Introduction
With continuous progress in modern medicine, the
overall survival of patients with malignant tumors has improved.
However, accompanying depression has become increasingly common
among cancer patients. A great deal of epidemiological studies have
shown that the incidence of depression in cancer patients is 2–4
times higher than that in the normal population, and up to 20–50%
of the patients are afflicted (1,2). Ovarian
cancer has the highest mortality rate among all gynecological
malignancies, which results in extreme anxiety and depression in
the patients. Bodurka reported a depression rate of 21% among the
patients with ovarian cancer (3),
while Price et al (4), found
clinical depression in 5.9% of 798 women with ovarian cancer in a
prospective cohort study.
Due to the belief that depression is a normal and
universal reaction to cancer, it is often underplayed in patients
with cancer. However, depression not only results in emotional
trauma, but more importantly also causes pathophysiological changes
in the patients (5,6). Neuroendocrine-immune modulation (NIM)
negative feedback network is one of the important
pathophysiological basis of clinical depression in patients with
cancer. Depression in pancreatic cancer patients has been linked to
the secretion of amines resistant to emotional excitement (7).
Various genes are involved in clinical depression,
and miRNA-mRNA interactions play an important role in regulating
its pathophysiological basis. Micro RNAs bind to the
3′-untranslated region (3′-UTR) of target genes involved in
cellular processes like proliferation, differentiation, apoptosis,
and immune responses (8). The
miRNA-mRNA regulatory network is of great significance in
identifying the mechanism of major depression disorder (MDD) in
ovarian cancer. In addition, the relevant miRNAs may be potential
diagnostic markers for the early detection of ovarian cancer
related depression, as well as prognostic indicators for treatment
response. Although a large number of epidemiological studies have
reported depression in cancer patients, few studies exist on the
miRNA-mRNA networks related to depression in cancer patients, which
can detect and diagnose MDD at early stages. Bioinformatics and
expression profiling techniques can help identify such networks in
various diseases.
In the present study, we analyzed the miRNA
expression profiles of patients with ovarian cancer or MDD, and the
mRNA expression profiles of depressed and non-depressed patients
with ovarian tumors. Using bioinformatics, we identified the miRNAs
and their target genes, and constructed an miRNA-mRNA-pathway
regulatory network.
Materials and methods
Acquisition of microarray data
Ovarian cancer and MDD associated gene expression
datasets, original data and platform records were acquired from the
Gene Expression Omnibus (GEO, http://www.ncbi.nlm nih.gov/geo/) database from the
National Center for Biotechnology Information (NCBI). The dataset
GSE61741 (ovarian cancer) based on GPL9040 platform (febit Homo
Sapiens miRBase 13.0) was submitted by Keller, and included 94
normal and 24 ovarian cancer samples. The MDD-associated dataset
GSE58105 based on GPL1873 platform [Agilent-021827 Human miRNA
Microarray (miRNA_107_Sep09)] was submitted by Lopez, and included
11 normal and 14 MDD samples. The GSE9116 dataset of depressed
patients with ovarian tumors based on GPL96 platform [(HG-U133A)
Affymetrix Human Genome U133A Array], was submitted by Cole, and
included data from 5 depressed patients and non-depressed patients
each, all with primary ovarian tumors.
Identification of differentially
expressed microRNAs (DEmiRs) or DEmRNAs and miRNA target genes
GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) is a useful
online microarray data analysis tool that allows users to compare
two or more groups of samples in a GEO series in order to identify
DEmRNAs. The miRNA or mRNA with P-value <0.05 were regarded as
DEmiRs or DEmRNAs respectively. The miRNA target genes were
obtained from the following 5 prediction tools: TargetScan,
microRNAorg, microT-CDS, miRDB and miRTarBase. Genes overlapping in
three or more prediction tools were selected as putative target
genes of miRNA.
