Introduction
The initiation and progression of hepatocellular
carcinoma (HCC) is regarded as a multi-step process entailing
genetic and epigenetic alterations (1–3).
Epigenetic modifications can affect gene expression at the cancer
transcriptome level more extensively than genetic changes.
Furthermore, the majority of epigenetic alterations are reversible,
and therefore could provide a prospective treatment for HCC
(4). Increasing evidence has
suggested that tumor-associated histone methylation is an important
determinant of initiation and the development of HCC, and
represents a promising therapeutic target (5).
Histone methylation can be divided into arginine
methylation and lysine methylation. SET domain-containing protein 8
(SET8) is a member of the SET domain-containing methyltransferase
family, which particularly catalyzes the monomethylation of histone
H4 Lys20 (6–8). SET8 has been reported to participate in
diverse biological processes, the regulation of gene transcription,
maintenance of genomic stability, formation of heterochromatin, and
cell cycle progression and development (9–15). Protein
arginine methyltransferase 5 (PRMT5), a type II arginine
methyltransferase, regulates gene transcription and modulates
chromatin structure through symmetrically dimethylating arginine
residues on target proteins (16,17). H3R8
and H4R3 are the key repressive histone methylation sites that are
methylated by PRMT5 (18,19). Accumulating evidence has suggested
that PRMT5 may act as an oncogene in multiple tumors via epigenetic
repression of several tumor suppressor genes or by
post-translational modification of signaling molecules (16,20,21).
Histone methylation has diverse functional
consequences depending on the type of residue and which site the
methylation modifies (22). Several
studies have revealed a complex interplay between the distinct
covalent modifications that occur on the histone tails, including
methylation of histone H3 and H4, which supported the histone code
hypothesis (23,24). The patterns of histone methylation
associated with progression of cancer have potential clinical
value. The present study revealed that there was a significant
correlation between distinct histone methylations, and their
combination to predict survival and tumor recurrence, particularly
for the prediction of early recurrence (ER).
Patients and methods
Patients and HCC tissue
A total of 195 HCC patients (175 male and 20 female)
who underwent curative resection by the same surgical team between
1999 and 2006 at the Liver Cancer Institute of Fudan University
(Shanghai, China) were included in this study. The mean age of
patients was 52 years (range, 26–78 years). The surgical techniques
for liver resection have been described previously (25). The inclusion and exclusion criterion
of patient cohort include (1) having
a distinctive pathologic diagnosis of HCC, (2) without preoperative anticancer treatment
and extrahepatic metastases, (3)
under primary and curative liver resection, and (4) with complete clinicopathologic and
follow-up data. Tumor tissue was collected with the written
informed consent of each patient and the study was approved by the
Ethics Committee of Zhongshan Hospital.
Patients in the cohort were followed up until March
2012, with a median observation time of 60 months (range, 2–142
months). The follow-up procedures were described in a previous
study (25), and treatment programs
following relapse were administered according to a uniform
guideline (26). Tumor staging was
determined according to the 2002 International Union Against Cancer
Tumor-Node-Metastasis (TNM) classification system. Overall survival
(OS) was defined as the interval between the dates of surgery and
patient mortality. Time to recurrence (TTR) was defined as the
interval between the dates of surgery and the first detection of
intrahepatic or distant recurrence. Early recurrence was defined as
any recurrence type that occurred within the time period of up to
and including 24 months (27). Data
were censored at the final follow-up for patients without
recurrence or mortality.
Tissue microarray (TMA) and
immunohistochemistry
Tissue microarrays were conducted as described
previously (25), and were embedded
by Shanghai Biochip Company Ltd. (Shanghai, China).
Immunohistochemistry for SET8 and PRMT5 was performed as described
previously (25). Briefly, sections
were deparaffinized in xylene (100%, three times, 5 min each) and
rehydrated by stepwise washes in decreasing ethanol solutions ratio
(100, 95, 80 and 75% for 5 min each) at room temperature. Slides
were treated with 3% hydrogen peroxide for 15 min at room
temperature. Antigen retrieval was achieved by boiling slides in
the autoclave (80 kpa and 100°C) for 1 min in 0.01 M sodium citrate
buffer (pH 6.0), followed by cooling to room temperature. Sections
were incubated with 10% normal goat serum (Boster Biological
Technology, Pleasanton, CA, USA) for 60 min at room temperature to
block nonspecific antigen sites. Slides were incubated with primary
antibodies against SET8 (1:200 dilution; Cell Signaling Technology,
Inc., Danvers, MA, USA) or PRMT5 (1:200 dilution; Abcam, Cambridge,
UK) overnight at 4°C, followed by incubation with a
biotin-conjugated secondary antibody, and horseradish
peroxidase-conjugated streptavidin for 60 min at room temperature.
The sections were stained with DAB for 1 min at room temperature
and counterstained with hematoxylin for 1 min at room temperature.
Negative controls were treated identically but with the primary
antibody omitted.
