8) | 4 (66.7) | 4 (20) | 6 | 0.0298 |
| APC | p.K1310M | 9 (29.0) | 4 (66.7) | 5 (20) | 1 | 0.0237 |
| BRAF | p.P75S | 10 (38.5) | 4 (66.7) | 6 (30) | 6 | 0.1054 |
| ERBB3 | p.Y86H | 5 (17.9) | 4 (66.7) | 1 (4.5) | 4 | 0.0004 |
| TP53 | p.T155I | 11 (39.3) | 5 (83.3) | 6 (27.3) | 4 | 0.0127 |
| AMER1 | p.A504V | 12 (44.4) | 5 (83.3) | 7 (33.3) | 5 | 0.0297 |
| AMER1 | p.V490D | 3 (11.5) | 3 (50) | 0 (0) | 6 | 0.0008 |
| AR | p.A700V | 9 (40.9) | 5 (83.3) | 4 (25) | 10 | 0.0132 |
The three most significant SNVs were TP53 p.T155I,
AMER1 p.A504 V and AR p.A700 V (Chi-square, P=0.0005). The results
showed that p53 exon 5 had a mutation present in 5 out of 6 (83.3%)
pathCR patients, comparing with none of the 10 non-pathCR patients.
AMER1 p.A504 V and AR p.A700 V exhibited a similar trend. P-values
of the six other significant SNVs ranged from 0.0029–0.0131. The
frequencies of the other gene mutations were not significantly
difference between pathCR and non-pathCR patients.
Validation of mutational status in the
pathCR and non-pathCR groups
To further validate the authenticity of the 9 SNVs,
the SNVs were assessed in the remaining 16 patients in the
non-pathCR group. There were three possible conditions for the
sites analyzed: Sites with <10× coverage, wild type sites with
≥10× coverage, mutant type sites with ≥10× coverage. For each loci,
we excluded patients with <10× coverage of the site (Table IIIB) and added the mutation status
of the remaining patients into the mutations status analysis of the
first 16 patients. Table IIIB shows
Chi-square analysis; 8 out of 9 SNVs passed this test, and one SNV
(BRAF, p.P75S) was eliminated according to the P-value
(P=0.1054).
The P-values changed with the addition of 16
patients. The most significant two SNVs were ERBB3 p.Y86H
(Chi-square, P=0.0004) and AMER1 p.V490D (Chi-square, P=0.0008).
ERBB3 is a member of the epidermal growth factor receptor (EGFR)
family of receptor tyrosine kinases genes. p.Y86H SNV in ERBB3 had
a 66.7% mutation frequency (4/6 patients) in the pathCR group,
compared with a 3.8% mutation frequency (1/22 patients) in the
non-pathCR group. p.V490D SNV in AMER1, a regulator of the
canonical Wnt signaling pathway, had a 50% mutation frequency in
the pathCR group (3/6 patients), compared with 0% mutation
frequency the in non-pathCR group (0/20 patients). The present
study also showed that p.T155I in TP53 had an 83.3% mutation
frequency in the pathCR group (5/6 patients), compared with a 27.3%
mutation frequency in the non-pathCR group (6/22 patients;
Chi-square, P=0.0127). Our study demonstrated that 8 SNVs were
significantly associated with pathCR, which is in accordance with
previous reports that mutations were correlated with increased OS
(17,18).
Discussion
Previous reports showed the gain of chromosome 8
copies was significantly correlated with lymph node metastasis
(19), and with advanced gastric
cancer, pancreatic cancer and lung cancer (20–22). In
the present study, we focused on whether aneuploidy of chromosome 8
and gene mutations of CTCs could assist in predicting the response
to preoperative CRT in rectal adenocarcinoma patients. Overall,
18.2% (6 out of 33) of patients achieved a pathCR following CRT.
Notably, ≥5 copies of chromosome 8 was associated with pathCR by
univariate logistic regression (P=0.042, Table II), which is in accordance with Chen
et al (23). Chen reported
that patients who achieved a pathCR had significantly fewer high
copy gains overall than non-pathCR patients (P=0.01) (23).