Risk pathways and survival curves of
target genes involved in risk pathways
Gene ontology (GO) is a widely used method for the
large-scale functional annotation of genes, and is based on certain
structured, defined and controlled terms (9). Kyoto Encyclopedia of Genes and Genomes
(KEGG) database is a collection of online databases of gene
functions, enzymatic pathways, and helps link genomic information
with higher-order functional information (10). The Database for Annotation,
Visualization and Integrated Discovery (https://david.ncifcrf.gov/, DAVID) provides a
comprehensive set of functional annotation tools to identify KEGG
pathways and biological process (11,12). To
determine the cellular pathways of target genes, the DAVID program
was used to identify KEGG pathways and biological process, with
P<0.05 as the threshold value. Kaplan-Meier Plotter for ovarian
cancer (13), an online analysis
tool, was used to assess the effect of the selected genes on
ovarian cancer. P<0.05 was considered to indicate a
statistically significant difference.
Construction of miRNA-mRNA-pathway
regulation network
Cytoscape (http://www.cytoscape.org/), an open source software
platform for complex network analysis and visualization, was used
to construct the miRNA-target genes-pathways network.
Results
Co-differentially expressed miRNA and
target genes in ovarian cancer and depression
After comparing 24 ovarian cancer and 94 normal
samples from the GSE61741 dataset, we identified 300 DEmiRs with
P<0.05. Similarly, 40 DEmiRs were identified from the GSE58105
