Introduction
Bladder cancer among the most common types of
urological neoplasms worldwide (1).
The aim of conventional therapies for bladder cancer, including
surgery, radiation and chemotherapy, is to eliminate cancer cells.
However, adverse effects and treatment failure are common (2–4). Numerous
studies have focused on the underlying molecular mechanisms of
pathogenesis in bladder cancer, and, although long non-coding RNAs
(lncRNAs) cannot be translated into proteins, they have emerged as
key regulators of the development of bladder cancer (5–7).
Therefore, cancer gene therapy via targeting of oncogenic lncRNAs
may be a future treatment option.
The lncRNA, PVT1, can promote the progression of
various types of tumor, including bladder cancer (8–10). The
lncRNA, ANRIL, is also involved in numerous diseases and has been
demonstrated to promote DNA methylation, which may be a perinatal
marker for subsequent adiposity (11). Overexpression of ANRIL has been
reported to accelerate cell invasion and suppress apoptosis in
osteosarcoma (12). ANRIL expression
is upregulated in bladder cancer and promotes disease progression
through the intrinsic pathway (13).
Taking into consideration the importance of these two lncRNAs in
bladder cancer, they were used as targets in the present study.
Gene editing can alter DNA sequences using
nucleases, which act as molecular scissors (14). The clustered regularly interspaced
short palindromic repeats (CRISPR)-associated (Cas) protein 9
system combines two components, guide RNA (gRNA) and Cas nuclease
(15). This system depends on gRNA
for specific cleavage (16). The
CRISPR/Cas9 system is considered a promising gene editing tool
(17), which can function in various
types of cells (18,19). Numerous methods based on this tool
have been created and used for cancer study (20,21). It
has been revealed that CRISPR/Cas9 can control gene expression by
generating loss-of-function or gain-of-function mutations in
oncogenes (22). However, due to
potential off-target effects of CRISPR/Cas9, the consistent and
safe use of this system remains a challenge. Artificially
controlling the ‘switch’ of this system may reduce the adverse
off-target cellular effects.
A tetracycline-inducible element was applied in the
present study, consisting of the tetracycline repressor protein
(TetR), a specific DNA-binding site, and the tetracycline operator
sequence (TetO). TetR is separated from TetO via a conformational
change, which is induced by tetracycline or its derivatives,
including doxycycline (DOX) (23).
The tetracycline-inducible switch controls the expression of Cas9.
The nontoxic inducer, DOX, is widely used in preclinical studies
(24). Cas9 was efficiently activated
when DOX was added to the system. Thus, constant expression of Cas9
nuclease could not have been achieved without the presence of
DOX.
In the present study, gRNAs were designed to target
oncogenes, PVT1 and ANRIL. The objective of the study was to
suppress the progression of bladder cancer by targeting multiple
sites using the CRISPR/Cas9 system. In addition, the present study
aimed to eliminate the off-target effects of this system by
utilizing the tetracycline-inducible element. The results indicated
that, although all vectors were transfected into cells, the
phenotype of the bladder cancer cells was not altered in the
absence of DOX. However, when DOX was added, the malignant behavior
of bladder cancer cells was significantly inhibited through this
tetracycline-inducible CRISPR/Cas9 system. Therefore, this system
could efficiently suppress the phenotype of bladder cancer cells
and also reduce the side effects of the CRISPR/Cas9 system.
Materials and methods
Cell lines and cell culture
The human bladder cancer cell lines, T24 and 5637,
were obtained from American Type Culture Collection (Manassas, VA,
USA). The T24 cells were cultured in DMEM (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) with 10% fetal bovine
serum (FBS; HyClone; GE Healthcare, Chicago, IL, USA). The 5,637
cells were maintained in RPMI-1640 media (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS and cultured at 37°C in
5% CO2.
Vectors
A total of 5 gRNA vectors targeting lncRNA PVT1 and
5 targeting lncRNA ANRIL were designed using CRISPR-ERA (http://crispr-era.stanford.edu/). The following
gRNA sequences were cloned into a plasmid vector (cat. no. 53188;
Addgene, Inc., Cambridge, MA, USA) using the restriction enzymes
sites Ndel and BIPl: PVT1-gRNA1: 5′-TCTCCAGAAGGACAGAATAA-3′;
PVT1-gRNA2: 5′-AAAAGAATTTAATAGACACG-3′; PVT1-gRNA3:
5′-TTGGTGGGGCTTGTGAATC-3′; PVT1-gRNA4: 5′-ACGAGGCCGGCCACGCCACG-3′;
PVT1-gRNA5: 5′-GATTCACAAGCCCCACCAAG-3′; ANRIL-gRNA1:
5′-GGGGCGCGGCCTCGGCGGAT-3′; ANRIL-gRNA2:
5′-CCGCTCCTCGGCCAAGTCCA-3′; ANRIL-gRNA3:
5′-CGCCGCGGCGCGGGGACTAG-3′; ANRIL-gRNA4:
5′-GCAGCAGCAGCTCCGCCACG-3′; ANRIL-gRNA5:
5′-ACGGCCAACGGTGGATTATC-3′. Tetracycline-inducible Cas9 (vector 1)
was purchased from SyngenTech Co., Ltd. (Beijing, China). The
vector 2 simultaneously expressing PVT1-gRNA3, PVT1-gRNA4,
ANRIL-gRNA2 and ANRIL-gRNA5 was constructed by SyngenTech Co., Ltd,
(Beijing, China).