The density of positive staining was measured with a
computerized image system. Briefly, images of the representative
fields were captured and analyzed with Image-Pro Plus version 6.2
software (Media Cybernetics, Inc., Rockville, MD, USA). The
integrated optical density of the positive staining for SET8 and
PRMT5 was measured, and the ratio to total area of each image was
defined as the SET8 and PRMT5 density. The final staining score was
classified into four grades: Score 1 (weak, less than the 25th
percentile value), score 2 (moderate, between 25th and 50th
percentile value), score 3 (strong, between 50th and 75th
percentile value), and score 4 (very strong, higher than 75th
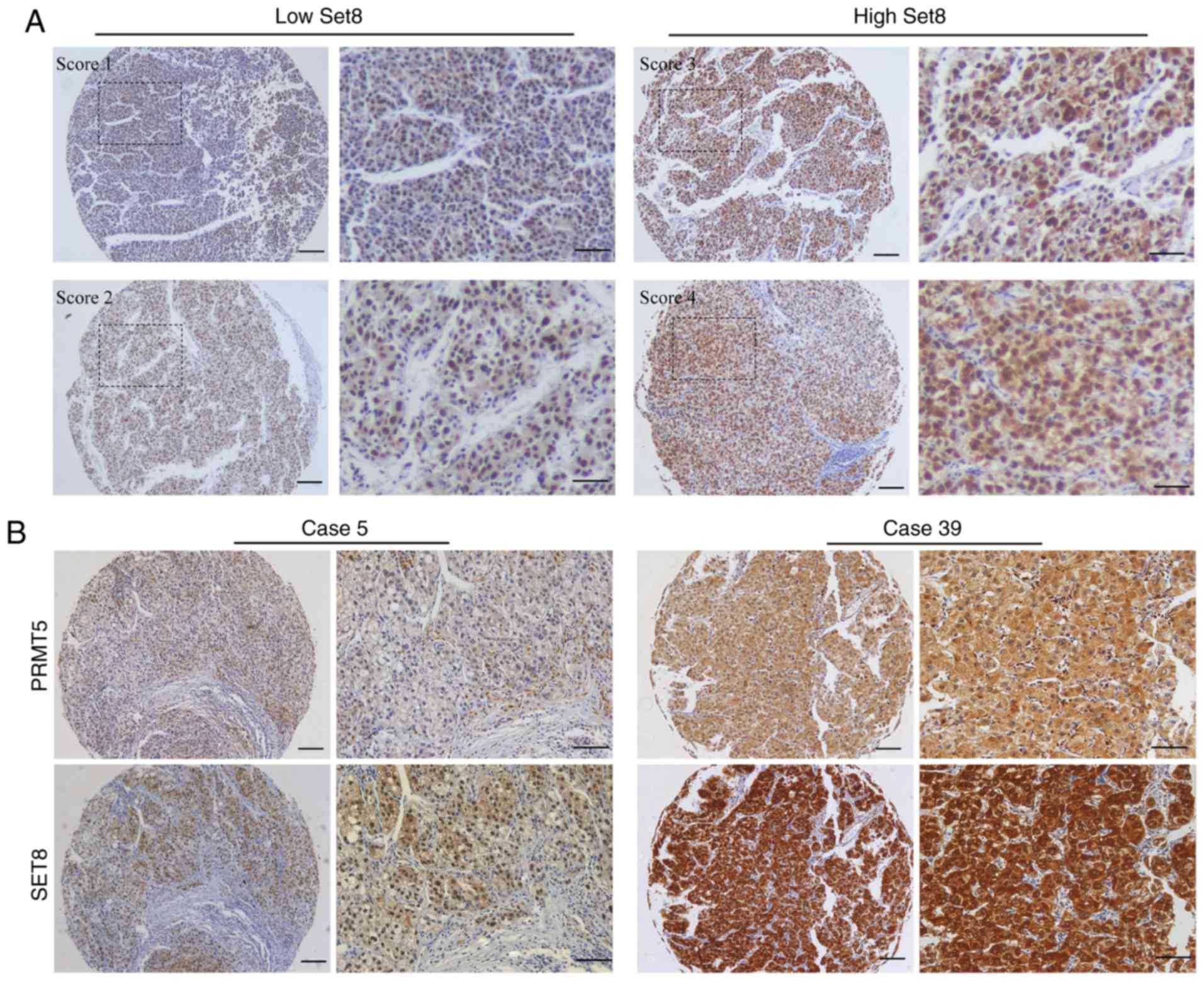
percentile value). HCC tissues with staining scores 1 and 2 were
defined as low expression, and those with scores 3 and 4 were
defined as high expression (representative figures are demonstrated
in Fig. 1).
Statistical analysis
Statistical analysis was performed with SPSS 13.0
for Windows (SPSS Inc., Chicago, IL, USA). The mean, standard
deviation and frequency were used to summarize patient variables
within the SET8/PRMT5 high and low expression groups. The Pearson's
χ2 test or Fisher's exact test were used to compare
categorical variables; and continuous variables were analyzed by
the Students t-test or Pearson's correlation test. SET8 and PRMT5
expression in tumor tissues was analyzed using the Spearman's rank
correlation test. Kaplan-Meier analysis was used to determine OS.
The log-rank test was used to compare OS between subgroups and the
Cox regression model was used to perform multivariate analyses.
Receiver operating characteristic (ROC) curve analysis was used to
determine the predictive value of the parameters. P<0.05 was
considered to indicate a statistically significant difference.
Results
Clinicopathological data
The present study included 195 patients with
surgically resected TNM stage I–IIIA HCC; comprising 123 stage I,
31 stage II and 41 stage IIIA patients. At the time of the last
follow-up, mortality was noted in 126 patients, including 11
patients who succumbed to liver failure or other diseases without
record of tumor recurrence; and 148 patients exhibited tumor
recurrence. Treatment programs following relapse included resection
(n=36), trans-arterial chemoembolization (n=79, ×1-10),
radiofrequency ablation and percutaneous ethanol injection (n=55)
and radiotherapy (n=6) according to a uniform guideline. Among
recurrent patients, six were unable to tolerate any antitumor
treatments due to severe liver dysfunction or weak general
performance. The 1-, 3- and 5-year OS rates were 79.8, 62.3 and
47.5%, respectively, and the 1-, 3- and 5-year probabilities of
recurrence were 27.4, 57.2 and 67.2%, respectively.
