Introduction
At present, colon cancer is the most common
malignant tumor of the digestive tract around the world. According
to a literature report (1,2), statistical studies released by the
American Cancer Society in 2015 pointed out that the incidence and
mortality rates of colon cancer show increasing trends year by year
in male and female tumor patients in the United States, both of
which rank third. Nowadays, the incidence rate of colon cancer in
China is also increased year by year due to the changes in
lifestyle and influence of aging of the population, seriously
threatening human health and safety. Despite the progress in
diagnosis and treatment levels in recent years, the 5-year survival
rate does not exceed 50% (3,4). The occurrence and development of colon
cancer is a multi-stage, multi-step and multi-factor process.
Therefore, further investigating the pathogenesis of colon cancer
and finding out the molecular biomarker for early diagnosis of
colon cancer have very important significance in clinical
research.
Micro ribonucleic acid (miRNA or miR) is a kind of
recently-discovered non-coding RNA with approximately 21–25
nucleotides in length. After transcription, miRNA exerts its
function of regulating the gene expression through enhancing the
degradation and inhibiting the translation of mRNA (5). In recent years, it has been found in
research on miRNA that it plays an important role in occurrence,
development, proliferation and invasion of tumor as well as
regulation of drug resistance of tumor cells (6–8). In this
experiment, the expression levels of miR-205 and miR-506 in tissues
and plasma of patients with colon cancer were detected, and the
relationships of the expression levels of miR-205 and miR-506 in
colon cancer patients with clinicopathological features were also
detected.
Materials and methods
Specimen collection
In this experiment, samples from 166 patients with
colon cancer treated in Weifang People's Hospital (Weifang, China)
from May 2008 to July 2012 were collected. Colon cancer tissues and
normal fresh tissues at 20 mm near the cancer resected during
operation were taken and immediately stored in liquid nitrogen, and
then they were transferred into a refrigerator at −80°C for standby
application. The plasma of patients before operation and the plasma
of 100 healthy subjects receiving physical examination in our
hospital were taken as the control group. There were no
statistically significant differences in age and sex between them.
Colon cancer tissues were diagnosed and staged according to the
seventh edition of tumor-node-metastasis (TNM) staging criteria of
the American Joint Committee on Cancer (AJCC). None of the patients
with colon cancer received chemotherapy, radiotherapy and special
treatment for cancer before operation. This study was approved by
the Ethics Committee of Weifang People's Hospital (Weifang, China),
and all patients and their families were informed and signed the
consent.
Plasma treatment
After 5 ml plasma specimen of patients and 5 ml
plasma specimen of the control group were placed into the
anticoagulant tube containing ethylene diamine tetraacetic acid
(EDTA; Beyotime Institute of Biotechnology, Shanghai, China), they
were let stand at room temperature for 30 min and centrifuged at
2,300 × g for 10 min at 4°C. The supernatant was transferred into
an RNase-free tube (Sangon Biotech, Shanghai, China) by using a
pipettor and centrifuged at 9,600 × g for 15 min at 4°C. Then the
supernatant was taken and placed into the RNase-free tube, after
which the supernatant was stored in a refrigerator at −80°C.
Clinical data of patients are shown in Table I.
 | Table I.Associations of miR-205 and miR-506
expression levels with clinicopathological data of colon cancer
patients. |
Table I.
Associations of miR-205 and miR-506
expression levels with clinicopathological data of colon cancer
patients.
