Introduction
Cancer is a major public health problem globally.
Cancer is a class of diseases that undergoes uncontrollable cell
proliferation and differentiation. Based on the 2015 cancer
statistics, it is currently the second leading cause of mortality
in numerous countries (including China, Europe and the USA), and is
expected to surpass heart diseases as the leading cause of
mortality in the near future (1).
Although the risk of succumbing to cancer has decreased by ~20%
from its maximum in 1991–2011 (1), it
must be diagnosed with high sensitivity and specificity in order to
determine the appropriate therapy and prognosis. Recently, a number
of biomarkers with diagnostic and prognostic potential value have
been demonstrated in numerous cancer types, including the tumor
markers human epididymis secretory protein 4 and cancer antigen 125
in endometrial (2) and ovarian cancer
types (3). Additionally, mutant genes
have been used in the selection of an appropriate therapy,
including epidermal growth factor receptor mutation in non-small
cell lung cancer (4), Kirsten rat
sarcoma viral oncogene homolog in colorectal cancer (5) and v-raf murine sarcoma viral oncogene
homolog B1 mutation in melanoma (6);
however, reliable and convenient biomarkers are required to
evaluate the diagnostic and prognostic significance of different
cancer types.
Classic biomarkers present with potentially limiting
factors, including cost, availability and reproducibility (7). Utility is compromised by different
disease heterogeneities, specific genetics and proteomics, and the
influence of lifestyle; therefore, a number of serum or tissue
biomarkers, including non-coding RNAs (ncRNAs), have been developed
for clinical experiments. ncRNAs have notable potential for future
biomarker approaches. Numerous studies have reported the use of
ncRNAs, including microRNAs and long ncRNAs (lncRNAs), in the early
detection and prognosis of various cancer types (8,9).
Previously, a number of studies focused on a novel class of ncRNAs
that is endogenously expressed as single-stranded,
covalently-closed circular molecules, also known as circular RNAs
(circRNAs) (10–12). circRNAs were demonstrated to be
antagonists of specific microRNAs by functioning as microRNA
sponges (10,13), and they are also known as stable
molecules, as demonstrated by their long half-lives in cells
(14). These observations resulted in
the consideration that circRNAs could serve as potential biomarkers
for the non-invasive diagnosis of numerous diseases, including
disorders of the central nervous system (15), cancer (16) and a number of forms of cardiovascular
diseases (17).
To determine if circRNA could serve as a sensitive
and specific biomarker for cancer, a systematic meta-analysis of
the published literature was performed in the present study, in
order to review the diagnostic efficiency of circRNA in patients
with cancer from the available data and to identify a novel
non-invasive biomarker for cancer diagnosis.
Materials and methods
Search strategy
This meta-analysis was conducted in accordance with
the guidelines of diagnostic meta-analysis as follows: Eligible
studies published up to November 30, 2017, on PubMed (https://www.ncbi.nlm.nih.gov/pubmed) and EMBASE
(https://www.elsevier.com/solutions/embase-biomedical-research),
were selected for the meta-analysis. Non-English studies were
excluded. No restriction was placed on the year of publication or
publishing status. The key words employed for literature retrieval
included the following: ‘circular RNA’ or ‘circRNA’, and ‘tumor’ or
‘neoplasm’, or ‘cancer’ or ‘carcinoma’. Additionally, the reference
lists of eligible articles were manually searched to obtain
additional sources.
Selection of publications
All studies were carefully and independently
reviewed by two researchers based on their titles and abstracts,
following which full texts were perused for potential eligibility.
Any disagreement was resolved by a full discussion, until consensus
was achieved. All publications included in the meta-analysis were
required to meet the following criteria: i) Studies should analyze
the association between circRNA and patients with any cancer type;
ii) studies should contain sensitivity and specificity data (or the
possibility of deriving such values from the data); and iii)
studies should have enrolled ≥20 patients and matched controls.
Studies were excluded if they involved any of the following
parameters: i) Duplicate studies; ii) letters, editorials, meeting
abstracts, case reports and reviews; iii) patients and control
subjects that did not qualify, in which the patients sample size
was low or the disease cannot be defined; iv) studies with missing
data, and v) No-English studies. If the same author reported that
their results were acquired from overlapping populations, only the
first study published or the most complete study was included.
Data extraction and quality
assessment
The following parameters were collected from each
study: Author name, publication year, country and ethnicity, sample
type, normalization control, sample size and data for two-by-two
tables (sensitivity and specificity). The Quality Assessment of
Diagnostic Accuracy Studies (QUADAS) checklist (http://www.bristol.ac.uk/population-health-sciences/projects/quadas/)
was used to systematically assess the quality of the articles
included in the diagnostic meta-analysis. Specifically, 14 items
from the QUADAS checklist were applied to each article, and an
answer of ‘Yes’, ‘No’ or ‘Unclear’ was determined. Only ‘Yes’
resulted in a score.