Expression of SET8 and PRMT5, and
their correlation with clinicopathological characteristics in
HCC
Immunohistochemical analysis revealed that the
staining of SET8 was mainly limited to the nucleus, whereas the
expression pattern of PRMT5 in the tumor cells was either diffuse
in the cytoplasm or concentrated in the nucleus. Representative low
staining and high staining of SET8 in HCC tissues are presented in
Fig. 1A. According to the staining
scores, 53.3% of HCC samples (104/195) revealed a high expression
of SET8, and 54.4% of HCC samples (106/195) revealed a high
expression of PRMT5 (data not shown). As is presented Fig. 1B, low expression of SET8 is associated
with low PRMT5 in the same HCC specimen, and high expression of
SET8 is also correlated with high PRMT5 in the same HCC specimen.
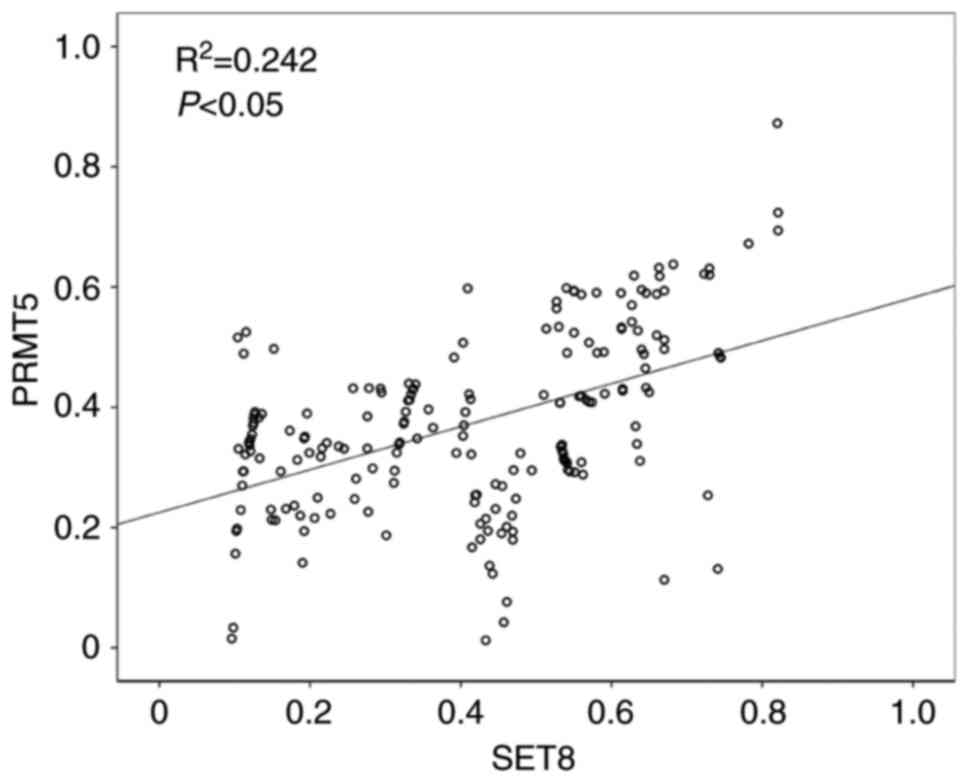
In addition, a statistically significant positive correlation
between SET8 and PRMT5 expression in tumor tissue was observed
(P<0.05; Fig. 2).
The present study assessed whether SET8 and PRMT5
expression were correlated with clinicopathological parameters,
including age, sex, Eastern Cooperative Oncology Group (ECOG)
performance score, liver cirrhosis, serum AFP level, encapsulation,
vascular invasion, tumor size, tumor number and TNM stage. It was
identified that SET8 and PRMT5 expression was not statistically
correlated with any clinicopathological characteristics (data not
shown).
Prognostic significance of the
expression of SET8 and PRMT5
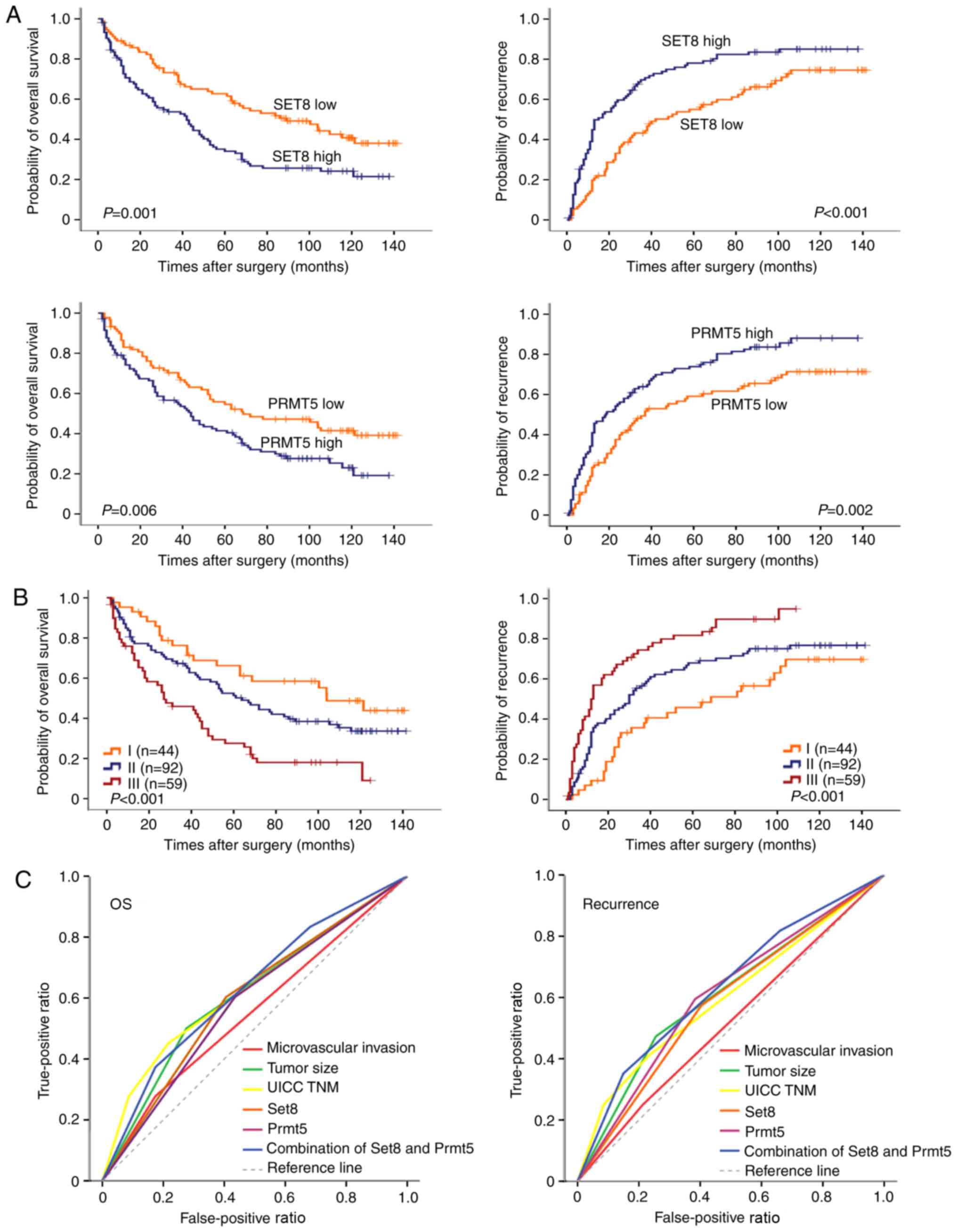
Kaplan-Meier survival curves revealed that high
expression of SET8 or PRMT5 was significantly associated with a
lower OS rate (P=0.001 and P=0.006, respectively; Fig. 3A) and TTR (P<0.001 and P=0.002,
respectively; Fig. 3A). In univariate
analysis, several clinicopathological factors, including AFP,
vascular invasion, tumor size, tumor number and TNM stage revealed
prognostic significance for OS and TTR (P<0.05 for all, Table I). Liver cirrhosis was also associated
with OS (P=0.038, Table I). In
addition, all clinicopathological factors revealing significance by
univariate analysis, with the exception of TNM stage, were adopted
when multivariate Cox proportional hazards analysis was performed
(Table I). The results suggested that
vascular invasion, tumor size, tumor number, and SET8 and PRMT5,
were independent risk factors for OS and TTR (P<0.05 for all,
Table I).
 | Table I.Association between
clinicopathological features, SET8, PRMT5 expression and
survival. |
Table I.
Association between
clinicopathological features, SET8, PRMT5 expression and