| Clinical feature | n | miR-205 expression
level | χ2 | P-value | miR-506 expression
level | χ2 | P-value |
|---|
| Age (years) |
| ≥55 | 84 | 1.894±0.447 | 0.219 | 0.812 | 3.417±1.267 | 1.307 | 0.264 |
|
<55 | 82 | 1.836±0.514 |
|
| 3.157±1.474 |
|
|
| Sex |
| Male | 97 | 1.819±0.681 | 1.684 | 0.627 | 3.387±1.678 | 0.354 | 0.437 |
|
Female | 69 | 1.864±0.544 |
|
| 3.574±1.461 |
|
|
| Smoking (years) |
| ≥10 | 88 | 2.074±0.374 | 2.146 | 0.061 | 3.671±1.257 | 1.814 | 0.296 |
|
<10 | 78 | 1.964±0.416 |
|
| 3.174±1.651 |
|
|
| Lymph node
metastasis |
| Yes | 100 | 2.341±0.241 | 4.227 | 0.037 | 3.754±1.058 | 4.519 | 0.031 |
| No | 66 | 1.841±0.539 |
|
| 3.271±1.487 |
|
|
| Degrees of
differentiation |
| Low
differentiation | 25 | 1.851±0.684 | 0.624 | 0.731 | 3.341±1.424 | 0.874 | 0.481 |
| Moderate
differentiation | 79 | 1.944±0.473 |
|
| 3.697±1.198 |
|
|
| High
differentiation | 62 | 2.181±0.314 |
|
| 3.864±0.874 |
|
|
| TNM staging |
| I | 13 | 1.830±0.846 | 4.847 | 0.029 | 3.074±1.384 | 2.234 | 0.056 |
| II | 87 | 1.924±0.681 |
|
| 3.438±1.108 |
|
|
|
III+IV | 66 | 2.324±0.437 |
|
| 3.733±0.798 |
|
|
Reagents and equipment
TRIzol reagent used for RNA extraction was purchased
from Shanghai Pufei Biotechnology Co., Ltd. (Shanghai, China);
mirVana™ PARIS™ kit and TaqMan MicroRNA assay were purchased from
ABI (Foster City, CA, USA); NanoDrop 2000 ultraviolet
spectrophotometer was purchased from Thermo Fisher Scientific, Inc.
(Waltham, MA, USA); Countess II FL Automated Cell Counter
fluorescence reverse transcription quantitative polymerase chain
reaction (RT-qPCR) instrument was purchased from ABI.
Methods
Total RNA extraction
Total RNA was extracted from colon cancer tissues
and para-carcinoma tissues by using TRIzol reagent (one-step
extraction method), and RNA was extracted from the centrifuged
plasma by using the mirVana™ PARIS™ kit in strict accordance with
the manufacturer's instructions. The concentration and purity of
extracted RNA were detected by using NanoDrop 2000 ultraviolet
spectrophotometer. The integrity of the extracted RNA was detected
via agarose gel electrophoresis.
Reverse transcription reaction
The reverse transcription primers of miR-205 and
miR-506 extracted were designed, and special circular structures
were made for the experiments. The primer sequences were designed
and synthesized by Takara Biotech (Beijing) Co., Ltd. (Beijing,
China). In this experiment, U6 was used as an internal reference
gene. A total of 15 µl reverse transcription system included 0.5 µl
reverse transcription primer, 0.5 µl reverse transcriptase, 2.0 µl
buffer solution and 2 µl RNA; diethylpyrocarbonate (DEPC)-treated
water was added if the volume was insufficient. Reaction
conditions: 37°C for 10 min, and 95°C for 5 min; after synthesis,
complementary DNA (cDNA) was stored at 4°C.
Detection of miR-205 and miR-506 via
RT-qPCR
A total of 20 µl RT-qPCR system was prepared by
using the TaqMan probe method according to the manufacturer's
instructions: 1 µl TaqMan MicroRNA assay (20*); 1.33 µl cDNA
(diluted at 1:15); 10 µl TaqMan 2× Universal PCR Master Mix II
(Beyotime Institute of Biotechnology); finally, DEPC-treated water
was added to make up for the volume to 20 µl. PCR amplification was
performed by using the Countess II FL Automated Cell Counter
RT-qPCR instrument. Reaction conditions: 95°C for 5 min, 95°C for
20 sec, and 60°C for 45 sec; a total of 45 cycles. The cycle
threshold (Cq) values were calculated by the 2−ΔΔCq
method (9) (Table II).
 | Table II.Primer sequences. |
Table II.