Statistical analysis
All statistical analyses were performed using the
STATA 13.0 statistical software (StataCorp LLC, TX, USA) and
Meta-DiSc 1.4 (Unit of Clinical Biostatistics, Ramón y Cajal
Hospital, Madrid, Spain). Data from each study (true-positives,
false-positives, true-negatives and false-negatives) were extracted
to obtain the pooled sensitivity, specificity, positive likelihood
ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio
(DOR) and their 95% confidence interval (CI), summary receiver
operator characteristic (SROC) curve and area under the curve
(AUC), in order to determine the overall performance of the
detection method. P<0.05 (two-sided) was considered to indicate
a statistically significant difference. Additionally, heterogeneity
across studies was assessed using Cochran's Q and I2
statistics, where I2>50% indicated the existence of
significant heterogeneity. Finally, evaluation of the threshold
effect (Spearman's rank correlation) and publication bias (funnel
plots) were also undertaken.
Results
Literature search
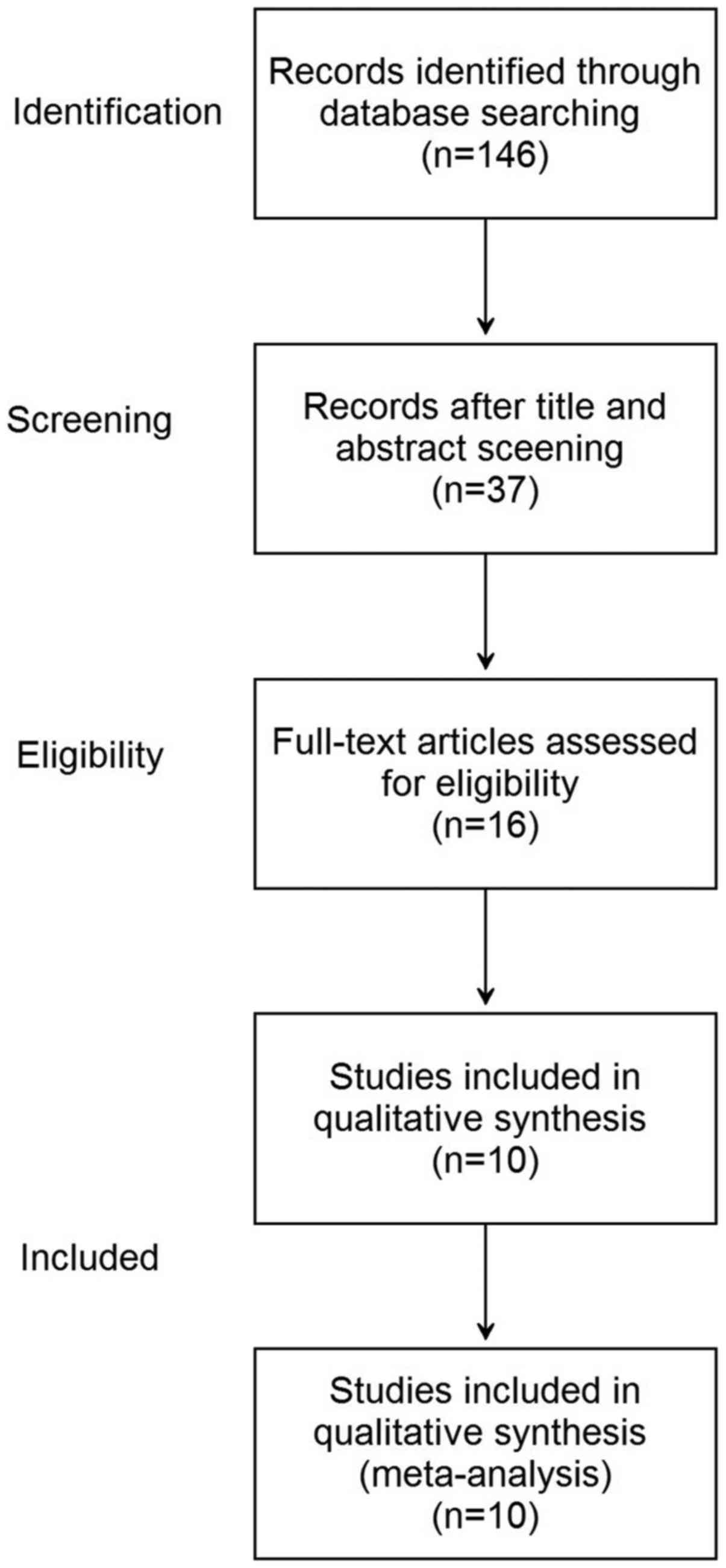
Electronic and manual searches yielded a total of
146 potentially eligible articles. The steps involved in screening
the articles for the meta-analysis is depicted as a flow chart in
Fig. 1. Screening titles and
abstracts resulted in the exclusion of 109 articles. A further 21
articles were excluded following more detailed assessment of the
full text. Finally, 10 eligible studies (12 tests) were included in
the meta-analysis (18–27).