To investigate the influence of gene mutations of
CTCs on response to preoperative chemoradiation, we developed a
workflow to genotype of CTCs from patients. In summary, we captured
CTCs, amplified the DNA and sequenced. The coverage breadth (sites
with ≥10× coverage) in pathCR group ranged from 50.20–73.68%. For
the issues of coverage breadth and the limited number of pathCR
patients, all six patients in pathCR group and ten patients in
non-pathCR group with coverage breadth (sites with ≥10× coverage)
>80% were used in initial analysis. Among all the covered
regions, 64 nonsynonymous SNVs were identified in the 16 patients.
Of the 64 nonsynonymous SNVs, the frequencies of 9 nonsyonymous
SNVs (Table IIIA) were
significantly different between the pathCR group and non-pathCR
group. The 9 SNVs were subsequently analyzed in the remaining 16
patients in the non-pathCR group and added the mutation status of
these patients to that of the first 16 patients; following this, 8
SNVs exhibited significant differences between the groups (Table IIIB). For example, p.T155I in TP53
was detected in 83.3% of patients in the pathCR group, compared
with 27.3% of the non-pathCR group (6/22 patient; Chi-square,
P=0.0127). Although the impact of p53 mutations on patient survival
remains unclear, previous work has showed that the Arg variant of
codon 72, located in exon 4 of p53 gene, enhances the pro-apoptotic
ability of the protein (24,25). Patients in the present study may have
a similar activating mutation in p53.
The other 7 SNVs had mutation frequencies of
50–83.3% in pathCR group compared with 0–33.3% in the non-pathCR
group (Chi-square, P≤0.0131). The present study showed that
mutations in 8 SNVs were significantly associated with pathCR,
which in in accordance with previous reports that various mutations
are correlated with increased OS (17,18).
Our current workflow has several limitations that
should be overcome in subsequent studies, including the WGA success
rate. Our results demonstrated that the WGA success rate [≥80%
coverage breadth (sites with ≥10× coverage)] of LCM-dissected CTC
samples was 30.3% (10 out of 33 CTCs) in 36-gene study; however,
when the MALBAC technique was used previously on live single cells,
and the WGA success rate was ~90% (18) (data not shown). The low WGA
amplification was possibly due to the poor DNA quality caused by
the workflow. However, the current CTCs method has high sensitivity
compared with other variable CTC methods and we are still improving
the protocol to reduce damage to CTCs and increase the WGA success
rate (26). Improvements in the
following aspects may improve the WGA amplification: Optimizing the
fixation conditions to reduce the degradation of DNA; and extending
the lysis time to uncover the DNA template.
In conclusion, the present study suggests that the
number of chromosome 8 copies and 8 nonsynonymous mutations (in
ARID1A, HDAC1, APC, ERBB3, TP53, AMER1and AR) in CTCs were
associated with pathCR. Out of 6 patients whose CTCs had <5
copies of chromosome 8, 3 had pathCR (3/6 patient, 50%) and of 27
patients whose CTCs had ≥5 copies of chromosome 8 only 3 had pathCR
(3/27 patients, 11.1%; Chi-square test, P=0.0255). The 8 SNVs were
reported had mutation frequencies of 50–83.3% in the pathCR group
compared with 0–33.3% mutation frequency in the non-pathCR group.
In the future we aim to perform larger, prospective studies and
analyze long-term follow-up survival data.
Acknowledgements
Not applicable.
Funding
The present study was funded by National Natural
Science Foundation of China (grant no. 81372432).
Availability of data and materials
The datasets used and analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JFW, XQL, JZ and ZZ conceived of and designed the
study. JFW, XQL, JZ, CYC, LFY, JZ, GCL, LPL, LJS, HZ, JL and YTZ
performed the analyses. JFW, XQL, JZ, CYC, LFY, JZ, GCL, LJS and HZ
prepared all the tables. JFW, CYC, JZ and XQL wrote the main
manuscript. All authors reviewed the manuscript.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were in accordance with the ethical standards of the
institutional and/or national research committee and with the 1964
Helsinki declaration and its later amendments or comparable ethical
standards. All patients signed informed consent and the Fudan
University Shanghai Cancer Center Ethics Review Board approved the
study.