survival.
|
| OS | TTR |
|---|
|
|
|
|
|---|
|
|
| Multivariate |
| Multivariate |
|---|
|
|
|
|
|
|
|---|
| Factor | Univariate P
-value | HR | 95% CI | P-value | Univariate
P-value | HR | 95% CI | P-value |
|---|
| Age (≤50 vs. >50
years) | 0.126 | |
| NA | 0.192 |
|
| NA |
| Sex (female vs.
male) | 0.105 |
|
| NA | 0.056 |
|
| NA |
| ECOG (0 vs. 1) | 0.39 |
|
| NA | 0.829 |
|
| NA |
| Liver cirrhosis
(yes vs. no) | 0.038 | 1.789 | 1.024–3.126 | 0.041 | 0.05 |
|
| NS |
| AFP (≤400 vs.
>400 ng/ml) | 0.029 |
|
| NS | 0.048 |
|
| NS |
| Encapsulation (yes
vs. no) | 0.284 |
|
| NA | 0.484 |
|
| NA |
| Vascular invasion
(yes vs. no) | <0.001 | 2.464 | 1.645–3.690 | <0.001 | 0.006 | 1.783 | 1.216–2.614 | 0.003 |
| Tumor size (≤5.8
vs. >5.8 cm) | <0.001 | 1.956 | 1.358–2.817 | <0.001 | <0.001 | 1.899 | 1.351–2.670 | <0.001 |
| Tumor number
(single vs. multiple) | 0.002 | 1.613 | 1.053–2.471 | 0.028 | 0.001 | 1.602 | 1.066–2.407 | 0.023 |
| UICC TNM stage (I
vs. II vs. IIIA) | <0.001 |
|
|
| <0.001 |
|
|
|
| Set8 (negative vs.
positive) | 0.001 | 2.003 | 1.385–2.897 | <0.001 | <0.001 | 1.894 | 1.356–2.646 | <0.001 |
| Prmt5 (negative vs.
positive) | 0.006 | 1.712 | 1.176–2.492 | 0.005 | 0.002 | 1.948 | 1.382–2.745 | <0.001 |
| Combine set8 and
Prmt5 | <0.001 | | |
| <0.001 | 1.908 | 1.513–2.407 |
|
Combination of SET8 and PRMT5
expression and ROC analysis
Patients were classified into three groups according
to their SET8 and PRMT5 expression: Group I (n=44), low expression
of SET8 and PRMT5; group II (n=92), high expression of SET8 or
PRMT5; and group III (n=59), high expression of SET8 and PRMT5.
Differences in OS (P<0.001) and TTR (P<0.001) were
significant among the three groups. The 5-year OS and TTR rates
were 67.1 and 55.3%, respectively, for group I, but only 27.5 and
18.3%, respectively, for group III (Fig.
3B).
Clinicopathological variables that revealed a
significant result in multivariate survival analyses and in
combination with SET8 and PRMT5 were incorporated in ROC analyses.
All the incorporated variables were significantly associated with
patient mortality and disease recurrence (P<0.05). The
predictive value of SET8 combined with PRMT5 for OS and TTR was
revealed to be more accurate than SET8 or PRMT5 alone. For patient
mortality and disease recurrence, the predictive value of the
combination of SET8 and PRMT5 was the most accurate (Fig. 3C). The area under the curve of this
combination was 0.635 [95% confidence interval (CI), 0.554–0.716;
P=0.002] for patient mortality and 0.641 (95% CI, 0.552–0.730;
P=0.004) for disease recurrence.