DNA sequencing
An amount of 1 µg/ml DOX was added to the
transfected cells and after 48 h the cells were harvested and
genomic DNA was extracted using EasyPure Genomic DNA kit (Beijing
Transgen Biotech Co., Ltd., Beijing, China), according to the
manufacturer's protocol. DNA sequencing was performed by Sangon
Biotech Co., Ltd. (Shanghai, China).
Cell transfection
The propagated vectors were extracted from E. coli
using Plasmid Midiprep kit (Promega Corporation, Madison, WI, USA),
according to the manufacturer's protocol. T24 and 5637 cells
(2×105) were transfected with 2 µg vectors for 48 h per
well in 6-well plates using 4 µl Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
number of cells seeded in the plates. Following transfection, T24
and 5637 cells were used for subsequent experiments
immediately.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from 2×106 T24 or
5637 cells following incubation with TRIzol®
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. Total RNA was reverse transcribed into
cDNA using a PrimeScript RT reagent kit with gDNA Eraser (Takara
Biotechnology Co., Ltd., Dalian, China), according to the
manufacturer's protocol. The mRNA expression levels of PVT1 and
ANRIL were measured by RT-qPCR by SYBR® Premix Ex Taq II
(cat. no. RR420A; Takara Biotechnology Co., Ltd.) using a Roche
LightCycler® 480 Real-Time PCR system. The following
thermocycling conditions were used: Initial incubation at 95°C for
1 min; 40 cycles at 95°C for 30 sec, 60°C for 30 sec and 72°C for
30 sec, and a final extension step at 72°C for 10 min. GAPDH was
used as the endogenous control. The following primer pairs were
used: PVT1, forward, 5′-GCCCCTTCTATGGGAATCACTA-3′, reverse,
5′-GGGGCAGAGATGAAATCGTAAT-3′; ANRIL, forward,
5′-CAACATCCACCACTGGATCTTAACA-3′, reverse,
5′-AGCTTCGTATCCCCAATGAGATACA-3′; GAPDH, forward,
5′-CGCTCTCTGCTCCTCCTGTTC-3′, reverse, 5′-ATCCGTTGACTCCGACCTTCAC-3′.
The comparative 2−ΔΔCq method (25) was used to analyze the relative
expression of PVT1 and ANRIL. All the experiments were performed at
least three times.
Proliferation assay
Cell Counting kit-8 (CCK-8; Beyotime Institute of
Biotechnology, Shanghai, China) was used to measure proliferation,
according to the manufacturer's protocol. Cells were seeded into a
96-well plate at 5,000 cells per well. Subsequently, 24, 48 or 72 h
after transfection, 10 µl of CCK-8 was added to each well and the
cells were incubated for 30 min. Absorbance was measured at a
wavelength of 450 nm using an ELISA microplate reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). All the assays were
performed at least in triplicate.
ELISA
A cell death detection ELISA kit (Roche Applied
Science, Penzberg, Germany) was used to determined apoptotic rate
by quantifying histone-complexed DNA fragments (nucleosomes) in the
cytoplasm, according to the manufacturer's protocols. The
absorbance was measured at 405 nm wavelength using a microplate
reader (Bio-Rad, Laboratories, Inc.). The experiment was performed
≥3 times.
Cell migration assay
T24 and 5637 cells were seeded in 6-well plates at
37°C and reached 90% confluence prior to transfection. They were
divided into negative control and experimental groups. A mass of 1
µg vector 1 and 1 µg vector 2 were transfected into the cells and a
sterile pipette tip was used to create a wound in the cell layer.
After 24 h of transfection, the migration distance was detected
using the software program, HMIAS-2000 (version 2.0; Wuhan Qianping
Imaging Technology Co., Ltd., Wuhan, China). The experiments were
repeated ≥3 times.
Western blotting
The transfected cells were washed with PBS and then
lysed in radioimmunoprecipitation assay buffer (Beyotime Institute
of Biotechnology). The assay was performed as previously described
(26). The specific primary antibody
against Cas9 (Streptococcus pyogenes) (clone no. D8Y4K) rabbit mAb
(cat. no. 65832) and GAPDH (clone no. D16H11) XP® Rabbit
mAb (cat. no. 5174) were purchased from Cell Signaling Technology,
Inc. (Danvers, MA, USA; both dilutions, 1:1,000). Incubation with
diluted primary antibodies, whilst shaken gently, was at 4°C
overnight. The peroxidase-conjugated secondary antibody anti-rabbit
IgG (cat. no. A0545) was bought from Sigma-Aldrich; Merck KGaA,
Darmstadt, Germany (1:10,000). Incubation with diluted secondary
antibody, whilst shaken, at room temperature for 1 h. The
experiments were repeated ≥3 times.