Primer sequences.
| Gene | Forward primers | Reverse primers |
|---|
| miRNA-205 |
5′-AATCCTTCATTCCACCGG-3′ |
5′-GTGCAGGGTCCGAGGT-3′ |
| miRNA-506 |
5′-TGCGGTAAGGCACCCTTCTGAGTAC-3′ |
5′-CCAGTGCAGGGTCCGAGGT-3′ |
| U6 |
5′-CTCGCTTCGGCAGCACA-3′ |
5′-AACGCTTCACGAATTTGCGT-3′ |
Statistical methods
In this study, Statistical Product and Service
Solutions (SPSS) 17.0 software package (SPSS, Inc., Chicago, IL,
USA) was used for data processing, and GraphPad Prism 5 software
(GraphPad Software, Inc., La Jolla, CA, USA) was used for image
processing. Enumeration data are presented as percentage (%), and
the Chi-square test was used for comparison between the two groups.
Measurement data are presented as mean ± standard deviation (mean ±
SD), and the least significant difference (LSD) method was used for
intragroup comparisons. Spearman analysis was used for correlation
analysis. P<0.05 suggested that the difference was statistically
significant.
Results
Expression levels of miR-205 and
miR-506 in colon cancer tissues and para-carcinoma tissues
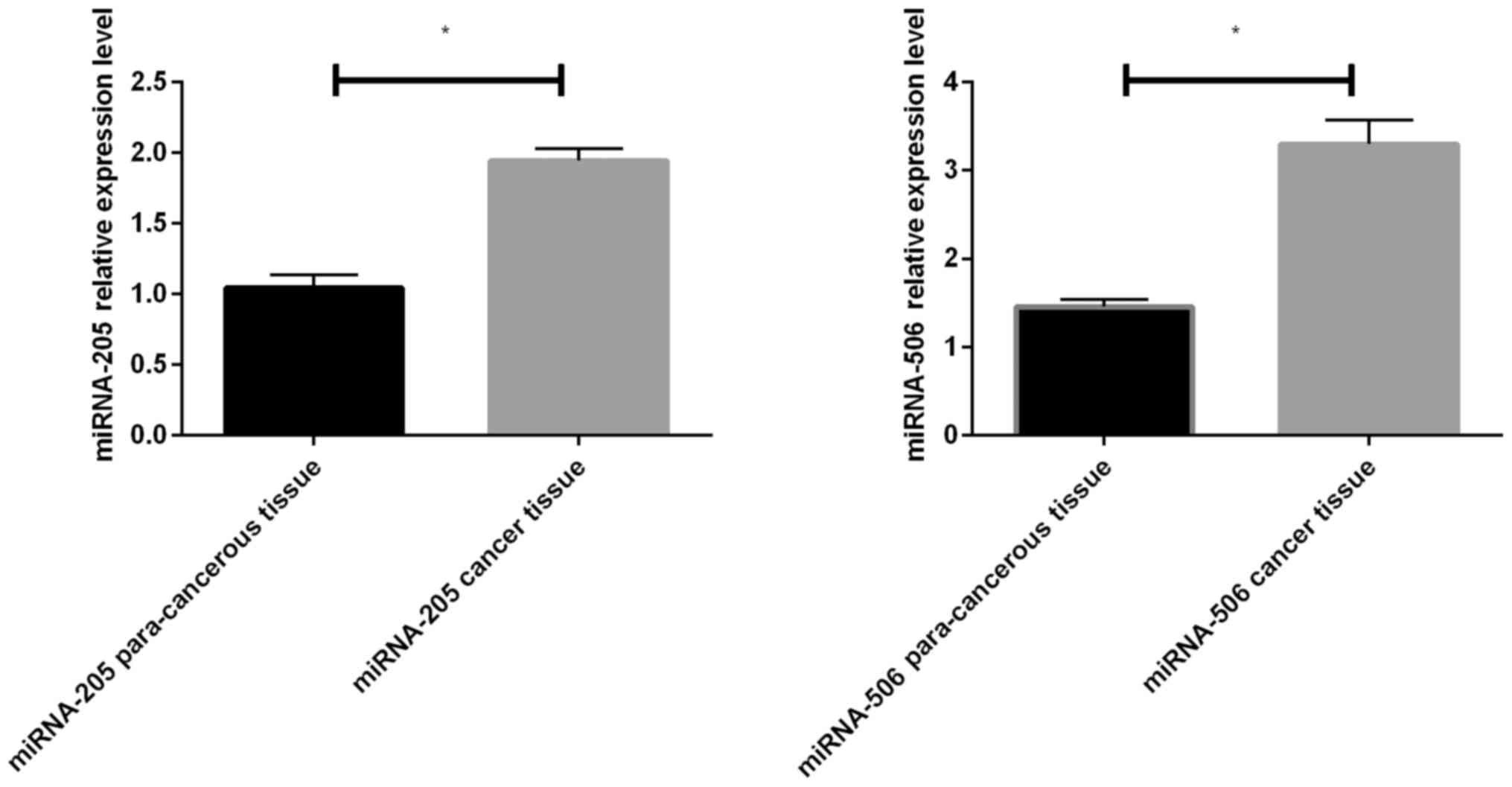
The results of RT-qPCR of miR-205 and miR-506 showed
that the expression levels of them in colon cancer tissues were
significantly increased compared with those in para-carcinoma
tissues, and the differences were statistically significant
(P<0.05) (Fig. 1).