dataset. Furthermore, overlapping DEmiRs from both datasets
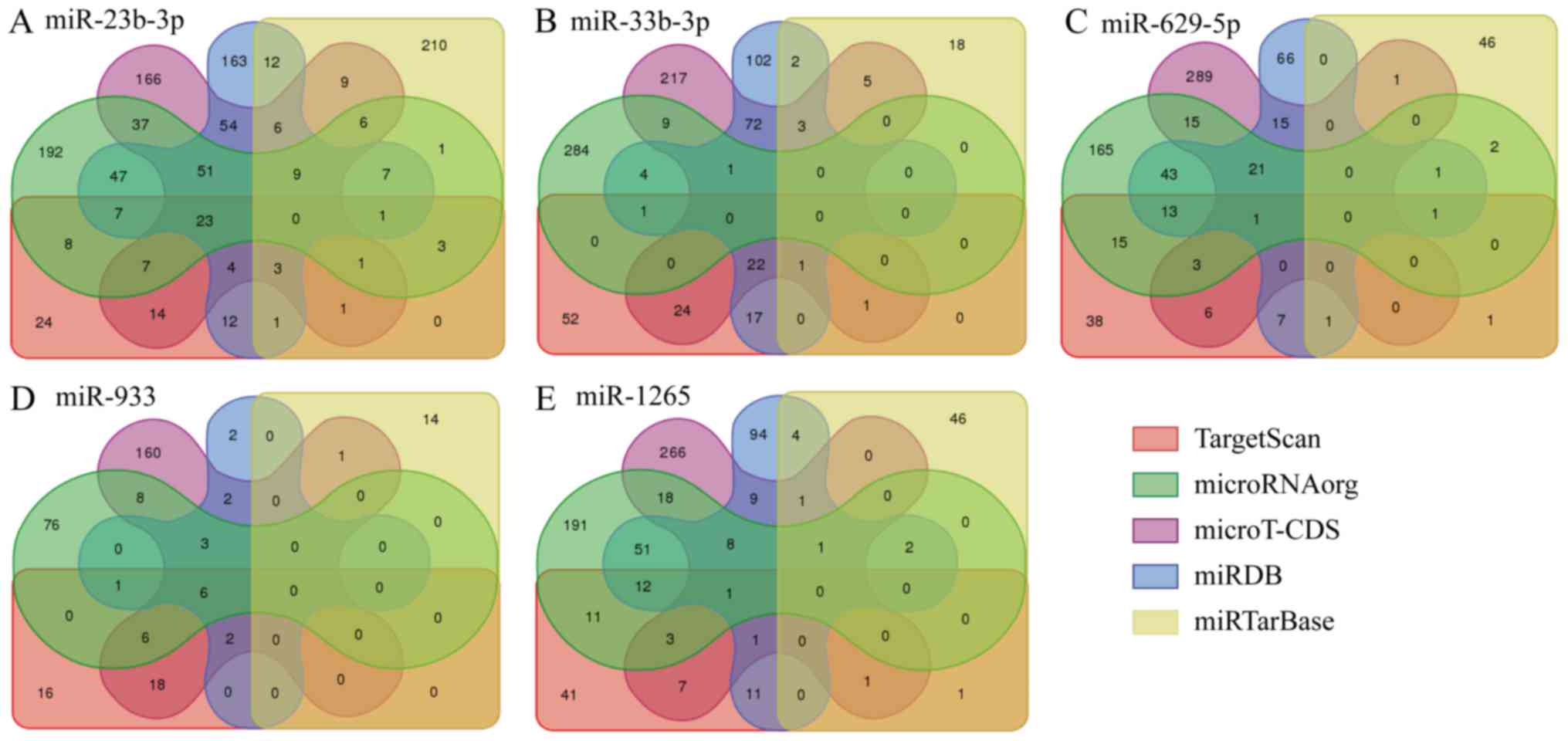
included 4 upregulated (miR-23b-3p, miR-33b-3p, miR-1265 and
miR-933) and 1 downregulated (miR-629-5p) DEmiRs. We used five gene
prediction programs on these DEmiRs, and selected the genes common
to three or more prediction tools as the target genes. There were
130, 29, 41, 18 and 30 target genes of miR-23b-3p, miR-33b-3p,
miR-1265, miR-933 and miR-33b-3p respectively, and 90, 14, 34, 12
and 22 of them respectively were also DEmRNAs in the GSE9116
dataset (Fig. 1).
Identification of the risk pathways,
risk genes and miRNA-risk gene-pathways regulation network
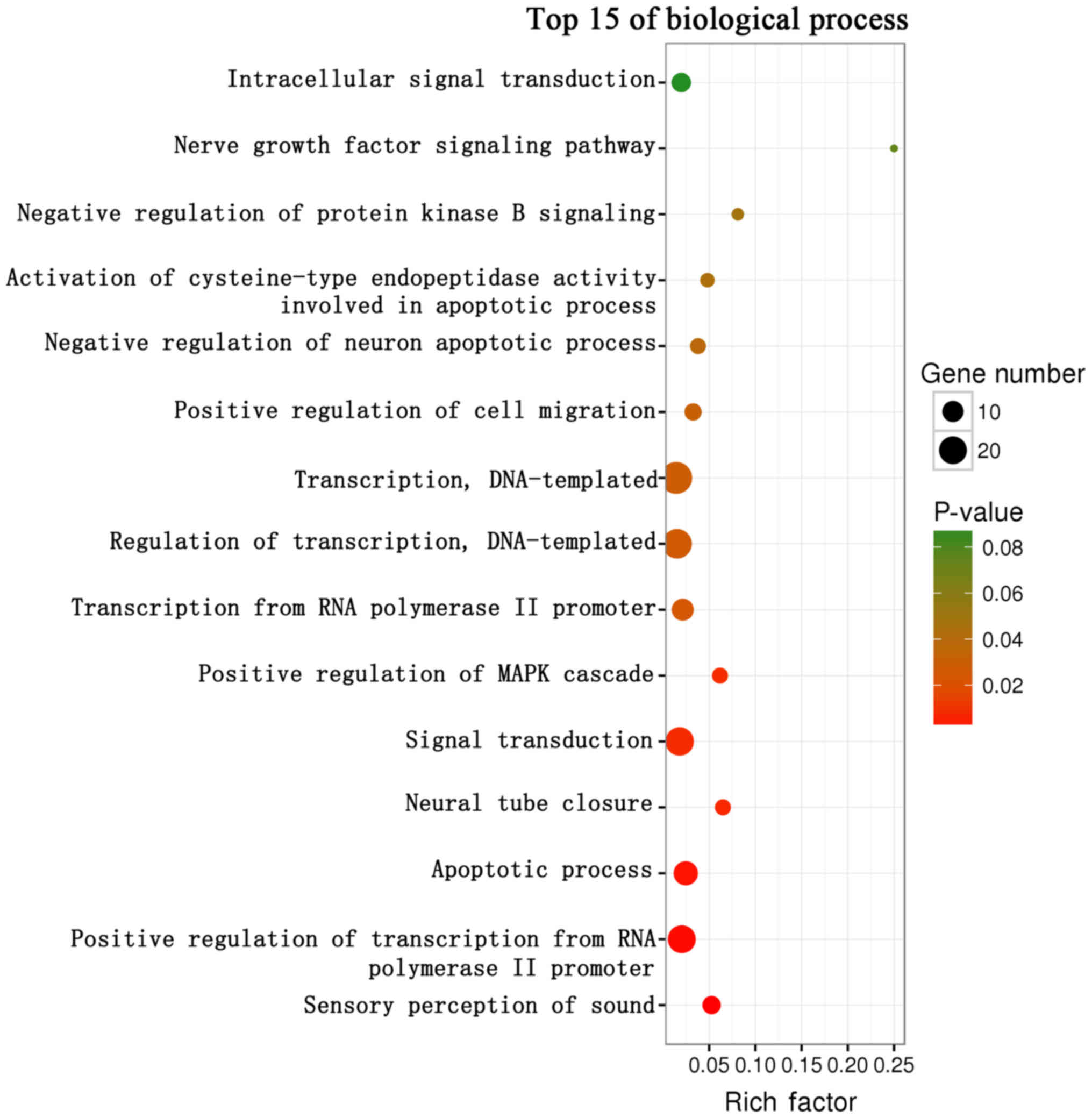
GO annotations showed that these target genes were