Statistical analysis
All statistical analyses were performed using SPSS
20.0 version software (IBM Corp., Armonk, NY, USA). The data are
presented as the mean ± standard deviation. Data was analyzed using
Student's t-test or analysis of variance with the
Least-Significant-Difference post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Shearing efficiency of CRISPR/Cas9,
analyzed by DNA sequencing
The bladder cancer cells were cultured in 6-well
plates and transfected with gRNA and tetracycline-inducible Cas9
vectors. DOX was used to control mRNA expression of Cas9. Genomic
DNA was extracted from the cells 48 h following transfection. The
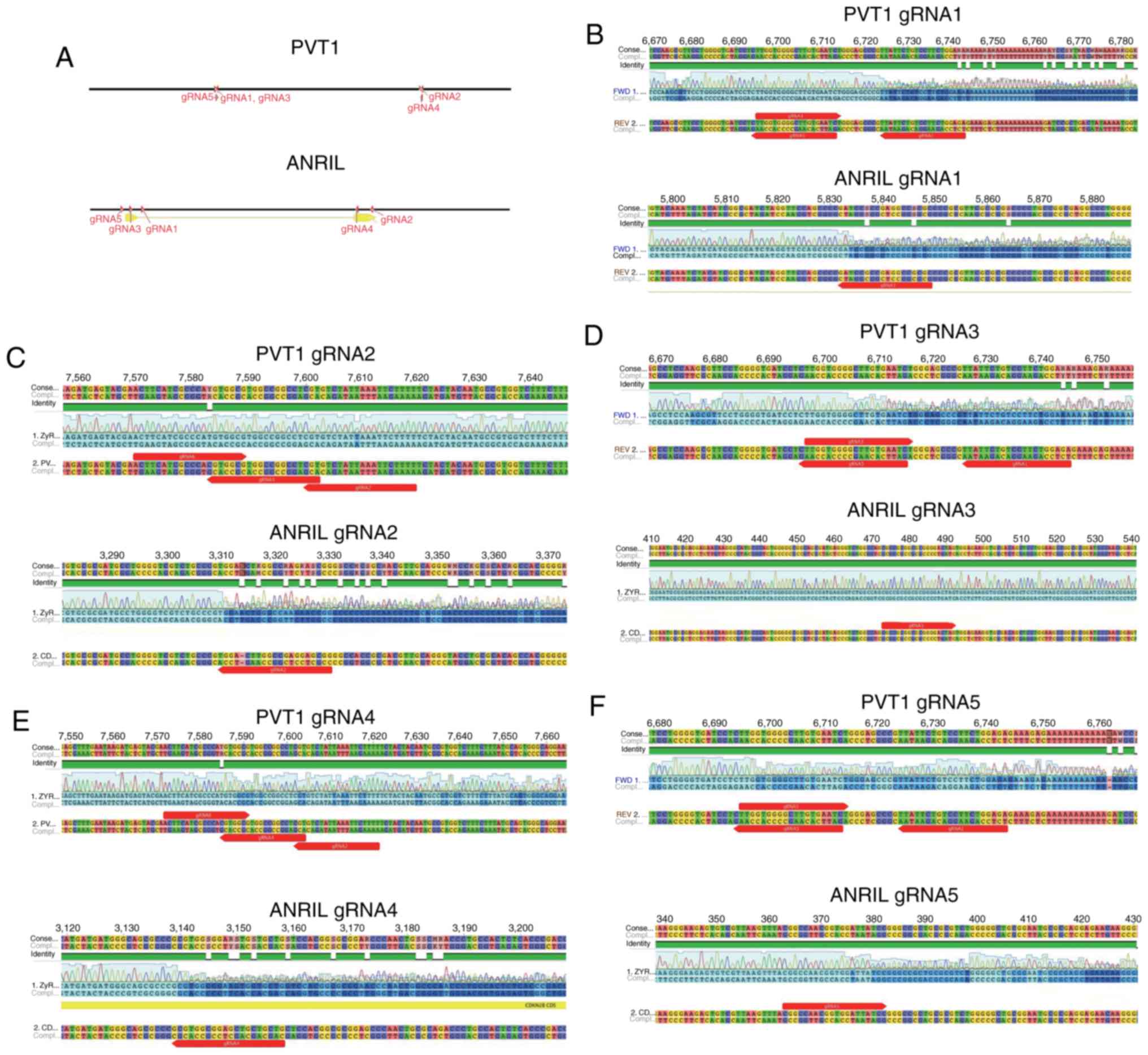
locations of different gRNAs targeting PVT1 or ANRIL are indicated
in Fig. 1A. A total of 5 gRNAs
targeting PVT1 and 5 targeting ANRIL were designed. When genomic
DNA was excised by CRISPR/Cas9, overlapped peaks were identified in
the DNA sequence. Overlapped peaks appeared when mutations were
generated in the PVT1 and ANRIL sequences using gRNA1 (Fig. 1B), gRNA2 9 (Fig. 1C), gRNA3 (Fig. 1D), gRNA4 (Fig. 1E) and gRNA5 (Fig. 1F). However, no overlapping peaks were
evident when gRNA2 was used to target PVT1 (Fig. 1C). In addition, gRNA3 targeting of
ANRIL was not effective in tetracycline-inducible CRISPR/Cas9
system (Fig. 1D). Therefore, the
effective gRNAs targeting of PVT1 (gRNA1, 3, 4 and 5) and the
effective gRNA-targeting of ANRIL (gRNA1, 2, 4 and 5) may guide the
system to excise PVT1 or ANRIL DNA (controlled by DOX).