Differential expression levels of
miR-205 and miR-506 in plasma of colon cancer patients and healthy
control group
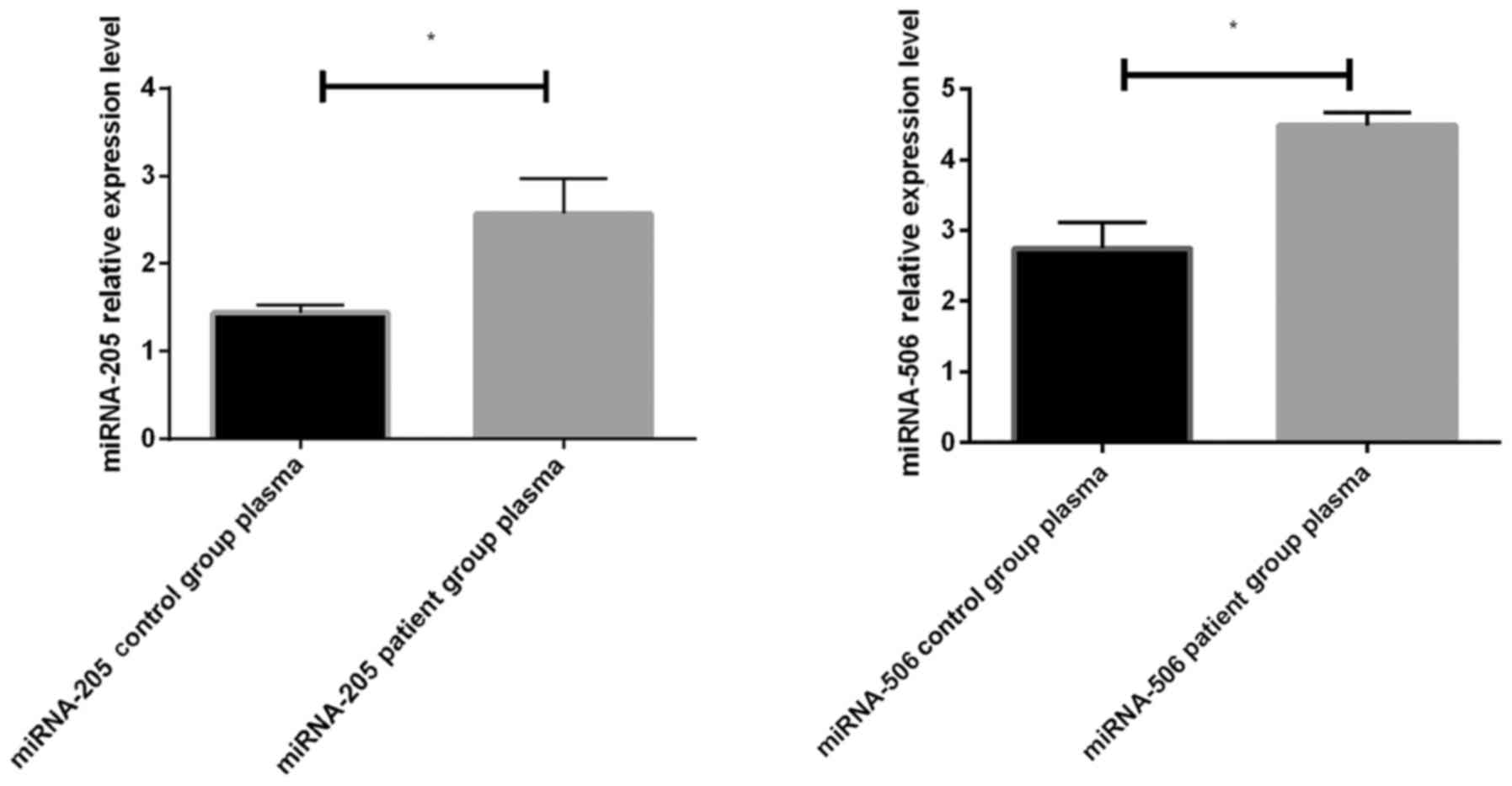
The expression levels of miR-205 and miR-506 in
plasma of colon cancer patients were significantly increased
compared with those in the control group, and the differences were
statistically significant (P<0.05) (Fig. 2).