involved in biological process including neural tube closure,
negative regulation of neuron apoptotic process, nerve growth
factor signaling pathway, positive regulation of cell migration,
and apoptosis (Fig. 2). The most
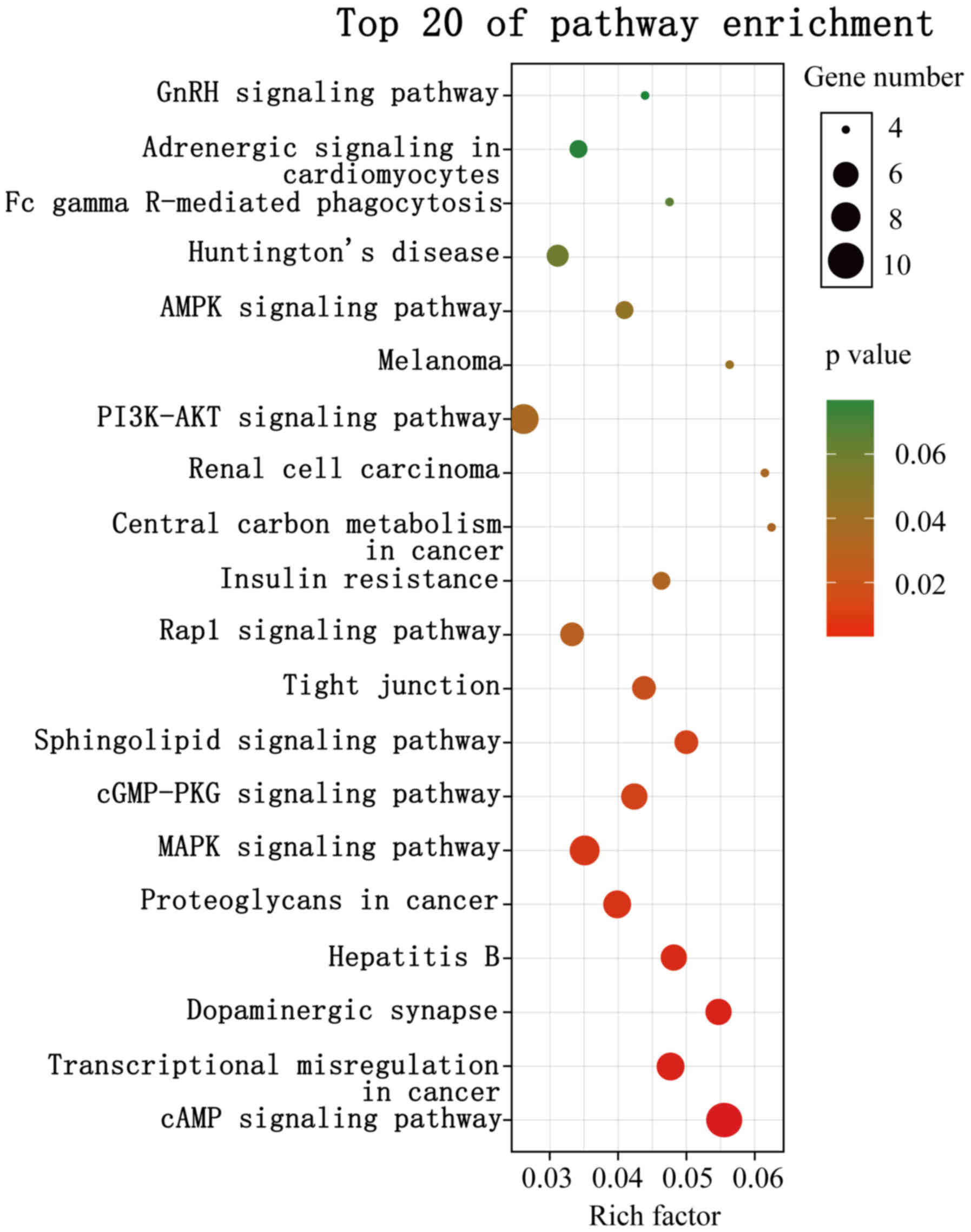
significantly enriched pathways of target genes which also exist as
DEmRNAs in the GSE9116 dataset are shown in Fig. 3. A P-value<0.05 indicated
statistically significant enrichment. Pathways associated with
ovarian cancer and MDD, considered risk pathways, included cAMP
signaling, transcriptional dysregulation in cancer, dopaminergic
synapse, hepatitis B, proteoglycans in cancer, MAPK signaling,
cGMP-PKG signaling, sphingolipid signaling, tight junction, rap1
signaling, central carbon metabolism in cancer, PI3K-Akt signaling,