Suppression efficiency of
tetracycline-inducible CRISPR/Cas9, analyzed by RT-qPCR and western
blot analysis
The expression levels of PVT1 and ANRIL in bladder
cancer cell lines, T24 and 5637, were measured by RT-qPCR. A total
of 5 gRNAs targeting PVT1 and 5 gRNAs targeting ANRIL were designed
and subcloned into plasmids. Their effects on PVT1 and ANRIL were
detected following transfection of each CRISPR/Cas9 system into
cells. Knockdown of PVT1 and ANRIL in T24 and 5637 cells was
achieved by 4 gRNAs targeting PVT1 (gRNA1, gRNA3, gRNA4 and gRNA5)
and 4 gRNAs targeting ANRIL (gRNA1, gRNA2, gRNA4 and gRNA5)
(Fig. 2A and B). The gRNA3 and gRNA4
targeting PVT1, gRNA2 and gRNA5 targeting ANRIL were determined to
induce maximal inhibition of PVT1 and ANRIL expression, compared
with the negative control in T24 and 5637 cells (Fig. 2A and B). gRNA3 and gRNA4 targeting
PVT1, and gRNA2 and gRNA5 targeting ANRIL were selected for further
examination. In order to achieve higher inhibition efficiency, the
sequences of these four gRNAs were inserted into one vector to
achieve simultaneous expression. A quantification analysis was
conducted in order to verify the tetracycline-inducible CRISPR/Cas9
system's ability to knock down PVT1 and ANRIL. The mRNA expression
levels of PVT1 and ANRIL were significantly suppressed following
addition of DOX to T24 cells (Fig.
2C; P<0.01) and 5637 cells (Fig.
2D; P<0.01). compared with the relative control cells. In
conclusion, the expression of Cas9 at the protein level was
verified. As indicated in Fig. 2E,
Cas9 was expressed in T24 and 5637 cells following DOX addition.
However, in the absence of DOX, no expression of Cas9 was
evident.
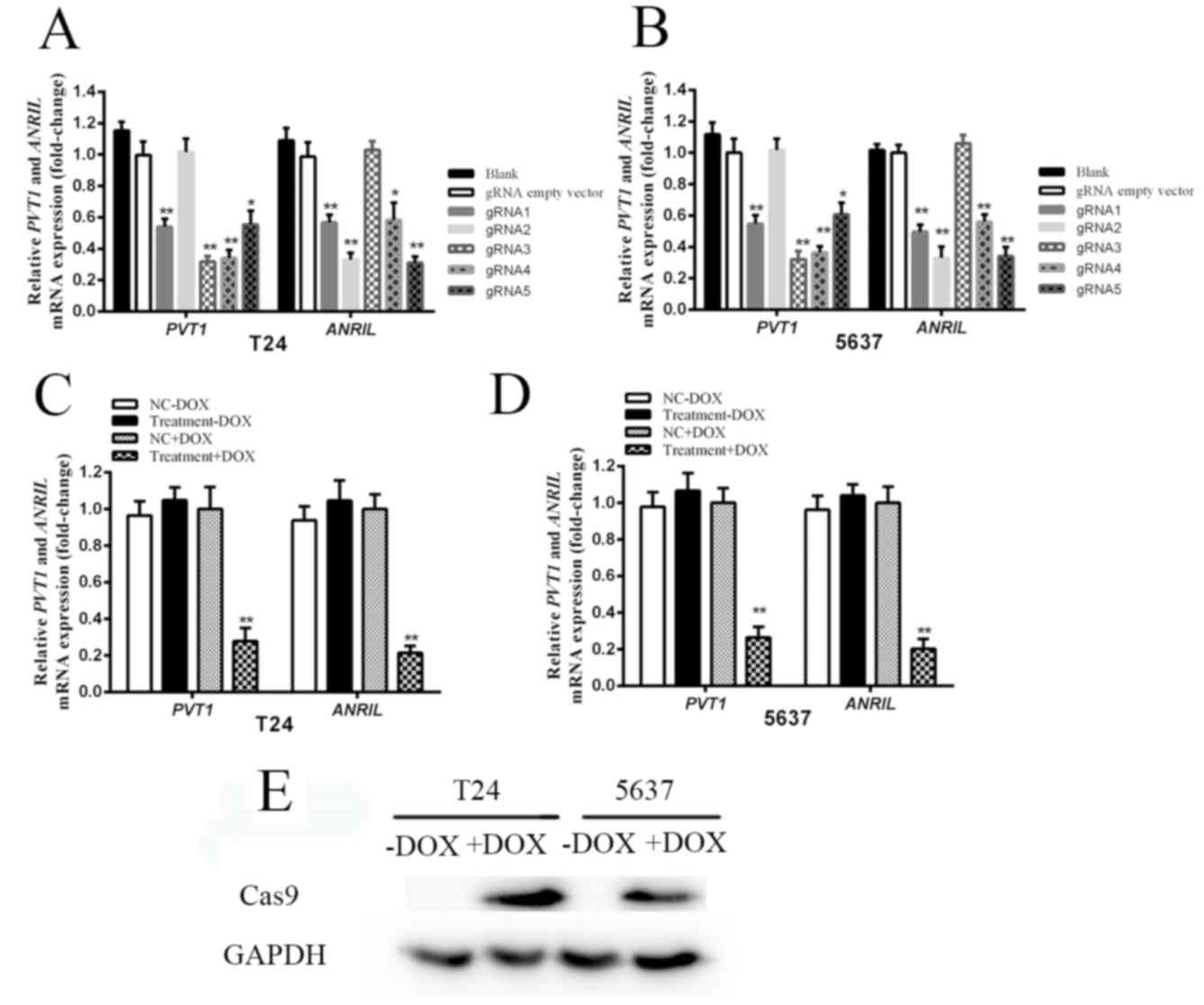 | Figure 2.The relative expression levels of
PVT1, ANRIL and Cas9 following transfection with
tetracycline-inducible CRISPR/Cas9. (A) The expression levels of