Associations of miR-205 and miR-506
expression levels with clinicopathological features of colon cancer
patients
Association analyses of clinicopathological features
with miR-205 and miR-506 showed that associations of them with sex
and age had no statistically significant differences (P>0.05).
The expression level of miR-205 was associated with TNM staging and
lymph node metastasis (P<0.05), while the expression level of
miR-506 was associated with lymph node metastasis (P<0.05)
(Table I).
Correlations of the expression levels
of the two miRNAs in plasma and colon cancer tissues
The experimental results showed that the expression
levels of miR-205 in colon cancer tissues and plasma in patients
had no significant correlation (r=0.467, P=0.081). There was a
positive correlation between the expression levels of miR-506 in
colon cancer tissues and plasma in patients, and the difference was
statistically significant (r=0.599, P=0.038) (Table III).
 | Table III.Correlations of the expression levels
of the two miRNAs in plasma and tissues of colon cancer
patients. |
Table III.
Correlations of the expression levels
of the two miRNAs in plasma and tissues of colon cancer
patients.
| Group | miRNA-205 expression
level | r | P-value | miR-506 expression
level | r | P-value |
|---|
| Plasma of
patients | 2.579±1.655 | 0.467 | 0.081 | 4.541±1.419 | 0.599 | 0.038 |
| Tissues of
patients | 1.806±0.424 |
|
| 3.147±0.441 |
|
|
Discussion
Colon cancer is a kind of malignant tumor of the
digestive tract with relatively high incidence and mortality rates,
seriously endangering human health and safety. It is estimated that
in 2012, there were approximately 1.4 million new cases of colon
cancer and 690,000 deaths in the world (10). miRNA is a kind of endogenous
non-coding microRNA with a length of 17–25 nucleotides. Incomplete
matching in non-coding region of 3′-untranslated region (UTR) of
its target genes blocks the mRNA translation of target genes, which
is closely related to cell apoptosis, proliferation, metastasis,
differentiation and other vital activities, as well as the
occurrence, development and progression of tumors. More and more
evidence has shown that the occurrence of colon cancer is a
multi-target and multi-step process (11–13). Some
miRNAs play an important role in the pathological process of colon
cancer. Different miRNAs, such as miR-141 (14), miR-145 (15), miR-231 (16) and miR-215 (17), have differential expression levels in
tissues.
In this experiment, the RT-qPCR technique was used
to detect the expression levels of miR-205 and miR-506 in colon
cancer tissues and para-carcinoma tissues. The differential
expression levels of them in the plasma of colon cancer patients
and healthy subjects were investigated, and the correlations of
their expression levels in tissues and plasma of colon cancer
patients were analyzed.