Study characteristics
The characteristics of the 10 eligible studies are
summarized in Table I (18–27). A
total of 799 patients with different cancer types and adjacent
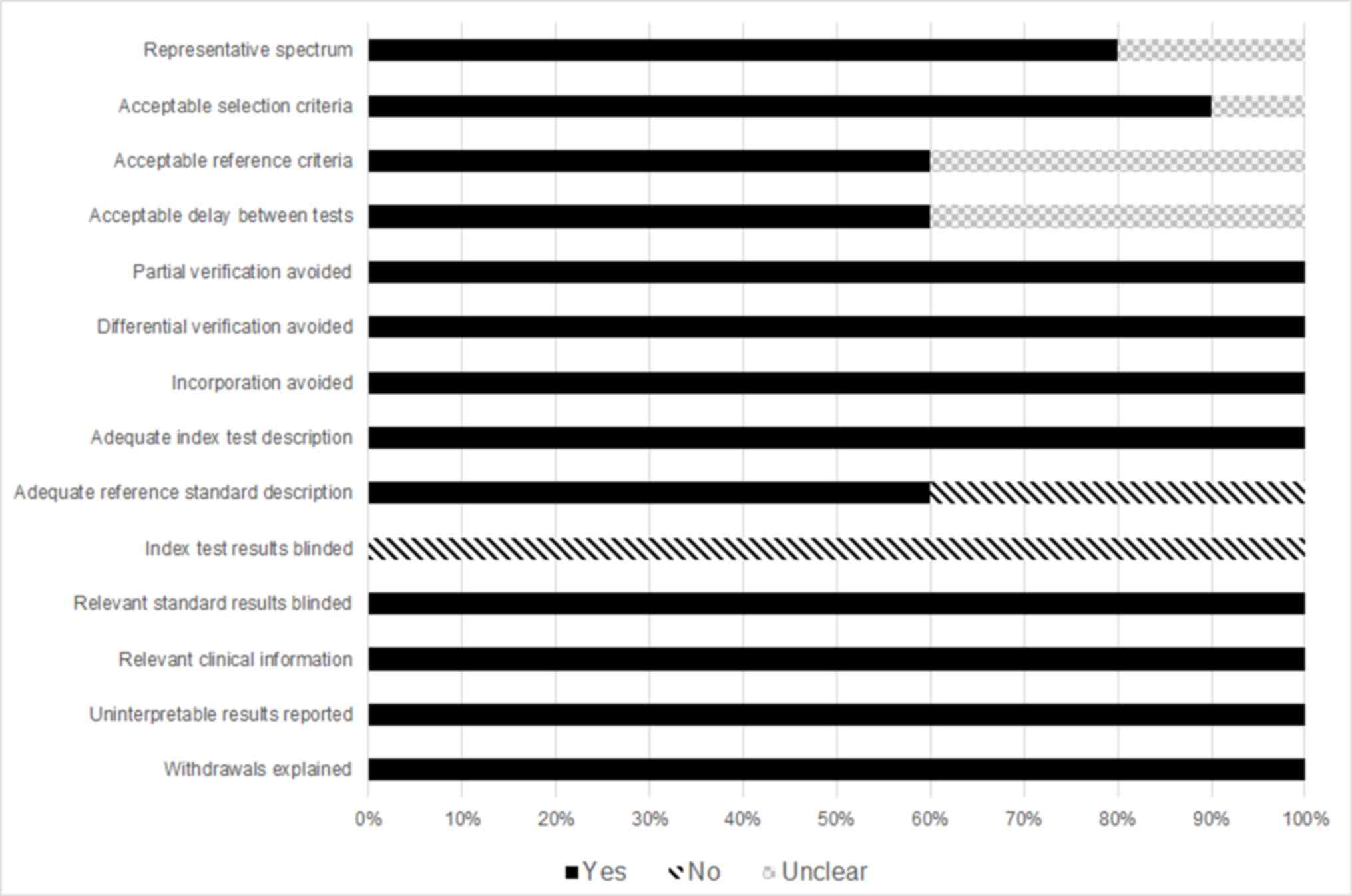
controls were involved in these 12 tests. Assessment using QUADAS
indicated that the studies were of high quality, with positive
results in 13/14 items (Fig. 2).
Additionally, the mean impact factor was calculated to be 2.59.
 | Table I.Characteristics of 10 studies included
in the meta-analysis. |
Table I.
Characteristics of 10 studies included
in the meta-analysis.