PVT1 and ANRIL in T24 cells following transfection with different
gRNAs and tetracycline-inducible Cas9. (B) The mRNA expression
levels of PVT1 and ANRIL in 5637 cells following transfection with
different gRNAs and tetracycline-inducible Cas9. The expression
levels of gRNA1, gRNA2, gRNA3, gRNA4 and gRNA5 were measured,
compared with the gRNA empty vector group. gRNA3 and gRNA4
targeting PVT1, and gRNA2 and gRNA5 targeting ANRIL significantly
inhibited PVT1 and ANRIL expression compared with the negative
control (gRNA empty vector group) in T24 and 5637 cells. (C) The
expression levels of PVT1 and ANRIL were significantly suppressed
following addition of DOX in T24 cells, compared with the NC+DOX
group cells (P<0.01). (D) The expression levels of PVT1 and
ANRIL were significantly suppressed following addition of DOX in
5637 cells, compared with the NC+DOX group cells (P<0.01). Error
bars represent the mean ± standard deviation. *P<0.05,
**P<0.01. (E) In the absence of DOX, Cas9 was not expressed in
T24 and 5637 cells. However, following DOX addition, expression of
Cas9 was evident. CRISPR, clustered regularly interspaced short
palindromic repeats; Cas9, CRISPR associated protein 9; gRNA, guide
RNA; DOX, doxycycline. |
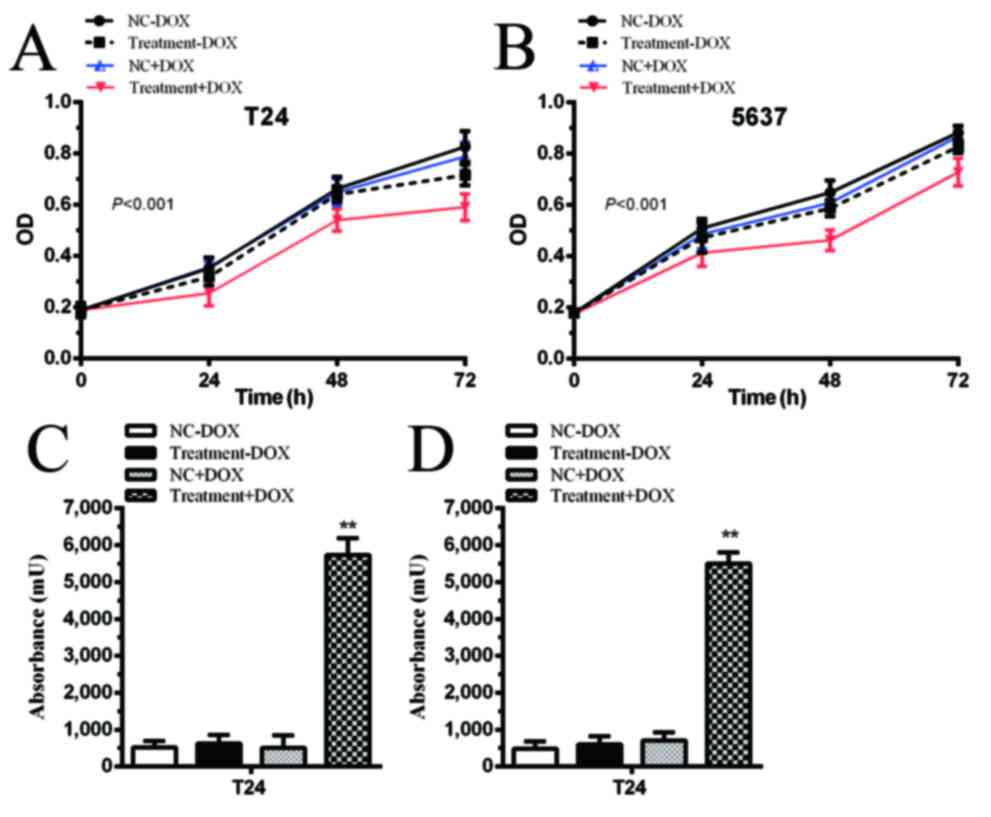
Proliferation is inhibited by
tetracycline-inducible CRISPR/Cas9 in bladder cancer cells
The cells transfected with gRNA vectors and
tetracycline-inducible Cas9 vectors were designated as the
Treatment group. No significant differences were identified in the
negative control (NC) and Treatment groups in T24 and 5637 cells
(Fig. 3A and B; P>0.05). When 1
µg/ml DOX was added to the medium, proliferation was significantly
inhibited in the Treatment+DOX group in T24 cells compared with the
NC+DOX group (Fig. 3A; P<0.001)
and 5637 cells (Fig. 3B; P<0.001).
