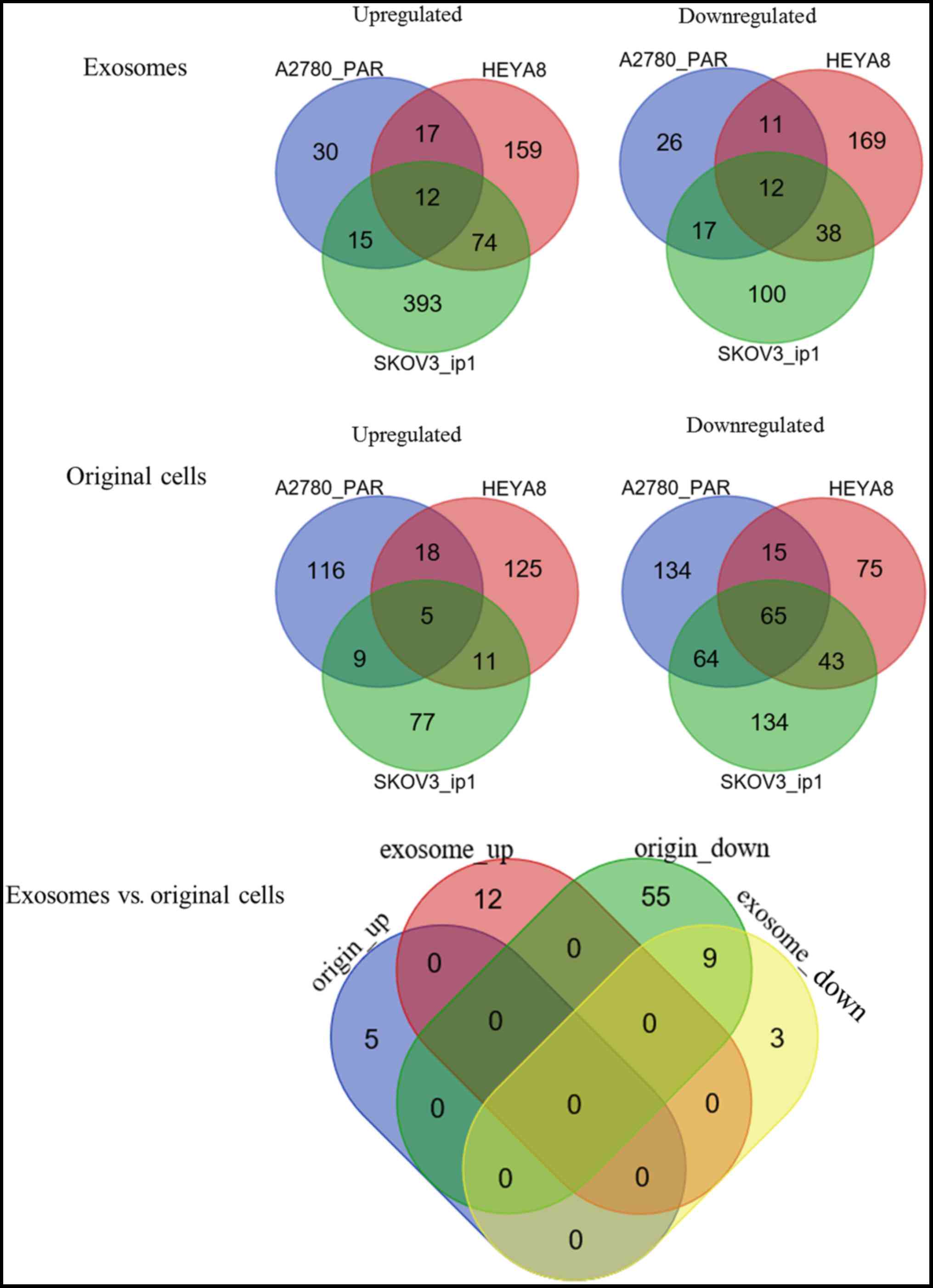
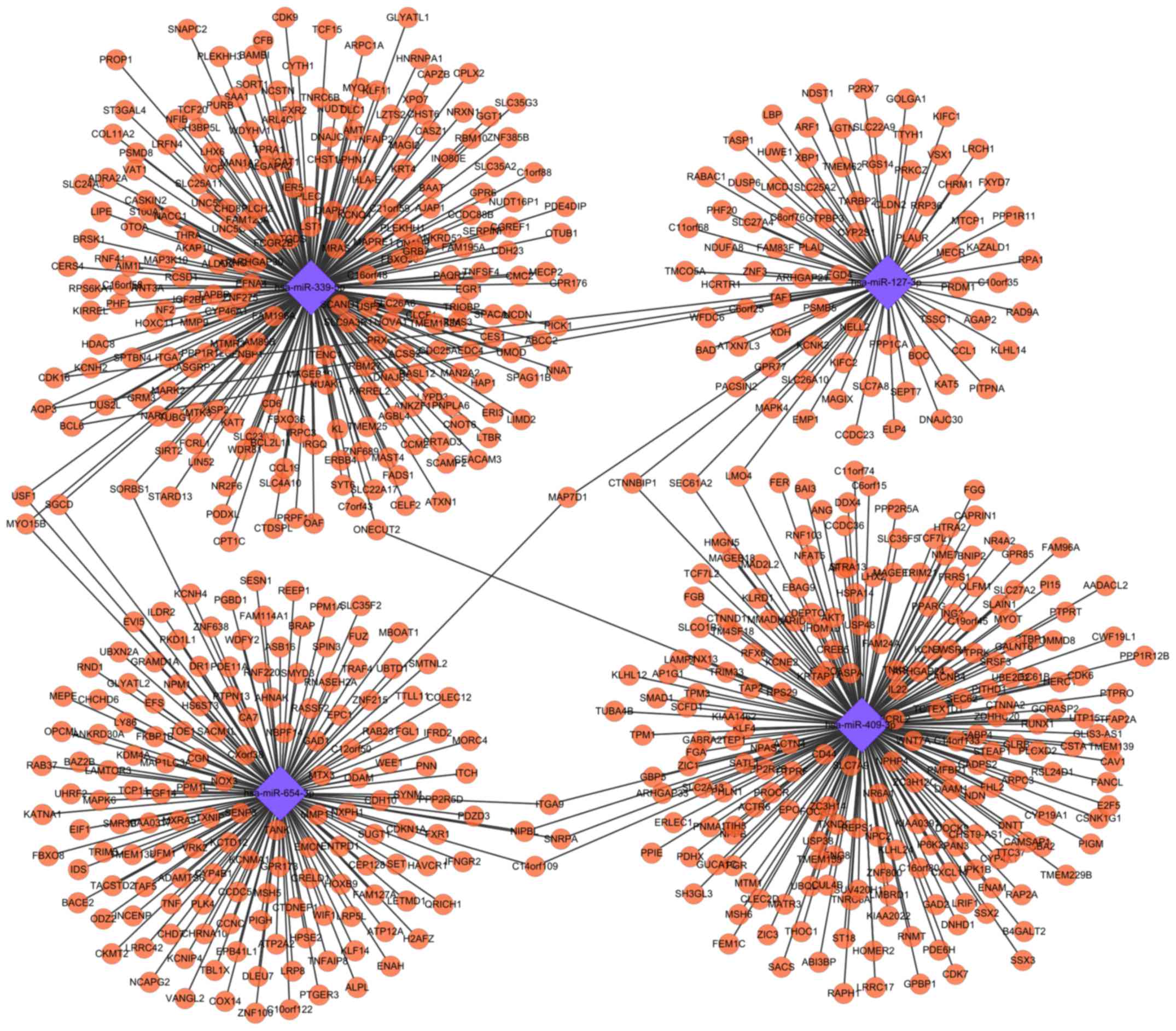
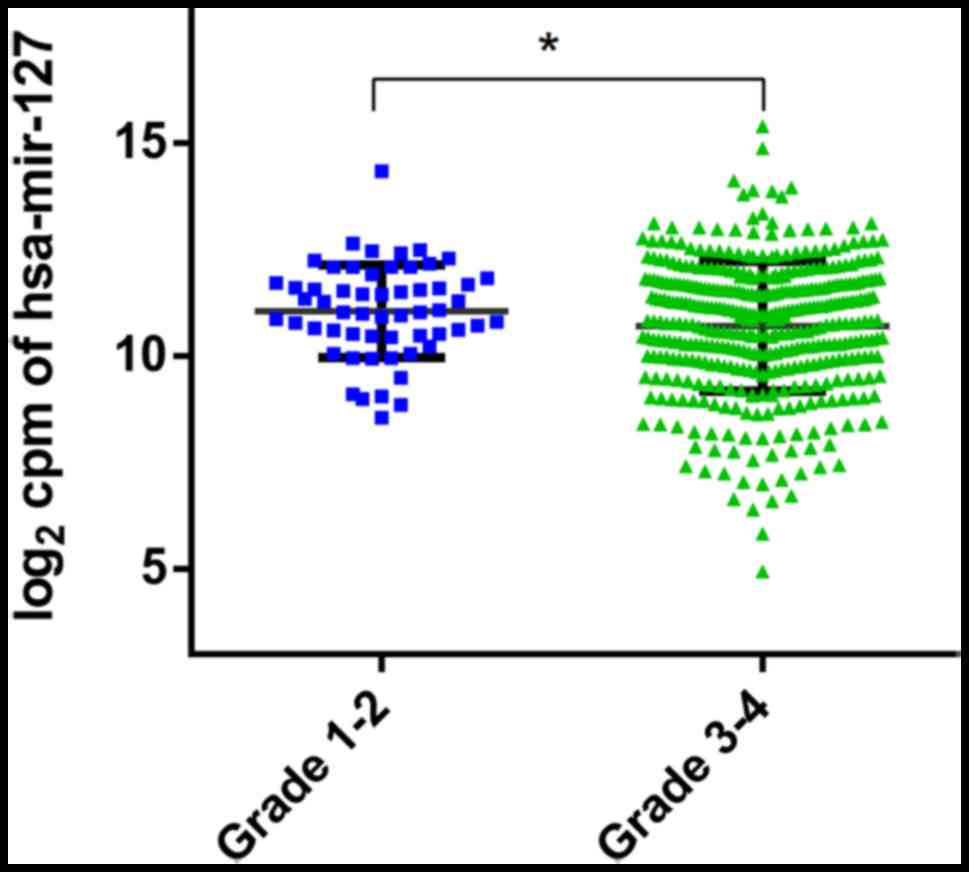
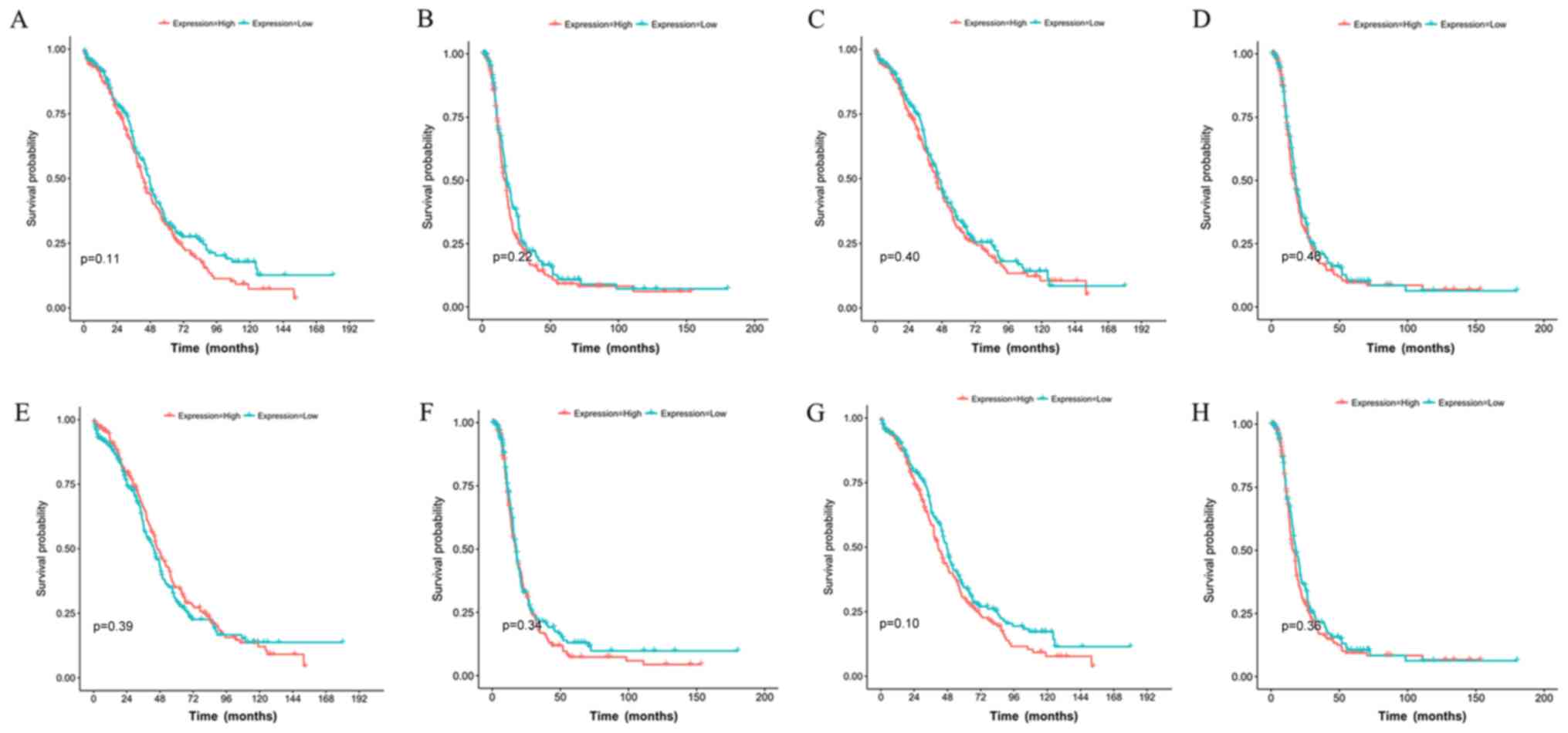
|
1
|
Chen W, Zheng R, Baade P, Zhang S, Zeng H,
Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China, 2015.
CA-Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cliby WA, Powell MA, Al-Hammadi N, Chen L,
Miller Philip J, Roland PY, Mutch DG and Bristow RE: Ovarian cancer
in the United States: Contemporary patterns of care associated with
improved survival. Gynecol Oncol. 136:11–17. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hausser J and Zavolan M: Identification
and consequences of miRNA-target interactions-beyond repression of
gene expression. Nat Rev Genet. 15:599–612. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang Z, Wang XL, Bai R, Liu WY, Li X, Liu
M and Tang H: miR-23a promotes IKKα expression but suppresses ST7L
expression to contribute to the malignancy of epithelial ovarian
cancer cells. Br J Cancer. 115:731–740. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shen W, Song M, Liu J, Qiu G, Li T, Hu Y
and Liu H: MiR-26a promotes ovarian cancer proliferation and
tumorigenesis. PLoS One. 9:e868712014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dong P, Xiong Y, Watari H, Hanley SJ,
Konno Y, Ihira K, Yamada T, Kudo M, Yue J and Sakuragi N: MiR-137
and miR-34a directly target Snail and inhibit EMT, invasion and
sphere-forming ability of ovarian cancer cells. J Exp Clin Cancer
Res. 35:1322016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Neviani P and Fabbri M: Exosomic microRNAs
in the tumor microenvironment. Front Med (Lausanne).