| First author | Year | Disease | circRNA used for
detection | Case no. | Control no. | Region | TP | FP | FN | TN | IF | (Refs.) |
|---|
| Li et al | 2015 | Gastric cancer | hsa_circ_002059 | 101 | 101 | China | 82 | 38 | 19 | 63 | 2.799 | (18) |
| Qin et al | 2016 | Hepatocellular
carcinoma | hsa_circ_0001649 | 89 | 89 | China | 72 | 28 | 17 | 61 | 1.736 | (19) |
| Wang et
al | 2015 | Colorectal
cancer | hsa_circ_001988 | 31 | 31 | China | 21 | 8 | 10 | 23 | 1.581 | (20) |
| Shang et
al | 2016 | Hepatocellular
carcinoma |
hsa_circ_0005075 | 30 | 30 | China | 25 | 3 | 5 | 27 | 2.133 | (21) |
| Chen et
al | 2017 | Gastric cancer |
hsa_circ_0000190 | 104 | 104 | China | 75 | 30 | 29 | 74 | 2.799 | (22) |
| Huang et
al | 2017 | Gastric cancer |
hsa_circ_0000745 | 60 | 60 | China | 51 | 33 | 9 | 27 | 3.365 | (23) |
| Fu et
al | 2017 | Hepatocellular
carcinoma |
hsa_circ_0003570 | 107 | 107 | China | 48 | 14 | 59 | 93 | 1.521 | (24) |
| Yin et
al | 2017 | Breast cancer |
hsa_circ_0001785 | 20 | 20 | China | 16 | 5 | 4 | 15 | 2.871 | (25) |
|
|
| Breast cancer |
hsa_circ_0108942 | 20 | 20 | China | 16 | 10 | 4 | 10 | 2.871 |
|
|
|
| Breast cancer |
hsa_circ_0068033 | 20 | 20 | China | 14 | 8 | 6 | 12 | 2.871 |