Combination of SET8 and PRMT5 in
predicting the prognosis of early stage HCC
Due to the high rates of recurrence following
hepatectomy, it is critical to identify reliable prognostic markers
to define patients at high risk of recurrence, particularly for
patients with early stage disease who do not have significant
vascular invasion or regional or distant metastasis. To fully
determine and validate the prognostic value of the combination of
SET8 and PRMT5 in patients with HCC, particularly for those with
early stage HCC, the prognostic value of the combination of SET8
and PRMT5 in the TNM stage I subgroup was further investigated
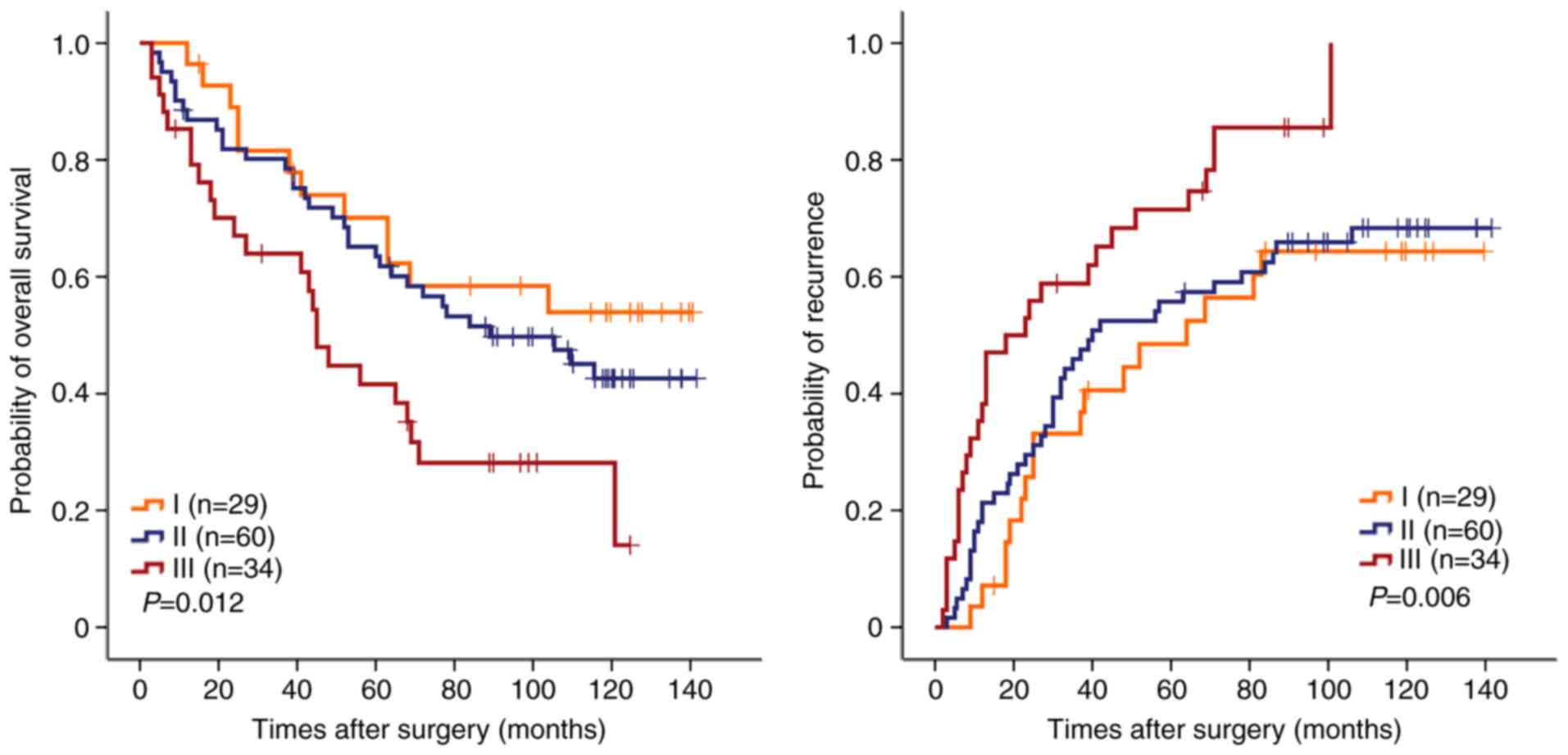
(n=123). The combination of SET8 and PRMT5 was demonstrated to be a
risk factor associated with OS (P=0.012) and TTR (P=0.006; Fig. 4). The median survival time was 45
months and mortality was observed in 24/34 patients. A high
expression of SET8 and PRMT5 was correlated with a median survival
time of >116 months. Mortality in 12/28 patients was correlated
with a low expression of SET8 and PRMT5. Furthermore, stratified
analyses according to serum AFP levels were also conducted to
investigate the prognostic value of individual expression levels of
SET8 and PRMT5 and their combination. Of note, in the lower AFP
subgroup (≤400 ng/ml), individual expression levels of SET8 and
PRMT5 and their combination were significantly associated with OS
and TTR in the subgroups (P<0.05, data not shown).