2:472015.PubMed/NCBI
|
|
8
|
Baroni S, Romero-Cordoba S, Plantamura I,
Dugo M, D'Ippolito E, Cataldo A, Cosentino G, Angeloni V, Rossini
A, Daidone MG and Iorio MV: Exosome-mediated delivery of miR-9
induces cancer-associated fibroblast-like properties in human
breast fibroblasts. Cell Death Dis. 7:e23122016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh R, Pochampally R, Watabe K, Lu Z and
Mo YY: Exosome-mediated transfer of miR-10b promotes cell invasion
in breast cancer. Mol Cancer. 13:2562014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kobayashi M, Salomon C, Tapia J, Illanes
SE, Mitchell MD and Rice GE: Ovarian cancer cell invasiveness is
associated with discordant exosomal sequestration of Let-7 miRNA
and miR-200. J Transl Med. 12:42014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ying X, Wu Q, Wu X, Zhu Q and Wang X,
Jiang L, Chen X and Wang X: Epithelial ovarian cancer-secreted
exosomal miR-222-3p induces polarization of tumor-associated
macrophages. Oncotarget. 7:43076–43087. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kanlikilicer P, Rashed MH, Bayraktar R,
Mitra R, Ivan C, Aslan B, Zhang X, Filant J, Silva AM,
Rodriguez-Aguayo C, et al: Ubiquitous release of exosomal tumor
suppressor miR-6126 from ovarian cancer cells. Cancer Res.
76:7194–7207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
De A, Powers B, De A, Zhou J, Sharma S,
Van Veldhuizen P, Bansal A, Sharma R and Sharma M: Emblica
officinalis extract downregulates pro-angiogenic molecules via
upregulation of cellular and exosomal miR-375 in human ovarian
cancer cells. Oncotarget. 7:31484–31500. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Smyth GK: Limma: Linear models for
microarray dataBioinform Comput Biol Solut Using R Bioconduct.
Gentleman R, Carey V, Dudoit S, Irizarry R and Huber W: Springer;
New York, NY: pp. 397–420. 2005
|
|
15
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
da Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kosary CL: FIGO stage, histology,
histologic grade, age and race as prognostic factors in determining
survival for cancers of the female gynecological system: An
analysis of 1973–87 SEER cases of cancers of the endometrium,
cervix, ovary, vulva, and vagina. Semin Surg Oncol. 10:31–46. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shimizu Y, Kamoi S, Amada S, Akiyama F and
Silverberg SG: Toward the development of a universal grading system
for ovarian epithelial carcinoma: Testing of a proposed system in a
series of 461 patients with uniform treatment and follow-up.
Cancer. 82:893–901. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46(D1): D956–D963. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang G, Liu Z, Xu H and Yang Q:
miR-409-3p suppresses breast cancer cell growth and invasion by
targeting Akt1. Biochem Biophys Res Commun. 469:189–195. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bi L, Yang Q, Yuan J, Miao Q, Duan L, Li F
and Wang S: MicroRNA-127-3p acts as a tumor suppressor in
epithelial ovarian cancer by regulating the BAG5 gene. Oncol Rep.
36:2563–2570. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shan W, Li J, Bai Y and Lu X: miR-339-5p
inhibits migration and invasion in ovarian cancer cell lines by
targeting NACC1 and BCL6. Tumour Biol. 37:5203–5211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Arend RC, Londoño-Joshi AI, Straughn JM Jr
and Buchsbaum DJ: The Wnt/β-catenin pathway in ovarian cancer: A
review. Gynecol Oncol. 131:772–779. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoshioka S, King ML, Ran S, Okuda H,
MacLean JA II, Mcasey ME, Sugino N, Brard L, Watabe K and Hayashi
K: WNT7A regulates tumor growth and progression in ovarian cancer
through the WNT/β-catenin pathway. Mol Cancer Res. 10:469–482.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Usongo M, Li X and Farookhi R: Activation
of the canonical WNT signaling pathway promotes ovarian surface
epithelial proliferation without inducing β-catenin/Tcf-mediated
reporter expression. Dev Dyn. 242:291–300. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Puvirajesinghe TM, Bertucci F, Jain A,
Scerbo P, Belotti E, Audebert S, Sebbagh M, Lopez M, Brech A,
Finetti P, et al: Identification of p62/SQSTM1 as a component of
non-canonical Wnt VANGL2-JNK signalling in breast cancer. Nat
Commun. 7:103182016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Deng Y, Deng H, Liu J, Han G, Malkoski S,
Liu B, Zhao R, Wang XJ and Zhang Q: Transcriptional down-regulation
of Brca1 and E-cadherin by CtBP1 in breast cancer. Mol Carcinog.