|
| Yao et
al | 2017 | Hepatocellular
carcinoma | cirZKSCAN1 | 102 | 102 | China | 84 | 28 | 18 | 74 | 5.314 | (26) |
| Zhao et
al | 2017 | Gastric cancer |
hsa_circ_0000181 | 115 | 115 | China | 62 | 17 | 53 | 98 | 1.521 | (27) |
Meta-analysis
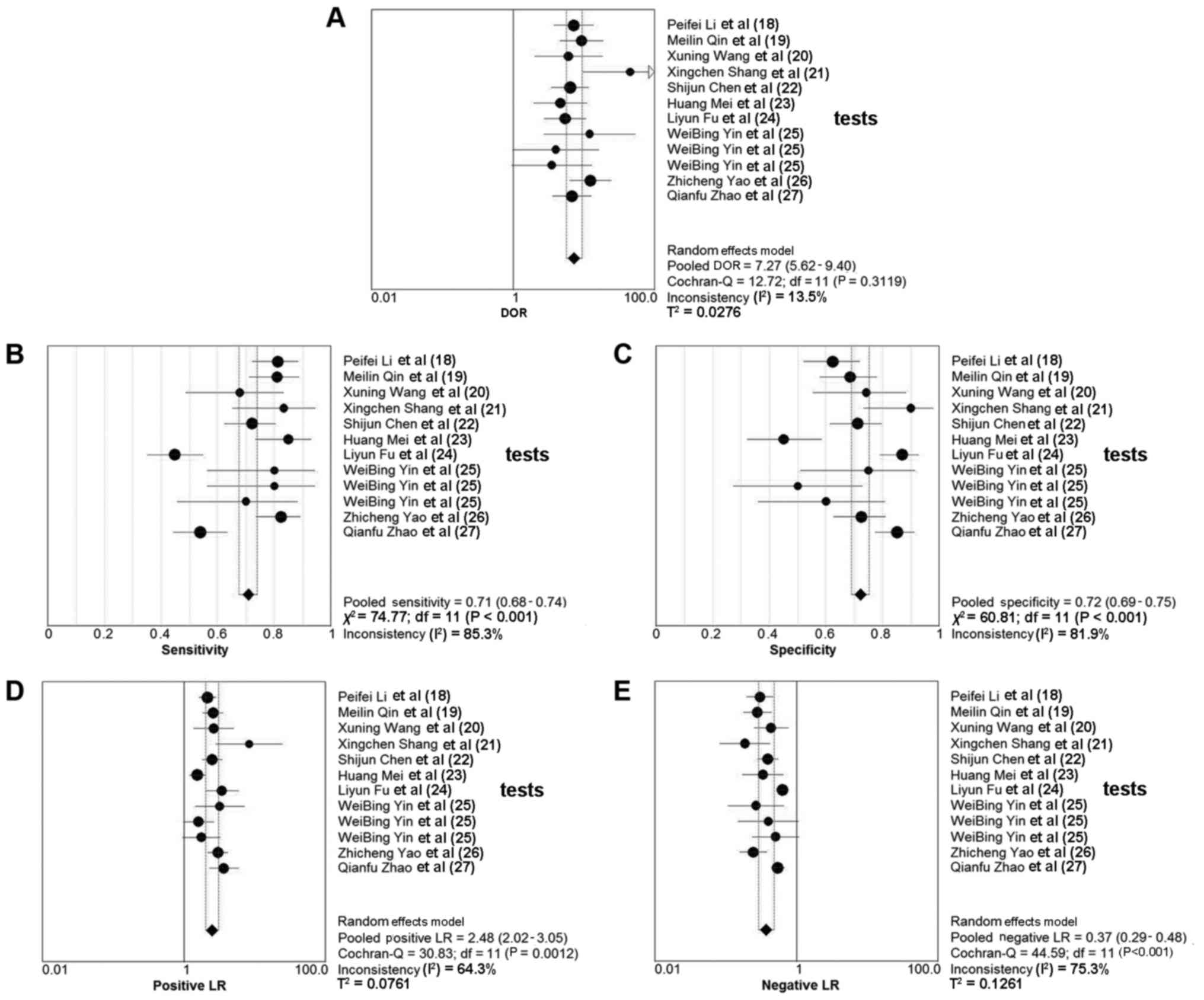
Overall, 10 studies involving 799 patients with
various cancer types reported the detection performances of circRNA
(Table II). The sensitivity of
circRNA detection testing ranged from 0.449–0.855, and the reported
specificity ranged from 0.450–0.900. The pooled DOR was 7.265 (95%
CI, 5.616–9.398; Q=12.72; P=0.312; I2=13.5%). The pooled
sensitivity was 0.708 (95% CI, 0.676–0.740; Q=74.77; P<0.001;
I2=85.3%) and the pooled specificity was 0.722 (95% CI,
0.690–0.753; Q=60.81; P<0.001; I2=81.9%). The PLR and
NLR were 2.483 (95% CI, 2.019–3.054; Q=30.83; P=0.001;
I2=64.3%) and 0.372 (95% CI, 0.289–0.479; Q=44.59;
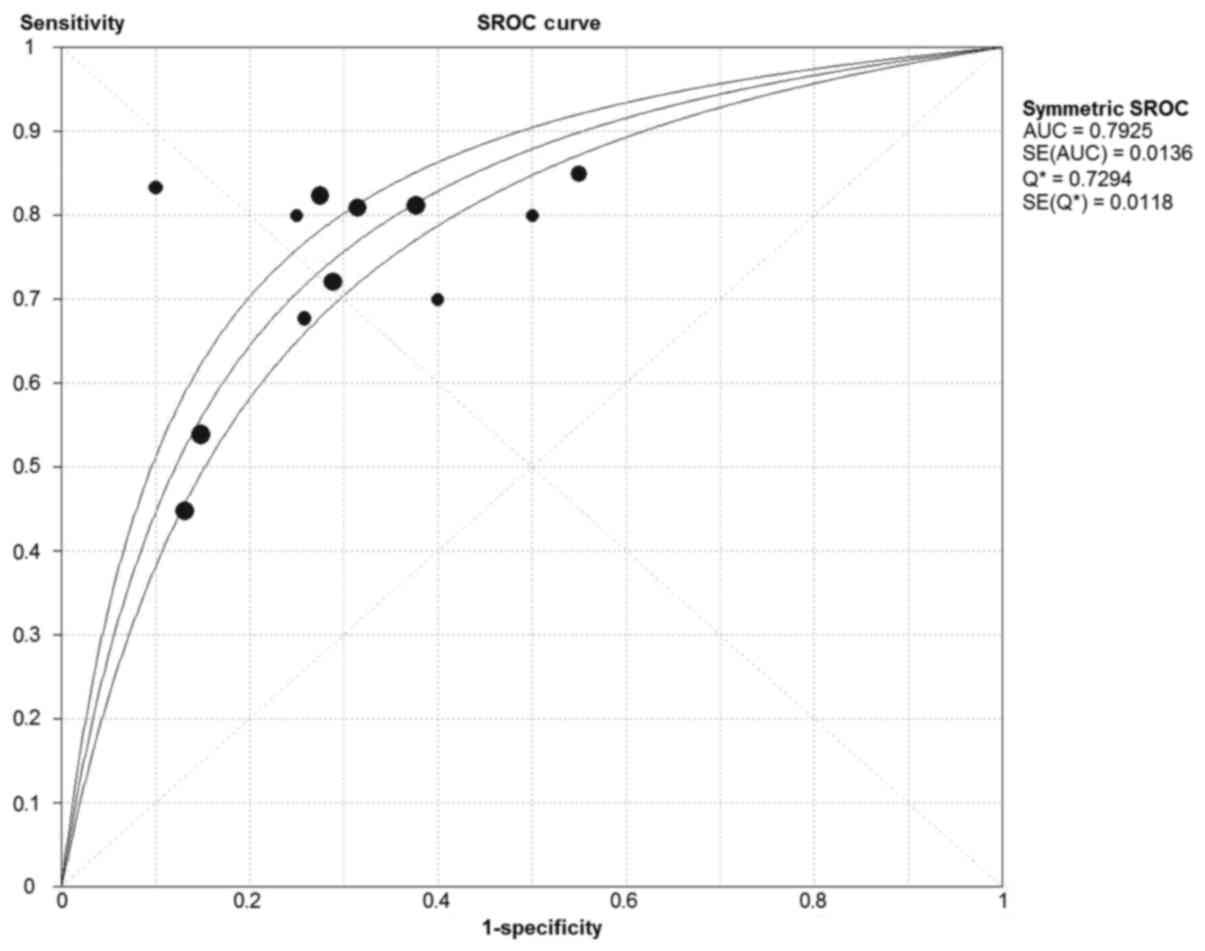
P<0.001; I2=75.3%), respectively. The AUC was 0.793.
The forest plots and SROC are depicted in Figs. 3 and 4,
respectively.
 | Table II.Detection performances of circular
RNA reported by 10 studies. |
Table II.
Detection performances of circular
RNA reported by 10 studies.