The prognostic value of SET8 and PRMT5
for ER
Recurrence, particularly early intrahepatic
recurrence (ER) within 2 years after hepatectomy, is the main cause
for the poor prognosis associated with HCC (28). The prognosis of early intrahepatic
recurrence is much worse than that of late intrahepatic recurrence,
due to the multi-centric occurrence of a new tumor. In the present
study, all the recurrences were divided into early and late groups,
with 24 months as the cut-off value, and the prognostic value of
SET8 and PRMT5 in the ER subgroup was investigated. The results
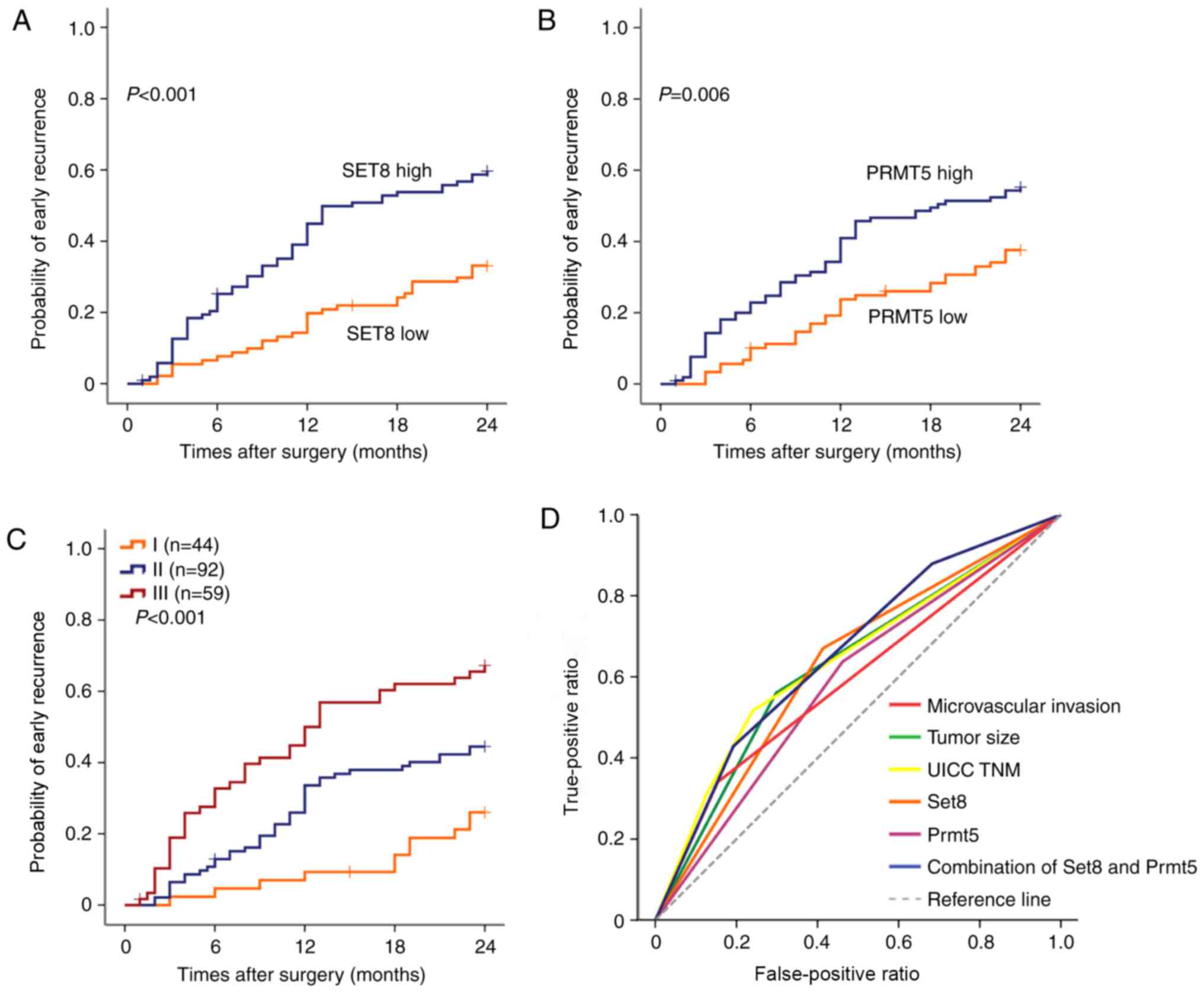
revealed that the patients with a high SET8 or PRMT5 expression
tended to have an ER (P<0.001 and P=0.006, respectively,
Fig. 5A and B). The patients with
both high expression of SET8 and PRMT5 in ER sub group III showed a
higher risk of early recurrence (Fig.
5C). When SET8 and PRMT5 were further combined, significant
differences between high and low combination of the two markers
were reached (Fig. 5D). The area
under the curve was up to 0.660 (95%CI, 0.584–0.736; P<0.001),
which was the largest among the adopted factors (data not
shown).
Discussion
Recurrence following hepatectomy remains the major
obstacle for further improving the outcomes of HCC (26). Currently, there are no effective
therapeutic strategies that can reduce recurrence as it is
difficult to determine which patients will have tumor recurrence
following hepatectomy. Global histone modification patterns can
predict the risk of cancer recurrence and clinical outcome,
representing a novel insight into therapeutic targets for cancer
(29). As histone methylations are
important determinants in the progression of HCC, identification of
biomarkers that focus on multiple histone methylation in tumor
tissue may aid in predicting the prognosis and recurrence of HCC.
In the present study, it was revealed that SET8 combined with PRMT5
was a promising predictor of OS and disease recurrence in HCC,
particularly for those with early stage HCC or low AFP levels.
The study of potential prognostic markers in large
numbers of clinical specimens is crucial in translating novel
findings from basic science into clinical practice. The ‘histone
code’ hypothesis proposes that distinct combinations of histone
modifications can generate unique transcriptional functions
(23,30). A previous study suggested that
different H3 and H4 tail modifications can act sequentially or in
combination to mediate distinct biological outcomes (23). Using high-throughput analyses, the
statistical results of TMA staining in the results of the present
study revealed that high expression of SET8 or PRMT5 was associated
with OS and TTR following resection of a primary tumor, and the
results of multivariate Cox proportional hazards analyses revealed
that they were independent risk factors for OS and TTR.