51:500–507. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang R, Asangani IA, Chakravarthi BV,
Ateeq B, Lonigro RJ, Cao Q, Mani RS, Camacho DF, McGregor N,
Schumann TE, et al: Role of transcriptional corepressor CtBP1 in
prostate cancer progression. Neoplasia. 14:905–914. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nishiyama M, Skoultchi AI and Nakayama KI:
Histone H1 recruitment by CHD8 is essential for suppression of the
Wnt-β-catenin signaling pathway. Mol Cell Biol. 32:501–512. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sawada G, Ueo H, Matsumura T, Uchi R,
Ishibashi M, Mima K, Kurashige J, Takahashi Y, Akiyoshi S, Sudo T,
et al: CHD8 is an independent prognostic indicator that regulates
Wnt/β-catenin signaling and the cell cycle in gastric cancer. Oncol
Rep. 30:1137–1142. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Menon T, Yates JA and Bochar DA:
Regulation of androgen-responsive transcription by the chromatin
remodeling factor CHD8. Mol Endocrinol. 24:1165–1174. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jones DH and Lin DI: Amplification of the
NSD3-BRD4-CHD8 pathway in pelvic high-grade serous carcinomas of
tubo-ovarian and endometrial origin. Mol Clin Oncol. 7:301–307.
2017.PubMed/NCBI
|
|
33
|
Shingleton JR and Hemann MT: The chromatin
regulator CHD8 is a context-dependent mediator of cell survival in
murine hematopoietic malignancies. PLoS One. 10:e01432752015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
MacLean JA II, King ML, Okuda H and
Hayashi K: WNT7A regulation by miR-15b in ovarian cancer. PLoS One.
11:e01561092016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Deng Y, Deng H, Bi F, Liu J, Bemis LT,
Norris D, Wang XJ and Zhang Q: MicroRNA-137 targets
carboxyl-terminal binding protein 1 in melanoma cell lines. Int J
Biol Sci. 7:133–137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang L, Madigan MC, Chen H, Liu F,
Patterson KI, Beretov J, O'Brien PM and Li Y: Expression of
urokinase plasminogen activator and its receptor in advanced
epithelial ovarian cancer patients. Gynecol Oncol. 114:265–272.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dorn J, Harbeck N, Kates R, Gkazepis A,
Scorilas A, Soosaipillai A, Diamandis E, Kiechle M, Schmalfeldt B
and Schmitt M: Impact of expression differences of
kallikrein-related peptidases and of uPA and PAI-1 between primary
tumor and omentum metastasis in advanced ovarian cancer. Ann Oncol.
22:877–883. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tang J, Wang J, Fan L, Li X, Liu N, Luo W,
Wang J and Wang Y and Wang Y: cRGD inhibits vasculogenic mimicry
formation by down-regulating uPA expression and reducing EMT in
ovarian cancer. Oncotarget. 7:24050–24062. 2016.PubMed/NCBI
|
|
39
|
Estrella VC, Eder AM, Liu S, Pustilnik TB,
Tabassam FH, Claret FX, Gallick GE, Mills GB and Wiener JR:
Lysophosphatidic acid induction of urokinase plasminogen activator
secretion requires activation of the p38MAPK pathway. Int J Oncol.