| First author | Diagnostic OR (95%
CI) | Sensitivity (95%
CI) | Specificity (95%
CI) | Positive LR (95%
CI) | Negative LR (95%
CI) | (Refs.) |
|---|
| Li et
al | 7.16
(3.77–13.59) | 0.81
(0.72–0.88) | 0.62
(0.52–0.72) | 2.16
(1.65–2.82) | 0.30
(0.20–0.46) | (18) |
| Qin et
al | 9.23
(4.62–18.44) | 0.81
(0.71–0.88) | 0.69
(0.58–0.78) | 2.57
(1.86–3.55) | 0.28
(0.18–0.44) | (19) |
| Wang et
al | 6.04
(2.01–18.17) | 0.68
(0.49–0.83) | 0.74
(0.55–0.88) | 2.63
(1.38–5.00) | 0.43
(0.25–0.75) | (20) |
| Shang et
al | 45.00
(9.73–208.08) | 0.83
(0.65–0.94) | 0.90
(0.73–0.98) | 8.33
(2.81–24.67) | 0.19
(0.08–0.42) | (21) |
| Chen et
al | 6.38
(3.49–11.66) | 0.72
(0.62–0.80) | 0.71
(0.61–0.80) | 2.50
(1.81–3.46) | 0.39
(0.28–0.55) | (22) |
| Huang et
al | 4.64
(1.94–11.09) | 0.85
(0.73–0.93) | 0.45
(0.32–0.58) | 1.55
(1.20–1.99) | 0.33
(0.17–0.65) | (23) |
| Fu et
al | 5.40
(2.70–53.33) | 0.45
(0.35–0.55) | 0.87
(0.79–0.93) | 3.43
(2.01–5.83) | 0.63
(0.53–0.76) | (24) |
| Yin et
al | 12.00
(2.70–53.33) | 0.80
(0.56–0.94) | 0.75
(0.51–0.91) | 3.20
(1.45–7.05) | 0.27
(0.11–0.66) | (25) |
|
| 4.00
(0.98–16.27) | 0.80
(0.56–0.94) | 0.50
(0.27–0.73) | 1.60
(0.98–2.61) | 0.40
(0.15–1.07) |
|
|
| 3.50
(0.94–12.97) | 0.70
(0.46–0.88) | 0.60
(0.36–0.81) | 1.75
(0.95–3.22) | 0.50
(0.23–1.07) |
|
| Yao et
al | 12.33
(6.31–24.09) | 0.82
(0.74–0.89) | 0.73
(0.63–0.81) | 3.00
(2.16–4.16) | 0.24
(0.16–0.38) | (26) |
| Zhao et
al | 6.74
(3.58–12.69) | 0.54
(0.44–0.63) | 0.85
(0.77–0.91) | 3.65
(2.28–5.84) | 0.54
(0.44–0.67) | (27) |
Investigation of the threshold
effect
Spearman's rank correlation was also performed to
confirm the threshold effect. No indication of a threshold effect
was determined in the studies [Spearman's correlation coefficient
(ρ), 0.340; P=0.280]. Additionally, the slope (b) of the regression
equation did not differ from zero (P=0.852), implying no
heterogeneity between the studies.
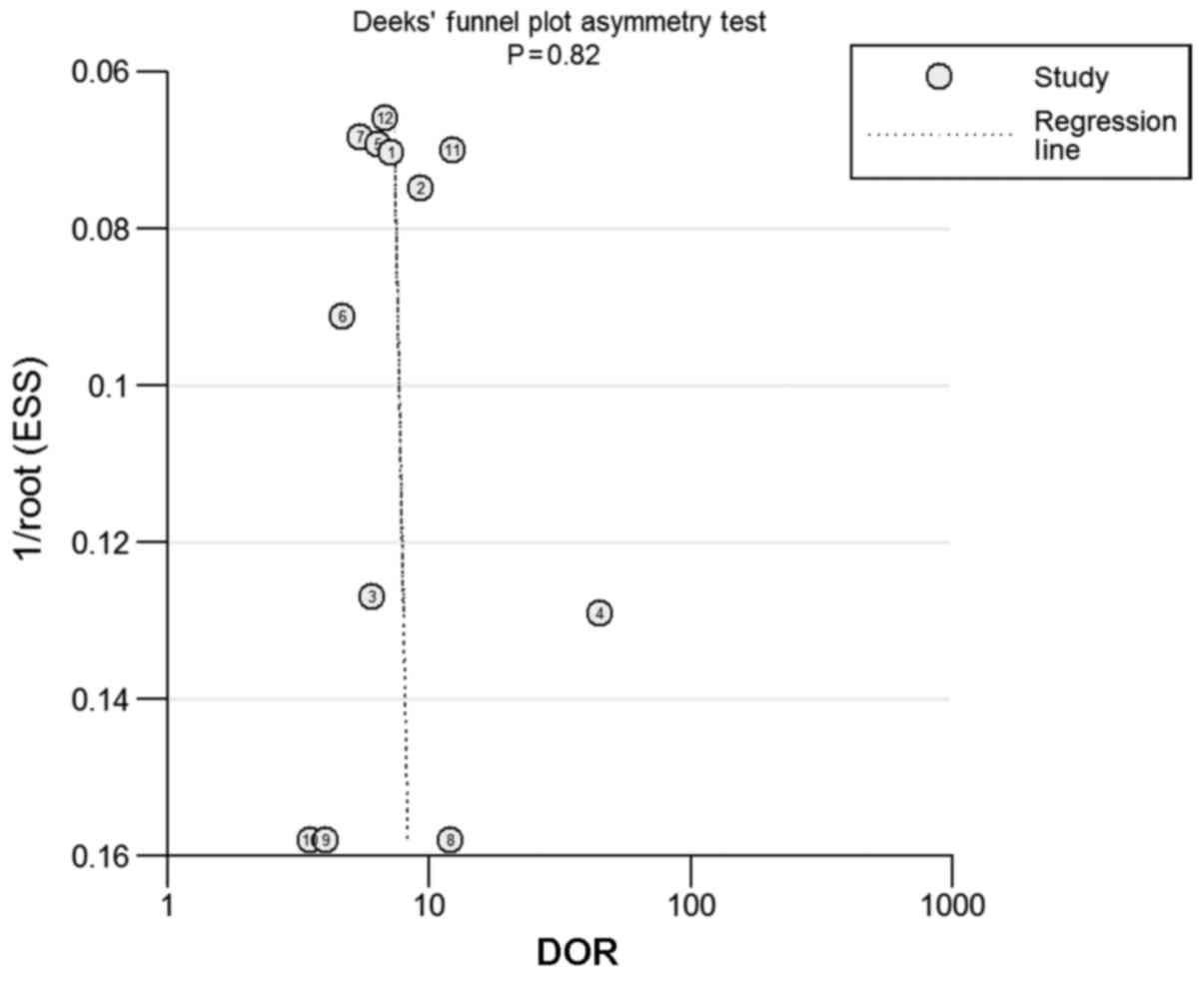
Publication bias
Finally, the presence of a statistically significant
slope coefficient (P<0.05) was considered to indicate a possible
publication bias. Funnel plots were produced (Fig. 5). No publication bias was observed in
the included studies (P=0.82) and the regression line represented a
symmetrical curve.