and AMPK signaling. Genes enriched in these risk pathways, i.e. the
risk genes, included 22 upregulated and 15 downregulated genes. The
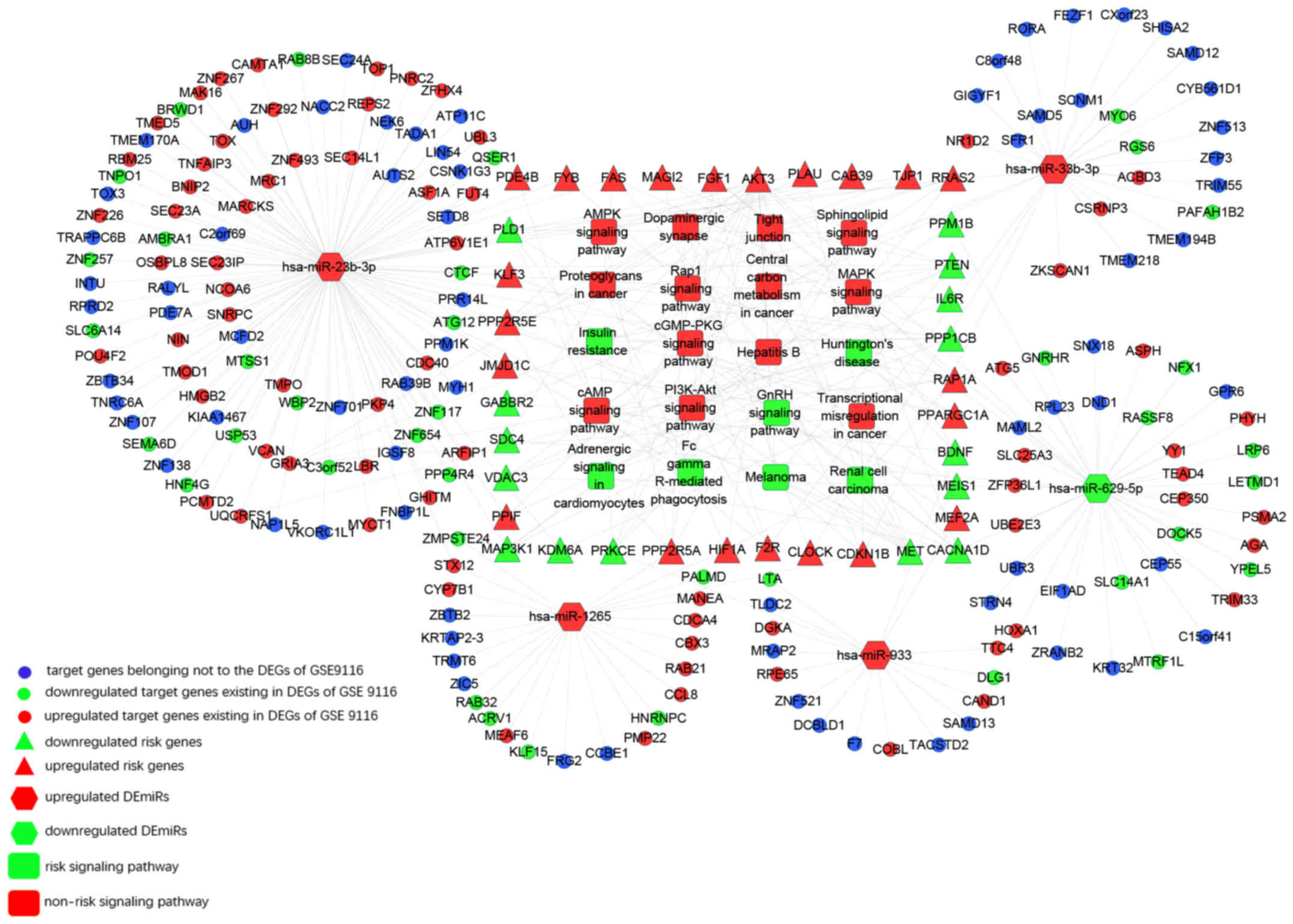
miRNA-mRNA regulatory network was constructed using 5 DEmiRs, 37
risk genes and 13 risk pathways (Fig.
4).
Identification of core genes in
ovarian cancer
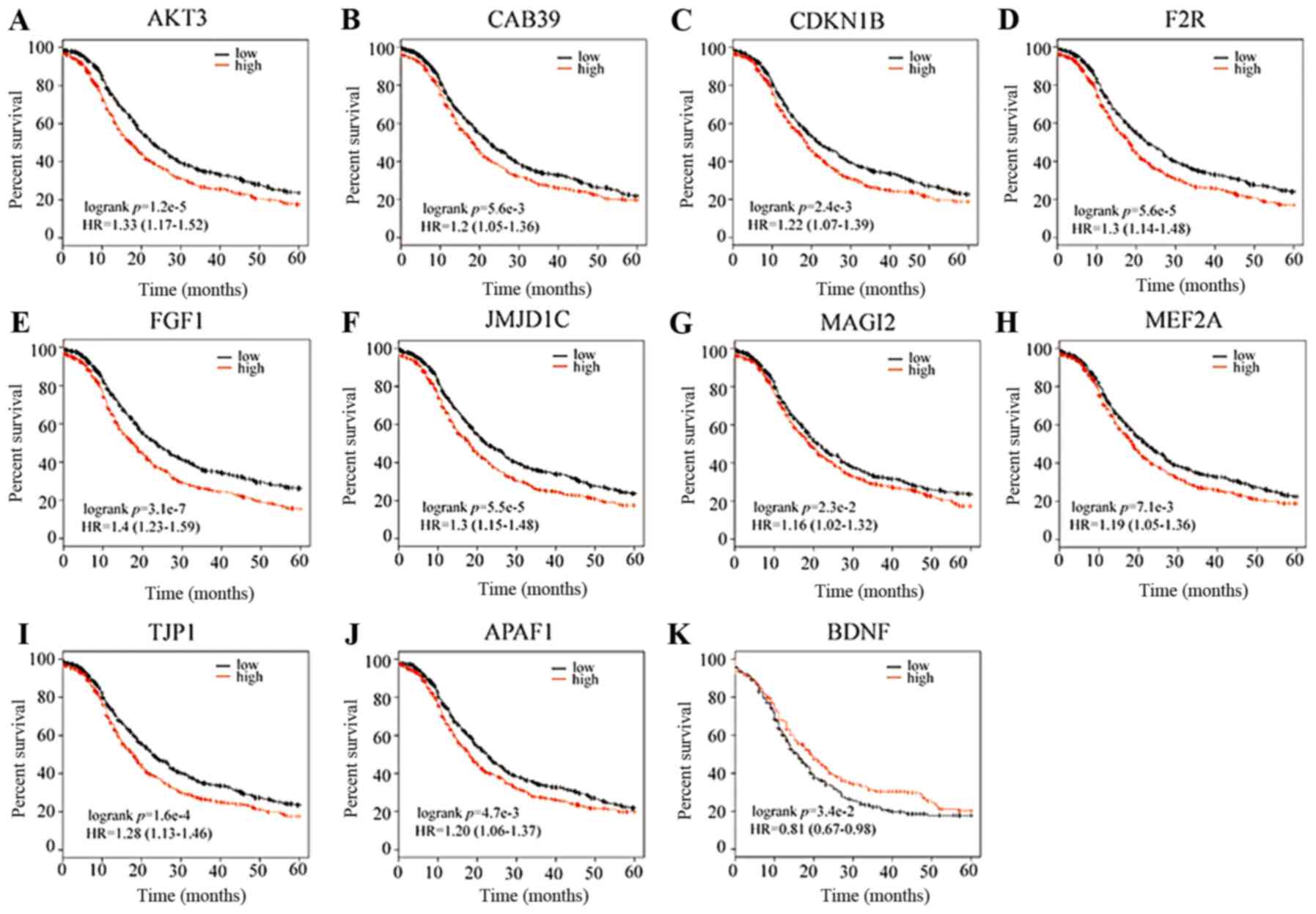
Upon overexpression of the 22 upregulated risk
genes, 10 risk genes could significantly increase the mortality of
ovarian cancer patients, while only BDNF overexpression among the
15 downregulated risk genes could significantly increase survival
rate. Taken together, 11 risk genes were identified as the core
genes of MDD development in ovarian cancer, which are also DEmRNAs
in GSE9116. The core genes include BDNF, MEF2A, FGF1, AKT3, MAGI2,