Proliferation was inhibited by controlling the expression of Cas9
via simultaneously targeting two oncogenic lncRNAs in the bladder
cancer cells.
Apoptotic rate is induced by
tetracycline-inducible CRISPR/Cas9 in bladder cancer cells
In the absence of DOX, apoptotic rate was not
significantly different between the NC and Treatment groups.
However, following the addition of DOX, Cas9 was expressed and
CRISPR/Cas9 was able to excise PVT1 and ANRIL. Apoptotic rate was
significantly increased in the Treatment+DOX group in T24 cells
(Fig. 3C; P<0.01) and 5637 cells
(Fig. 3D; P<0.01) compared with
the NC+DOX group.
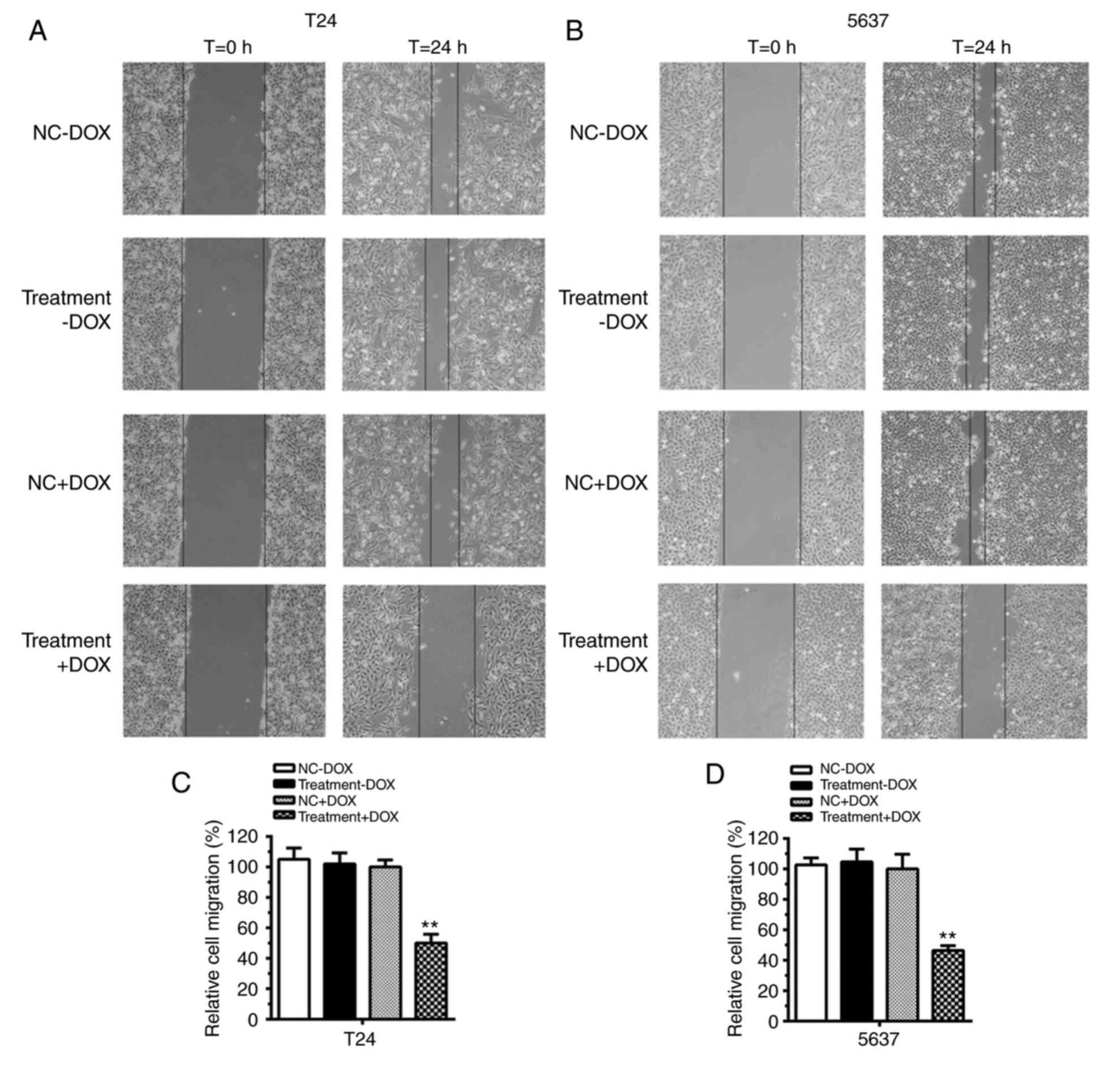
Cell migration was suppressed by
tetracycline-inducible CRISPR/Cas9 in bladder cancer cells
In the absence of DOX, no difference in cell
migration was evident between the NC and Treatment groups in T24
(Fig. 4A) and 5637 cells (Fig. 4B). Quantification analysis verified
that, in the absence of DOX, no significant differences in cell
migration were identified in T24 cells and 5637 cells compared with
the control (Fig. 4C and D;
P>0.05). However, with the addition of DOX to the medium, cell
migration was repressed in the Treatment+DOX group in T24 and 5637
cells compared with the NC+DOX group. Cell migration was inhibited
by almost 50% in T24 cells (Fig. 4C;
P<0.01) and 60% in 5637 cells (Fig.