Furthermore, it was revealed that SET8 expression was positively
associated with PRMT5 expression, and the combination of SET8 and
PRMT5 detection also improved the prognostic capacity. The results
indicated that histone methylation in multiple sites within tumor
tissue may have a synergistic effect on the progression of HCC.
Clinically, it remains difficult to predict the
prognosis of patients with early stage HCC. In the present study, a
significant trend of ER within 24 months after surgery was
discovered in patients with high SET8 and PRMT5 expression. The
combination of SET8 and PRMT5 expression could improve the
predictive accuracy for OS and ER, which revealed the largest AUC
for all clinicopathological variables investigated. Similarly, the
prognostic value of SET8 and PRMT5 in those patients with early
stage HCC (TNM stage I) and with a low AFP level (≤400 ng/ml) was
further validated. The OS and TTR rates of patients with early
stage HCC or with low serum AFP and high SET8 and PRMT5 expression
were significantly poorer than that of those with low expression of
the two. Therefore, more curative therapy, including liver
transplantation, and a close follow-up should be considered in
these high-risk patients.
The histone methyltransferases can not only serve as
a prognostic tool but may also have a therapeutic role associated
with inhibitors to enzymes that regulate histone methylation. A
number of studies have focused on molecule targeting inhibitors of
histone methyltransferase (31). For
example, arginine methyltransferase inhibitor 1 (AMI-1) was
generally considered to be a selective small-molecule inhibitor of
type I PRMTs, while a previous study found it could inhibit HCC
growth through inhibiting PRMT5. It was also indicated that PRMT5
is a potential pharmacologic therapeutic target for HCC (32). The present study demonstrated that the
expression level of SET8 and PRMT5 was not correlated with
hepatitis-related variables (serum ALT, presence of hepatitis B
antigen or cirrhosis), or the tumor size or number. Therefore, it
could be possible that the baseline expression level of histone
methyltransferase is associated with specific heterogeneity among
different individuals, including peritumoral environment. Due to
the retrospective nature of the present study, the molecular
mechanisms of the role of the peritumoral environment on
HCC-associated histone methyltransferase was not addressed, and a
selection bias may be present. However, postoperative adjuvant
therapies targeting residual tumor cells and peritumoral
environment may obtain improved therapeutic effects. Inhibiting
multiple sites of histone methylation may be a novel therapeutic
strategy for HCC.
In conclusion, high expression of SET8 combined with
PRMT5 was correlated with high recurrence and poor OS rates in
patients with HCC. The methylation of lysine and arginine in the
tumor tissue represents a novel insight into tumor progression and
recurrence, and therapeutic targets for HCC.
Acknowledgements
Not applicable.
Funding
The present study was jointly supported by three
grants (grant nos. 81672848 and 81372654) from the National Natural
Science Foundation of China, and China National Science and
Technology Major Project for Prevention and Treatment of Infectious
Diseases (grant no. 2017ZX10203207).
Availability of data and materials
The datasets analyzed during the current study are
available from the corresponding author on reasonable request.
Authors' contributions
JZ and JC designed and funded the project. ZL, HJ
and LH conducted the experiments. ZL and JZ wrote the manuscript.
YZ, WS, XR helped to collect the data from the patients. WZ, LL and
ML analyzed the data. All authors read and approved the manuscript
and agree to be accountable for all aspects of the research in
ensuring that the accuracy or integrity of any part of the study
are appropriately investigated and resolved.
Ethics approval and consent to
participate
This retrospective study was carried out in
accordance with the ethical standards of the institutional research
committee and with the 1964 Helsinki declaration and its later
amendments or ethical standards. The requirement to obtain informed
consent was waived as this was a retrospective study. All patient
data were treated in accordance with the local privacy
regulations.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
SET8
|
SET domain-containing protein 8
|
|
PRMT5
|
protein arginine methyltransferase
5
|
|
ER
|
early intrahepatic recurrence
|
|
TNM
|
Tumor-Node-Metastasis
|
|
TMA
|
tissue microarray
|
|
DAB
|
3,3′-diaminobenzidine
|
|
OS
|
overall survival
|
|
TTR
|
time to recurrence
|
|
ROC
|
receiver operating characteristic
|
|
CI
|
confidence interval
|
|
HR
|
hazard ratio
|
|
NA
|
not adopted
|
|
NS
|
not significant
|
|
AFP
|
α-fetoprotein
|
|
UICC
|
International Union Against Cancer
|
References
|
1
|
Liu M, Jiang L and Guan XY: The genetic
and epigenetic alterations in human hepatocellular carcinoma: A
recent update. Protein Cell. 5:673–691. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ma L, Chua MS, Andrisani O and So S:
Epigenetics in hepatocellular carcinoma: An update and future
therapy perspectives. World J Gastroenterol. 20:333–345. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Herath NI, Leggett BA and MacDonald GA:
Review of genetic and epigenetic alterations in
hepatocarcinogenesis. J Gastroenterol Hepatol. 21:15–21. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wahid B, Ali A, Rafique S and Idrees M:
New insights into the epigenetics of hepatocellular carcinoma.
Biomed Res Int. 2017:16095752017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pogribny IP and Rusyn I: Role of
epigenetic aberrations in the development and progression of human
hepatocellular carcinoma. Cancer Lett. 342:223–230. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fang J, Feng Q, Ketel CS, Wang H, Cao R,
Xia L, Erdjument-Bromage H, Tempst P, Simon JA and Zhang Y:
Purification and functional characterization of SET8, a nucleosomal
histone H4-lysine 20-specific methyltransferase. Curr Biol.