This study showed that the expression levels of
miR-205 and miR-506 in colon cancer tissues were higher than those
of para-carcinoma tissues (P<0.05), indicating that the two
miRNAs are closely related to the occurrence of colon cancer and
may participate in the pathological process of colon cancer as
proto-oncogenes. The associations of miR-205 and miR-506 with
clinicopathological data were further analyzed, and the results
showed that the expression level of miR-205 was related to TNM
staging and lymph node metastasis, and the expression level of
miR-506 was related to lymph node metastasis. The expression of
miR-205 was positively associated with TNM staging of patients with
colon cancer, and the higher the staging was, the higher the
expression level of miR-205 would be. de Carvalho et al
(18) pointed out in his study that
miR-205 has comparatively good sensitivity, specificity and
accuracy in differentiating the metastasis of patients with head
and neck squamous carcinoma, indicating that miR-205 may
participate in the metastasis of cancer cells to a large extent, so
it may serve as a potential biological target for whether tumor
metastasis occurs. This study showed that miR-205 was highly
expressed in colon cancer patients with lymph node metastasis,
which promoted cell metastasis in colon cancer patients, indicating
that miRNA is expressed identically in different tissues. Besides,
miR-506 is located in chromosome Xq27.3. Studies have shown that
miR-506 can be used as a target for a variety of target genes,
silence the target gene expression levels and influence the tumor
proliferation, invasion, metastasis and other processes (19). In the study of Zhang et al
(20), the expression level of
miR-506 is downregulated in colon cancer tissues, and it is
negatively associated with tumor size, lymph node metastasis and
TNM staging. However, the results in this experiment were opposite
to the above conclusions, which may be caused by the differences in
regions and races.
At present, colonoscope is mainly used in the
primary screening of colon cancer, which is the most effective
screening tool. However, it has certain dangers to the patients,
and its cost is high, so it is difficult to be widely used in
clinical practice. In particular, the sensitivity of occult blood
test in detection is low, and patients are strictly required to
control the diet and life to a certain degree, so a kind of more
effective and convenient biomarker is urgently needed for the
detection of colon cancer. miRNA can be detected in serum, plasma
and body fluids (21), suggesting
that plasma miRNA can be used as a biomarker for detection. Many
studies indicated that (22,23) specific miRNA exists in the patient's
tumor tissues and plasma. This experiment showed that the
expression levels of miR-205 and miR-506 in plasma were higher than
those in the normal control group, so miR-205 and miR-506 may be
used as potential molecular markers for screening colon cancer.
However, whether the two miRNAs can become tumor markers requires
the big-data test to verify the results.
With the improvement of living standards and changes
in the dietary structure, colon cancer, as a malignant tumor of the
digestive tract, has become one of the main factors threatening
human health. Therefore, improving the survival rate and the
quality of life of patients is a problem to be solved urgently.
In conclusion, the differential expression levels of
miR-205 and miR-506 in colon cancer and para-carcinoma tissues of
patients in this study suggest that they play an important role in
the occurrence and development of colon cancer, and it was found
that the expression levels of miR-205 and miR-506 in tissues of
colon cancer patients were associated with the clinicopathological
indexes. Moreover, changes in the miRNA expression level in plasma
can be used as potential tumor markers. As therapeutic targets,
miR-205 and miR-506 may act as new treatment means for colon
cancer.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
NJ was responsible for the extraction of total RNA.
LY and JS were responsible for the reverse transcription reaction.
ZC and WM performed PCR. NJ edited and WM revised the manuscript.
NJ was responsible for the data collection; LY was responsible for
the data analysis. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Weifang People's Hospital (Weifang, China), and all patients and
their families were informed and signed the consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 5:7–30. 2015.
|
|
2
|
Huang Z, Huang D, Ni S, Peng Z, Sheng W
and Du X: Plasma microRNAs are promising novel biomarkers for early
detection of colorectal cancer. Int J Cancer. 127:118–126. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Terzić J, Grivennikov S, Karin E and Karin
M: Inflammation and colon cancer. Gastroenterology. 138(2101–2114):
e52010.