4D; P<0.01). These results suggested that the
tetracycline-inducible CRISPR/Cas9 system could function as an
inhibitor of cell migration in bladder cancer cells.
Discussion
CRISPR/Cas9 is used as a robust genome editing tool
to induce specific genomic modifications in mammalian cells
(27,28). CRISPR/Cas9 can induce mutations in
genomes when multiple gRNAs are integrated with Cas9 in an array
(27). This tool has been applied in
transcription regulation and gene therapy (15). In a previous study, a catalytically
defective Cas9 mutant (dCas9) was created and co-expressed with
gRNA to generate a recognition complex, which was indicated to
control gene expression at a transcriptional level by interfering
with RNA polymerase, elongation and transcription factor binding
(29). In addition, CRISPR/Cas9 has
been used to target the long-terminal repeat promoter of HIV-1 to
inhibit HIV-1 expression in infected human cells (30).
However, there are certain adverse effects of the
CRISPR/Cas9 system, including off-target effects, protospacer
adjacent motif dependence and gRNA production (15). Researchers have attempted to eliminate
the off-target mutations of CRISPR/Cas9 (31,32). The
dosage of CRISPR/Cas9 has been demonstrated to affect the
off-target effects (15) and, in the
present study, a tetracycline switch was used to control the
expression of Cas9 to regulate the dosage of CRISPR/Cas9.
Numerous long non-coding RNAs (lncRNAs) serve
important roles in the development of different types of cancer and
may be potential biomarkers (33).
Oncogenic lncRNAs in bladder cancer were selected as targets in the
present study. With the use of CRISPR/Cas9 to simultaneously knock
down ≥2 lncRNAs, higher suppression efficiency may be achieved.
Previous studies revealed that overexpression of PVT1 and ANRIL
promoted the progression of bladder cancer (9,13).
Therefore, these 2 lncRNAs were selected for targeting by gRNAs
In the present study, a tetracycline-inducible
CRISPR/Cas9 system was constructed targeting lncRNAs to reduce
off-target effects and inhibit the malignant behavior of bladder
cancer cells. The results indicated that this
tetracycline-inducible system had no effects in the absence of DOX.
However, with the addition of DOX, this system could significantly
repress the malignant phenotype of bladder cancer cells.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LP, PP and JC performed the experiments. JC wrote
the paper. XY analyzed the data. JW and YC designed the project. YC
provided financial support for the project.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of
interest.
References
|
1
|
Kaufman DS, Shipley WU and Feldman AS:
Bladder cancer. Lancet. 374:239–249. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Marta GN, Hanna SA, Gadia R, Correa SF,
Silva JL and Carvalho A: The role of radiotherapy in urinary
bladder cancer: Current status. Int Braz J Urol. 38:144–153. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Racioppi M, Agostino DD, Totaro A, Pinto
F, Sacco E, D'Addessi A, Marangi F, Palermo G and Bassi PF: Value
of current chemotherapy and surgery in advanced and metastatic
bladder cancer. Urol Int. 88:249–258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Amit D and Hochberg A: Development of
targeted therapy for bladder cancer mediated by a double promoter
plasmid expressing diphtheria toxin under the control of H19 and
IGF2-P4 regulatory sequences. J Transl Med. 8:1342010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Droop J, Szarvas T, Schulz WA, Niedworok
AC, Niegisch G, Scheckenbach K and Hoffmann MJ: Diagnostic and
prognostic value of long noncoding RNAs as biomarkers in urothelial
carcinoma. PloS One. 12:e01762872017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Berrondo C, Flax J, Kucherov V, Siebert A,
Osinski T, Rosenberg A, Fucile C, Richheimer S and Beckham CJ:
Expression of the long non-coding RNA HOTAIR correlates with
disease progression in bladder cancer and is contained in bladder
cancer patient urinary exosomes. PloS One. 11:01472362016.
View Article : Google Scholar
|
|
7
|
Heubach J, Monsior J, Deenen R, Niegisch
G, Szarvas T, Niedworok C, Schulz WA and Hoffmann MJ: The long
noncoding RNA HOTAIR has tissue and cell type-dependent effects on
HOX gene expression and phenotype of urothelial cancer cells. Mol
Cancer. 14:1082015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Song J, Wu X, Liu F, Li M, Sun Y, Wang Y,
Wang C, Zhu K, Jia X, Wang B and Ma X: Long non-coding RNA PVT1
promotes glycolysis and tumor progression by regulating miR-497/HK2
axis in osteosarcoma. Biochem Biophys Res Commun. 490:217–224.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhuang C, Li J, Liu Y, Chen M, Yuan J, Fu
X, Zhan Y, Liu L, Lin J, Zhou Q, et al: Tetracycline-inducible
shRNA targeting long non-coding RNA PVT1 inhibits cell growth and
induces apoptosis in bladder cancer cells. Oncotarget.