12:1086–1099. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiao B, Jing C, Kelly G, Walker PA,
Muskett FW, Frenkiel TA, Martin SR, Sarma K, Reinberg D, Gamblin SJ
and Wilson JR: Specificity and mechanism of the histone
methyltransferase Pr-Set7. Genes Dev. 19:1444–1454. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nishioka K, Rice JC, Sarma K,
Erdjument-Bromage H, Werner J, Wang Y, Chuikov S, Valenzuela P,
Tempst P, Steward R, et al: PR-Set7 is a nucleosome-specific
methyltransferase that modifies lysine 20 of histone H4 and is
associated with silent chromatin. Mol Cell. 9:1201–1213. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Houston SI, McManus KJ, Adams MM, Sims JK,
Carpenter PB, Hendzel MJ and Rice JC: Catalytic function of the
PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for
mitotic entry and genomic stability. J Biol Chem. 283:19478–19488.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oda H, Okamoto I, Murphy N, Chu J, Price
SM, Shen MM, Torres-Padilla ME, Heard E and Reinberg D:
Monomethylation of histone H4-lysine 20 is involved in chromosome
structure and stability and is essential for mouse development. Mol
Cell Biol. 29:2278–2295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Z, Nie F, Wang S and Li L: Histone H4
Lys 20 monomethylation by histone methylase SET8 mediates Wnt
target gene activation. Proc Natl Acad Sci USA. 108:3116–3123.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu S, Wang W, Kong X, Congdon LM, Yokomori
K, Kirschner MW and Rice JC: Dynamic regulation of the PR-Set7
histone methyltransferase is required for normal cell cycle
progression. Genes Dev. 24:2531–2542. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jørgensen S, Elvers I, Trelle MB, Menzel
T, Eskildsen M, Jensen ON, Helleday T, Helin K and Sørensen CS: The
histone methyltransferase SET8 is required for S-phase progression.
J Cell Biol. 179:1337–1345. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Congdon LM, Houston SI, Veerappan CS,
Spektor TM and Rice JC: PR-Set7-mediated monomethylation of histone
H4 lysine 20 at specific genomic regions induces transcriptional
repression. J Cell Biochem. 110:609–619. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Abbas T, Shibata E, Park J, Jha S, Karnani
N and Dutta A: CRL4(Cdt2) regulates cell proliferation and histone
gene expression by targeting PR-Set7/Set8 for degradation. Mol
Cell. 40:9–21. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stopa N, Krebs JE and Shechter D: The
PRMT5 arginine methyltransferase: Many roles in development, cancer
and beyond. Cell Mol Life Sci. 72:2041–2059. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Y and Bedford MT: Protein arginine
methyltransferases and cancer. Nat Rev Cancer. 13:37–50. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pal S, Vishwanath SN, Erdjument-Bromage H,
Tempst P and Sif S: Human SWI/SNF-associated PRMT5 methylates
histone H3 arginine 8 and negatively regulates expression of ST7
and NM23 tumor suppressor genes. Mol Cell Biol. 24:9630–9645. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pal S, Baiocchi RA, Byrd JC, Grever MR,
Jacob ST and Sif S: Low levels of miR-92b/96 induce PRMT5
translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO
J. 26:3558–3569. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Karkhanis V, Hu YJ, Baiocchi RA, Imbalzano
AN and Sif S: Versatility of PRMT5-induced methylation in growth
control and development. Trends Biochem Sci. 36:633–641. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wei TY, Juan CC, Hisa JY, Su LJ, Lee YC,
Chou HY, Chen JM, Wu YC, Chiu SC, Hsu CP, et al: Protein arginine
methyltransferase 5 is a potential oncoprotein that upregulates G1
cyclins/cyclin-dependent kinases and the phosphoinositide
3-kinase/AKT signaling cascade. Cancer Sci. 103:1640–1650. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bernstein BE, Meissner A and Lander ES:
The mammalian epigenome. Cell. 128:669–681. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Strahl BD and Allis CD: The language of
covalent histone modifications. Nature. 403:41–45. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Y and Reinberg D: Transcription
regulation by histone methylation: Interplay between different
covalent modifications of the core histone tails. Genes Dev.
15:2343–2360. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qian YB, Zhang JB, Wu WZ, Fang HB, Jia WD,
Zhuang PY, Zhang BH, Pan Q, Xu Y, Wang L, et al: P48 is a
predictive marker for outcome of postoperative interferon-alpha
treatment in patients with hepatitis B virus infection-related
hepatocellular carcinoma. Cancer. 107:1562–1569. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bruix J and Sherman M: Practice Guidelines
Committee, American Association for the Study of Liver Diseases:
Management of hepatocellular carcinoma. Hepatology. 42:1208–1236.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Imamura H, Matsuyama Y, Tanaka E, Ohkubo
T, Hasegawa K, Miyagawa S, Sugawara Y, Minagawa M, Takayama T,
Kawasaki S and Makuuchi M: Risk factors contributing to early and
late phase intrahepatic recurrence of hepatocellular carcinoma
after hepatectomy. J Hepatol. 38:200–207. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Budhu A, Forgues M, Ye QH, Jia HL, He P,
Zanetti KA, Kammula US, Chen Y, Qin LX, Tang ZY and Wang XW:
Prediction of venous metastases, recurrence, and prognosis in
hepatocellular carcinoma based on a unique immune response
signature of the liver microenvironment. Cancer Cell. 10:99–111.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Seligson DB, Horvath S, Shi T, Yu H, Tze
S, Grunstein M and Kurdistani SK: Global histone modification
patterns predict risk of prostate cancer recurrence. Nature.
435:1262–1266. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jenuwein T and Allis CD: Translating the
histone code. Science. 293:1074–1080. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cheng D, Yadav N, King RW, Swanson MS,
Weinstein EJ and Bedford MT: Small molecule regulators of protein
arginine methyltransferases. J Biol Chem. 279:23892–23899. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang B, Dong S, Li Z, Lu L, Zhang S, Chen
X, Cen X and Wu Y: Targeting protein arginine methyltransferase 5
inhibits human hepatocellular carcinoma growth via the
downregulation of beta-catenin. J Transl Med. 13:3492015.
View Article : Google Scholar : PubMed/NCBI
|