Discussion
There are an increasing number of molecular
biomarkers, including microRNAs and lncRNAs, being used in cancer
diagnostics. circRNAs are widely expressed in human cells (28). Highly conserved sequences and a high
degree of stability in mammalian cells are two of their most
important properties (10,13); thus, circRNAs have the potential to be
ideal biomarkers in the diagnosis of cancer. Numerous studies have
evaluated the performance of circRNAs in cancer diagnosis (18–27);
however, no systematic evaluation of circRNAs has been performed.
The differences in the performances were too large and hence, to
the best of our knowledge, the present study is the first
meta-analysis to provide precise and controlled data on the
diagnostic performance of circRNAs in cancer.
A total of 10 eligible, high-quality studies were
included in the present meta-analysis. The present study
demonstrated the varying sensitivities and specificities of
circRNAs in the diagnosis of cancer; however, the range of their
sensitivity and specificity was large and their diagnostic
performance cannot be evaluated. The pooled sensitivity and
specificity were observed to be slightly high (70.8 and 72.2%),
which demonstrated that circRNAs could be used as assistant
indicators in the diagnosis of cancer. The SROC curve and DOR
indicated that circRNAs exhibited a moderate diagnostic
performance. The pattern of the data points in the SROC curve did
not indicate a ‘shoulder-arm’ shape, which indicates no threshold
effect was determined in these studies, and the AUC of the SROC was
0.793. Cumulatively, these results indicated that circRNA had a
moderate level of overall diagnostic accuracy for cancer
diagnosis.
In the present study, heterogeneity was not
determined in the pooled DOR of the circRNAs (P=0.852).
Furthermore, publication bias and Spearman's rank correlation were
also performed. No statistical difference was determined using
Spearman's rank correlation, which meant that no threshold effect
among these studies was observed. No publication bias was observed
either in the included studies.
However, a number of limitations in this
meta-analysis should be noted. Firstly, all included studies were
reported by Chinese researchers. For this reason, the diagnostic
performance of circRNAs may be not be all-sided, in spite of the
absence of heterogeneity, threshold effect and publication bias;
Therefore, further research regarding circRNAs, particularly in
relation to the other countries' projects, as a biomarker in cancer
diagnosis is required. Secondly, only the integral diagnostic
performance of circRNAs on cancer was evaluated. The performance
may be cursory on a specific type of circRNA for specific cancer
types. Since the aim of the present study was to evaluate the
likelihood of circRNAs performing for the diagnosis of cancer, the
integral performance of circRNA in cancer was sufficient. Finally,
the moderate levels of circRNA sensitivity and specificity could be
attributed to technological, instrumental and staffing limitations;
however, there is not sufficient data to evaluate these parameters.
The cut-off value of circRNA efficiency in different cancer types
remains controversial, and investigating its clinical significance
may improve the diagnostic performance of circRNAs.
In conclusion, circRNA is a moderately effective
assistant diagnostic biomarker for cancer; however, its diagnostic
performance remains to be determined and further research of
specific circRNA types for specific cancer types is required in
order to determine this.
Acknowledgements
The authors thank Dr Si Chen (Department of Clinical
Laboratory, Beijing Anzhen Hospital) and Dr Chuiwen Deng
(Department of Rheumatology and Clinical Immunology, Peking Union
Medical College Hospital) who gave positive advice regarding this
study.
Funding
The present study was supported by the Talent
Training Project of High Level Health Technology from Beijing
Health System (grant no. 2015-3-052) and the National Natural
Science Foundation of China (grant no. 81500332).
Availability of data and materials
All data generated or analyzed during this study
are included in this published article.
Authors' contributions
YL and XZ carried out the conception and design,
acquisition of data, analysis of data, and drafting the manuscript.