6:41194–41203. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li T, Meng XL and Yang WQ: Long noncoding
RNA PVT1 acts as a ‘sponge’ to inhibit microRNA-152 in gastric
cancer cells. Dig Dis Sci. 62:3021–3028. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lillycrop K, Murray R, Cheong C, Teh AL,
Clarke-Harris R, Barton S, Costello P, Garratt E, Cook E, Titcombe
P, et al: ANRIL promoter DNA methylation: A perinatal marker for
later adiposity. EBioMedicine. 19:60–72. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wei X, Wang C, Ma C, Sun W, Li H and Cai
Z: Retraction note: Long noncoding RNA ANRIL is activated by
hypoxia-inducible factor-1α and promotes osteosarcoma cell invasion
and suppresses cell apoptosis upon hypoxia. Cancer Cell Int.
17:602017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu H, Li X, Song Y, Zhang P, Xiao Y and
Xing Y: Long non-coding RNA ANRIL is up-regulated in bladder cancer
and regulates bladder cancer cell proliferation and apoptosis
through the intrinsic pathway. Biochem Biophys Res Commun.
467:223–228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Clement F, Grockowiak E, Zylbersztejn F,
Fossard G, Gobert S and Maguer-Satta V: Stem cell manipulation,
gene therapy and the risk of cancer stem cell emergence. Stem Cell
Investig. 4:672017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang F, Wen Y and Guo X: CRISPR/Cas9 for
genome editing: Progress, implications and challenges. Hum Mol
Genet. 23:R40–46. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jinek M, Chylinski K, Fonfara I, Hauer MJ,
Doudna A and Charpentier E: A programmable dual-RNA-guided DNA
endonuclease in adaptive bacterial immunity. Science. 337:816–821.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen X, Janssen JM, Liu J, Maggio I, 't
Jong AEJ, Mikkers HMM and Goncalves MAFV: In trans paired nicking
triggers seamless genome editing without double-stranded DNA
cutting. Nat Commun. 8:6572017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hruscha A, Krawitz P, Rechenberg A,
Heinrich V, Hecht J, Haass C and Schmid B: Efficient CRISPR/Cas9
genome editing with low off-target effects in zebrafish.
Development. 140:4982–4987. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Shen B, Zhang J, Wu H, Wang J, Ma J, Li Z,
Zhang X, Zhang P and Huang X: Generation of gene-modified mice via
Cas9/RNA-mediated gene targeting. Cell Res. 23:720–723. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen Y, Zeng S, Hu R, Wang X, Huang W, Liu
J, Wang L, Liu G, Cao Y and Zhang Y: Using local chromatin
structure to improve CRISPR/Cas9 efficiency in zebrafish. PloS One.
12:e01825282017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang X, Zhuang C, Zhuang C, Xiong T, Li Y
and Gui Y: An enhanced hTERT promoter-driven CRISPR/Cas9 system
selectively inhibits the progression of bladder cancer cells. Mol
BioSyst. 13:1713–1721. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sanchez-Rivera FJ and Jacks T:
Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev
Cancer. 15:387–395. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vilaboa N and Voellmy R: Regulatable gene
expression systems for gene therapy. Curr Gene Ther. 6:421–438.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Goverdhana S, Puntel M, Xiong W, Zirger
JM, Barcia C, Curtin JF, Soffer EB, Mondkar S, King GD, Hu J, et
al: Regulatable gene expression systems for gene therapy
applications: Progress and future challenges. Mol Ther. 12:189–211.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhuang C, Huang X, Zhuang C, Luo X, Zhang
X, Cai Z and Gui Y: Synthetic regulatory RNAs selectively suppress
the progression of bladder cancer. J Exp Clin Cancer Res.
36:1512017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cong L, Ran FA, Cox D, Lin S, Barretto R,
Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA and Zhang F:
Multiplex genome engineering using CRISPR/Cas systems. Science.
339:819–823. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jinek M, East A, Cheng A, Lin S, Ma E and
Doudna J: RNA-programmed genome editing in human cells. Elife.
2:e004712013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Qi LS, Larson MH, Gilbert LA, Doudna JA,
Weissman JS, Arkin AP and Lim WA: Repurposing CRISPR as an
RNA-guided platform for sequence-specific control of gene
expression. Cell. 152:1173–1183. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ebina H, Misawa N, Kanemura Y and Koyanagi
Y: Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1
provirus. Sci Rep. 3:25102013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fu Y, Foden JA, Khayter C, Maeder ML,
Reyon D, Joung JK and Sander JD: High-frequency off-target
mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat
Biotechnol. 31:822–826. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mali P, Aach J, Stranges PB, Esvelt KM,
Moosburner M, Kosuri S, Yang L and Church GM: CAS9 transcriptional
activators for target specificity screening and paired nickases for
cooperative genome engineering. Nat Biotechnol. 31:833–838. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li J, Li Z, Zheng W, Li X, Wang Z, Cui Y
and Jiang X: LncRNA-ATB: An indispensable cancer-related long
noncoding RNA. Cell Prolif. 50:2017. View Article : Google Scholar
|