31:441–449. 2007.PubMed/NCBI
|
|
40
|
Li XF, Yan PJ and Shao ZM: Downregulation
of miR-193b contributes to enhance urokinase-type plasminogen
activator (uPA) expression and tumor progression and invasion in
human breast cancer. Oncogene. 28:3937–3948. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li J, Kong F, Wu K, Song K, He J and Sun
W: miR-193b directly targets STMN1 and uPA genes and suppresses
tumor growth and metastasis in pancreatic cancer. Mol Med Rep.
10:2613–2620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xie C, Jiang XH, Zhang JT, Sun TT, Dong
JD, Sanders AJ, Diao RY, Wang Y, Fok KL, Tsang LL, et al: CFTR
suppresses tumor progression through miR-193b targeting urokinase
plasminogen activator (uPA) in prostate cancer. Oncogene.
32(2282–2291): 2291. e1–e7. 2013.
|
|
43
|
Kawabe T, Muslin AJ and Korsmeyer SJ:
HOX11 interacts with protein phosphatases PP2A and PP1 and disrupts
a G2/M cell-cycle checkpoint. Nature. 385:454–458. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Dessauge F, Cayla X, Albar JP, Fleischer
A, Ghadiri A, Duhamel M and Rebollo A: Identification of PP1alpha
as a caspase-9 regulator in IL-2 deprivation-induced apoptosis. J
Immunol. 177:2441–2451. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Hsu LC, Huang X, Seasholtz S, Potter DM
and Gollin SM: Gene amplification and overexpression of protein
phosphatase 1alpha in oral squamous cell carcinoma cell lines.
Oncogene. 25:5517–5526. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nohata N, Hanazawa T, Kikkawa N, Sakurai
D, Fujimura L, Chiyomaru T, Kawakami K, Yoshino H, Enokida H,
Nakagawa M, et al: Tumour suppressive microRNA-874 regulates novel
cancer networks in maxillary sinus squamous cell carcinoma. Br J
Cancer. 105:833–841. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shastry AH, Thota B, Srividya MR,
Arivazhagan A and Santosh V: Nuclear Protein Phosphatase 1 α (PP1A)
expression is associated with poor prognosis in p53 expressing
glioblastomas. Pathol Oncol Res. 22:287–292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Noskova V, Ahmadi S, Asander E and Casslén
B: Ovarian cancer cells stimulate uPA gene expression in
fibroblastic stromal cells via multiple paracrine and autocrine
mechanisms. Gynecol Oncol. 115:121–126. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lin F, Wang HJ, Li CX, Li H, Wang T, Nan
P, Qian HL and Zhan QM: Effects of esophageal cancer cell-derived
exosomes on cancer cell migration and invasion and its mechanism
research. Med J Chin PLA. 42:307–313. 2017.
|
|
50
|
Dempsey E, Dervin F and Maguire PB:
Platelet derived exosomes are enriched for specific microRNAs which
regulate WNT signalling in endothelial cells. Blood.
124:27602014.PubMed/NCBI
|
|
51
|
Zhang P, Garnett J, Creighton CJ, Al
Sannaa GA, Igram DR, Lazar A, Liu X, Liu C and Pollock RE:
EZH2-miR-30d-KPNB1 pathway regulates malignant peripheral nerve
sheath tumour cell survival and tumourigenesis. J Pathol.
232:308–318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li B, Jiang S, Yu X, Cheng C, Chen S,
Cheng Y, Yuan JS, Jiang D, He P and Shan L: Phosphorylation of
trihelix transcriptional repressor ASR3 by MAP KINASE4 negatively
regulates Arabidopsis immunity. Plant Cell. 27:839–856. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rashed MH, Kanlikilicer P,
Rodriguez-Aguayo C, Pichler M, Bayraktar R, Bayraktar E, Ivan C,
Filant J, Silva A, Aslan B, et al: Exosomal miR-940 maintains
SRC-mediated oncogenic activity in cancer cells: A possible role
for exosomal disposal of tumor suppressor miRNAs. Oncotarget.
8:20145–20164. 2017. View Article : Google Scholar : PubMed/NCBI
|