HY performed the acquisition of data, and the drafting and revising
of the manuscript. JH and SZ aided with acquisition of data. HC
aided with the statistical analysis. QS and NJ participated in the
design and coordination of the study and helped to revise the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
circRNAs
|
circular RNAs
|
|
QUADAS
|
quality assessment of diagnostic
accuracy studies
|
|
PLR
|
positive likelihood ratio
|
|
NLR
|
negative likelihood ratio
|
|
DOR
|
diagnostic odds ratio
|
|
CI
|
confidence interval
|
|
SROC
|
summary receiver operator
characteristic
|
|
AUC
|
area under the curve
|
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bian J, Sun X, Li B and Ming L: Clinical
significance of serum HE4, CA125, CA724, and CA19-9 in patients
with endometrial cancer. Technol Cancer Res Treat. 16:435–439.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Karakaya BK, Başer E, Bildacı B, Cömert
EÇ, Bayraktar N, Dursun P, Kuşçu E and Ayhan A: Alternative tumor
markers in the diagnosis of ovarian cancer. Ginekol Pol.
87:565–769. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Brugger W, Triller N, Blasinska-Morawiec
M, Curescu S, Sakalauskas R, Manikhas GM, Mazieres J, Whittom R,
Ward C, Mayne K, et al: Prospective molecular marker analyses of
EGFR and KRAS from a randomized, placebo-controlled study of
erlotinib maintenance therapy in advanced non-small-cell lung
cancer. J Clin Oncol. 29:4113–4120. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van Cutsem E, Köhne CH, Hitre E, Zaluski
J, Chien Chang CR, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G,
et al: Cetuximab and chemotherapy as initial treatment for
metastatic colorectal cancer. N Engl J Med. 360:1408–1417. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gear H, Williams H, Kemp EG and Roberts F:
BRAF mutations in conjunctival melanoma. Invest Ophthalmol Vis Sci.
45:2484–2488. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bodzin AS and Busuttil RW: Hepatocellular
carcinoma: Advances in diagnosis, management, and long term
outcome. World J Hepatol. 7:1157–1167. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hu AX, Huang ZY, Zhang L and Shen J:
Potential prognostic long non-coding RNA identification and their
validation in predicting survival of patients with multiple
myeloma. Tumour Biol. 39:10104283176945632017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Song T, Liang Y, Cao Z, Du W and Li Y:
Computational analysis of specific MicroRNA biomarkers for
noninvasive early cancer detection. Biomed Res Int.
2017:46806502017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bachmayr-Heyda A, Reiner AT, Auer K,
Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW,
Zeillinger R and Pils D: Correlation of circular RNA abundance with
proliferation-exemplified with colorectal and ovarian cancer,
idiopathic lung fibrosis, and normal human tissues. Sci Rep.
5:80572015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lukiw WJ: Circular RNA (circRNA) in
Alzheimer's disease (AD). Front Genet. 4:3072013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z
and Sharpless NE: Expression of linear and novel circular forms of
an INK4/ARF-associated non-coding RNA correlates with
atherosclerosis risk. PLoS Genet. 6:e10012332010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li P, Chen S, Chen H, Mo X, Li T, Shao Y,
Xiao B and Guo J: Using circular RNA as a novel type of biomarker
in the screening of gastric cancer. Clin Chim Acta. 444:132–136.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z,
Yang J, Fan J, Liu L and Qin W: Hsa_circ_0001649: A circular RNA
and potential novel biomarker for hepatocellular carcinoma. Cancer
Biomark. 16:161–169. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Zhang Y, Huang L, Zhang J, Pan F,
Li B, Yan Y, Jia B, Liu H, Li S and Zheng W: Decreased expression
of hsa_circ_001988 in colorectal cancer and its clinical
significances. Int J Clin Exp Pathol. 8:16020–16025.
2015.PubMed/NCBI
|
|
21
|
Shang X, Li G, Liu H, Li T, Liu J, Zhao Q
and Wang C: Comprehensive circular RNA profiling reveals that
hsa_circ_0005075, a new circular RNA biomarker, is involved in
hepatocellular crcinoma development. Medicine (Baltimore).
95:e38112016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen S, Li T, Zhao Q, Xiao B and Guo J:
Using circular RNA hsa_circ_0000190 as a new biomarker in the
diagnosis of gastric cancer. Clin Chim Acta. 466:167–171. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang M, He YR, Liang LC, Huang Q and Zhu
ZQ: Circular RNA hsa_circ_0000745 may serve as a diagnostic marker
for gastric cancer. World J Gastroenterol. 23:6330–6338. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fu L, Wu S, Yao T, Chen Q, Xie Y, Ying S,
Chen Z, Xiao B and Hu Y: Decreased expression of hsa_circ_0003570
in hepatocellular carcinoma and its clinical significance. J Clin
Lab Anal. 32:2018. View Article : Google Scholar
|
|
25
|
Yin WB, Yan MG, Fang X, Guo JJ, Xiong W
and Zhang RP: Circulating circular RNA hsa_circ_0001785 acts as a
diagnostic biomarker for breast cancer detection. Clin Chim Acta:
S0009-8981(17)30407-2. 2017. View Article : Google Scholar
|
|
26
|
Yao Z, Luo J, Hu K, Lin J, Huang H, Wang
Q, Zhang P, Xiong Z, He C, Huang Z, et al: ZKSCAN1 gene and its
related circular RNA (circZKSCAN1) both inhibit hepatocellular
carcinoma cell growth, migration, and invasion but through
different signaling pathways. Mol Oncol. 11:422–437. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhao Q, Chen S, Li T, Xiao B and Zhang X:
Clinical values of circular RNA 0000181 in the screening of gastric
cancer. J Clin Lab Anal. 32:e223332018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|