Consent for publication
Informed consent was obtained from all individual
participants included in the present study.
Competing interests
The authors declare no competing interests.
Glossary
Abbreviations
Abbreviations:
|
pathCR
|
pathologic complete response
|
|
CTCs
|
circulating tumor cells
|
|
CRT
|
preoperative chemoradiotherapy
|
|
LARC
|
locally advanced rectal cancer
|
|
TME
|
total mesorectal excision
|
|
SNVs
|
single nucleotide variants
|
|
LCM
|
laser capture microdissection
|
|
MALBAC
|
multiple annealing and looping-based
amplification cycles
|
References
|
1
|
Aschele C, Cionini L, Lonardi S, Pinto C,
Cordio S, Rosati G, Artale S, Tagliagambe A, Ambrosini G, Rosetti
P, et al: Primary tumor response to preoperative chemoradiation
with or without oxaliplatin in locally advanced rectal cancer:
Pathologic results of the STAR-01 randomized phase III trial. J
Clin Oncol. 29:2773–2780. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gérard JP, Azria D, Gourgou-Bourgade S,
Martel-Laffay I, Hennequin C, Etienne PL, Vendrely V, François E,
de La Roche G, Bouche O, et al: Comparison of two neoadjuvant
chemoradiotherapy regimens for locally advanced rectal cancer:
Results of the phase III trial ACCORD 12/0405-Prodige 2. J Clin
Oncol. 28:1638–1644. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rödel C, Liersch T, Becker H, Fietkau R,
Hohenberger W, Hothorn T, Graeven U, Arnold D, Lang-Welzenbach M,
Raab HR, et al: Preoperative chemoradiotherapy and postoperative
chemotherapy with fluorouracil and oxaliplatin versus fluorouracil
alone in locally advanced rectal cancer: Initial results of the
German CAO/ARO/AIO-04 randomised phase 3 trial. Lancet Oncol.
13:679–687. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Park IJ, You YN, Agarwal A, Skibber JM,
Rodriguez-Bigas MA, Eng C, Feig BW, Das P, Krishnan S, Crane CH, et
al: Neoadjuvant treatment response as an early response indicator
for patients with rectal cancer. J Clin Oncol. 30:1770–1776. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Martens MH, Maas M, Heijnen LA, Lambregts
DM, Leijtens JW, Stassen LP, Breukink SO, Hoff C, Belgers EJ,
Melenhorst J, et al: Long-term outcome of an organ preservation
program after neoadjuvant treatment for rectal cancer. J Natl
Cancer Inst. 108(pii): djw1712016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Renehan AG, Malcomson L, Emsley R, Gollins
S, Maw A, Myint AS, Rooney PS, Susnerwala S, Blower A, Saunders MP,
et al: Watch-and-wait approach versus surgical resection after
chemoradiotherapy for patients with rectal cancer (the OnCoRe
project): A propensity-score matched cohort analysis. Lancet Oncol.
17:174–183. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kalady MF, de Campos-Lobato LF, Stocchi L,
Geisler DP, Dietz D, Lavery IC and Fazio VW: Predictive factors of
pathologic complete response after neoadjuvant chemoradiation for
rectal cancer. Ann Surg. 250:582–589. 2009.PubMed/NCBI
|
|
8
|
Russo AL, Ryan DP, Borger DR, Wo JY,
Szymonifka J, Liang WY, Kwak EL, Blaszkowsky LS, Clark JW, Allen
JN, et al: Mutational and clinical predictors of pathologic
complete response in the treatment of locally advanced rectal
cancer. J Gastrointest Cancer. 45:34–39. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zong C, Lu S, Chapman AR and Xie XS:
Genome-wide detection of single-nucleotide and copy-number
variations of a single human cell. Science. 338:1622–1626. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Armaghany T, Wilson JD, Chu Q and Mills G:
Genetic alterations in colorectal cancer. Gastrointest Cancer Res.