|
|
4
|
Buunen M, Veldkamp R, Hop WC, Kuhry E,
Jeekel J, Haglind E, Påhlman L, Cuesta MA, Msika S, Morino M, et
al: Colon Cancer Laparoscopic or Open Resection Study Group:
Survival after laparoscopic surgery versus open surgery for colon
cancer: Long-term outcome of a randomised clinical trial. Lancet
Oncol. 10:44–52. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Schetter AJ, Leung SY, Sohn JJ, Zanetti
KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et
al: MicroRNA expression profiles associated with prognosis and
therapeutic outcome in colon adenocarcinoma. JAMA. 299:425–436.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shen Z, Zhan G, Ye D, Ren Y, Cheng L, Wu Z
and Guo J: MicroRNA-34a affects the occurrence of laryngeal
squamous cell carcinoma by targeting the antiapoptotic gene
survivin. Med Oncol. 29:2473–2480. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiao CZ, Wei W, Guo ZX, Zhang MY, Zhang
YF, Wang JH, Shi M, Wang HY and Guo RP: MicroRNA-34c-3p promotes
cell proliferation and invasion in hepatocellular carcinoma by
regulation of NCKAP1 expression. J Cancer Res Clin Oncol.
143:263–273. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fattore L, Sacconi A, Mancini R and
Ciliberto G: MicroRNA-driven deregulation of cytokine expression
helps development of drug resistance in metastatic melanoma.
Cytokine Growth Factor Rev. 36:39–48. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Douaiher J, Ravipati A, Grams B, Chowdhury
S, Alatise O and Are C: Colorectal cancer-global burden, trends,
and geographical variations. J Surg Oncol. 115:619–630. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Munteanu CR, Magalhães AL, Uriarte E and
González-Díaz H: Multi-target QPDR classification model for human
breast and colon cancer-related proteins using star graph
topological indices. J Theor Biol. 257:303–311. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cheng H, Zhang L, Cogdell DE, Zheng H,
Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR and Zhang W:
Circulating plasma MiR-141 is a novel biomarker for metastatic
colon cancer and predicts poor prognosis. PLoS One. 6:e177452011.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yu Y, Nangia-Makker P, Farhana L, Rajendra
SG, Levi E and Majumdar AP: miR-21 and miR-145 cooperation in
regulation of colon cancer stem cells. Mol Cancer. 14:982015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cekaite L, Rantala JK, Bruun J, Guriby M,
Agesen TH, Danielsen SA, Lind GE, Nesbakken A, Kallioniemi O, Lothe
RA, et al: MiR-9, −31, and −182 deregulation promote proliferation
and tumor cell survival in colon cancer. Neoplasia. 14:868–879.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karaayvaz M, Pal T, Song B, Zhang C,
Georgakopoulos P, Mehmood S, Burke S, Shroyer K and Ju J:
Prognostic significance of miR-215 in colon cancer. Clin Colorectal
Cancer. 10:340–347. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
de Carvalho AC, Scapulatempo-Neto C, Maia
DC, Evangelista AF, Morini MA, Carvalho AL and Vettore AL: Accuracy
of microRNAs as markers for the detection of neck lymph node
metastases in patients with head and neck squamous cell carcinoma.
BMC Med. 13:1082015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ruiz AJ and Russell SJ: MicroRNAs and
oncolytic viruses. Curr Opin Virol. 13:40–48. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang Y, Lin C, Liao G, Liu S, Ding J,
Tang F, Wang Z, Liang X, Li B, Wei Y, et al: MicroRNA-506
suppresses tumor proliferation and metastasis in colon cancer by
directly targeting the oncogene EZH2. Oncotarget. 6:32586–32601.
2015.PubMed/NCBI
|
|
21
|
Tölle A, Jung M, Rabenhorst S, Kilic E,
Jung K and Weikert S: Identification of microRNAs in blood and
urine as tumour markers for the detection of urinary bladder
cancer. Oncol Rep. 30:1949–1956. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Agaoglu Yaman F, Kovancilar M, Dizdar Y,
Darendeliler E, Holdenrieder S, Dalay N and Gezer U: Investigation
of miR-21, miR-141, and miR-221 in blood circulation of patients
with prostate cancer. Tumour Biol. 32:583–588. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schwarzenbach H, Milde-Langosch K,
Steinbach B, Müller V and Pantel K: Diagnostic potential of
PTEN-targeting miR-214 in the blood of breast cancer patients.
Breast Cancer Res Treat. 134:933–941. 2012. View Article : Google Scholar : PubMed/NCBI
|