5:19–27. 2012.PubMed/NCBI
|
|
11
|
Kaz AM and Brentnall TA: Genetic testing
for colon cancer. Nat Clin Pract Gastroenterol Hepatol. 3:670–679.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Martin M: Cutadapt removes adapter
sequences from high-throughput sequencing reads. EMBnet J.
17:10–12. 2011. View Article : Google Scholar
|
|
13
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li H, Handsaker B, Wysoker A, Fennell T,
Ruan J, Homer N, Marth G, Abecasis G and Durbin R; Genome Project
Data Processing S: The sequence alignment/map format and SAMtools.
Bioinformatics. 25:2078–2079. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
DePristo MA, Banks E, Poplin R, Garimella
KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA,
Hanna M, et al: A framework for variation discovery and genotyping
using next-generation DNA sequencing data. Nature Genet.
43:491–498. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Leung ML, Wang Y, Waters J and Navin NE:
SNES: Single nucleus exome sequencing. Genome Biol. 16:552015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gibson MK, Abraham SC, Wu TT, Burtness B,
Heitmiller RF, Heath E and Forastiere A: Epidermal growth factor
receptor, p53 mutation and pathological response predict survival
in patients with locally advanced esophageal cancer treated with
preoperative chemoradiotherapy. Clin Cancer Res. 9:6461–6468.
2003.PubMed/NCBI
|
|
18
|
Villafranca E, Okruzhnov Y, Dominguez MA,
García-Foncillas J, Azinovic I, Martinez E, Illarramendi JJ, Arias
F, Martinez Monge R, Salgado E, et al: Polymorphisms of the
repeated sequences in the enhancer region of the thymidylate
synthase gene promoter may predict downstaging after preoperative
chemoradiation in rectal cancer. J Clin Oncol. 19:1779–1786. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hao JJ, Yao HQ, Dai GY, Kang W, Jia XM, Xu
X, Cai Y, Zhan QM, Wang GQ and Wang MR: Chromosomal aneuploidies
and combinational fluorescence in situ hybridization probe panels
are useful for predicting prognosis for esophageal squamous cell
carcinoma. J Gastroenterol. 50:155–166. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Y, Zhang X, Ge S, Gao J, Gong J, Lu M,
Zhang Q, Cao Y, Wang DD, Lin PP, et al: Clinical significance of
phenotyping and karyotyping of circulating tumor cells in patients
with advanced gastric cancer. Oncotarget. 5:6594–6602. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gao Y, Zhu Y, Zhang Z, Zhang C, Huang X
and Yuan Z: Clinical significance of pancreatic circulating tumor
cells using combined negative enrichment and
immunostaining-fluorescence in situ hybridization. J Exp Clin
Cancer Res. 35:662016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu C, Hao H, Li L, Zhou X, Guo Z, Zhang L,
Zhang X, Zhong W, Guo H, Bremner RM, et al: Preliminary
investigation of the clinical significance of detecting circulating
tumor cells enriched from lung cancer patients. J Thorac Oncol.
4:30–36. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Z, Liu Z, Li W, Qu K, Deng X, Varma
MG, Fichera A, Pigazzi A and Garcia-Aguilar J: Chromosomal copy
number alterations are associated with tumor response to
chemoradiation in locally advanced rectal cancer. Genes Chromosomes
Cancer. 50:689–699. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dumont P, Leu JI, Della Pietra AC III,
George DL and Murphy M: The codon 72 polymorphic variants of p53
have markedly different apoptotic potential. Nat Genet. 33:357–365.
2003. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Johnstone RW, Ruefli AA and Lowe SW:
Apoptosis: A link between cancer genetics and chemotherapy. Cell.
108:153–164. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin PP: Integrated EpCAM-independent
subtraction enrichment and iFISH strategies to detect and classify
disseminated and circulating tumors cells. Clin Transl Med.
4:382015. View Article : Google Scholar : PubMed/NCBI
|