TJP1, JMJD1C, APAF1, CAB39, CDKN1B and F2R (Fig. 5).
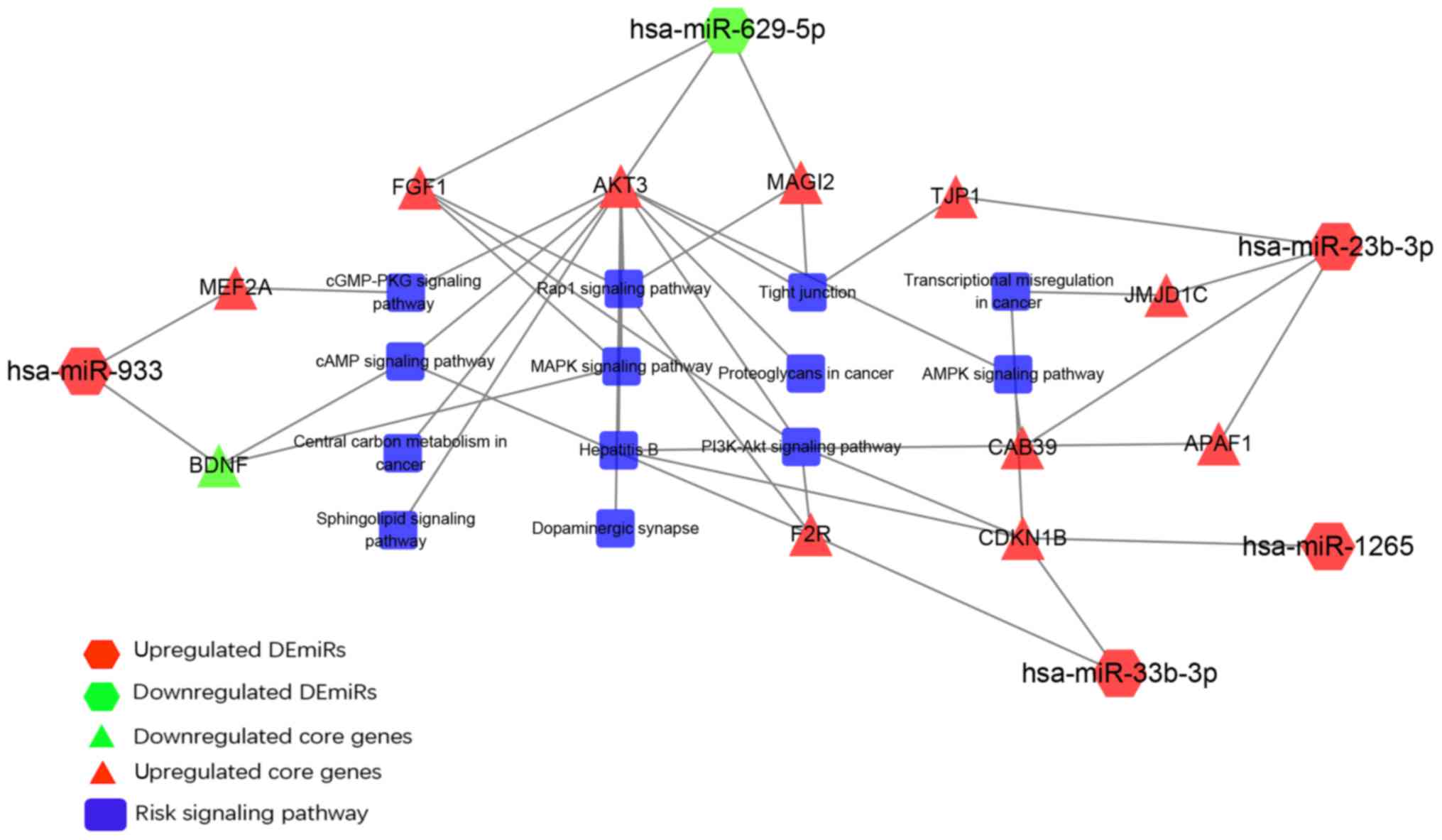
Analysis of miRNA-core gene-pathway
regulation network
We confirmed 12 pairs of miRNA-mRNA interaction
including miR-629-5p-FGF1, miR-629-5p-AKT3,
miR-629-5p-MAGI2, miR-933-BDNF, miR-933-MEF2A,
miR-23b-3p-TJP1, miR-23b-3p-JMJD1,
miR-23b-3p-APAF1, miR-23b-3p-CAB39,
miR-1265-CDKN1B, miR-33b-3p-CDKN1B, and
miR-33b-3p-F2R (Fig. 6).
Discussion
MDD is a relatively common, yet frequently
overlooked, co-morbidity of cancer. It is essential to study this
condition, since comorbidities often complicate the treatment of
cancer and may lead to poor clinical outcome. Although MDD occurs
in a considerable proportion of patients with malignant tumors, its
specific mechanism and pathophysiological basis are still unclear.
Previous studies have linked the pathogenesis of malignant cancer
associated MDD with psychological stress. Changes in the gene
regulatory network underlies the interaction of MDD and cancer
(5). However, few studies have
reported any role of the miRNA-mRNA regulatory network in MDD
accompanying cancer. Using bioinformatics analysis of the miRNA and
mRNA expression profiles, we constructed an miRNA-mRNA interaction
network with a putative role in the occurrence of MDD in ovarian
cancer patients.
We analyzed the miRNA expression profiles of ovarian
cancer patients with depression, and selected the differentially
expressed miRNAs (DEmiRs) with P<0.05. Target gene prediction
suggested multiple target gene regulation by a single miRNA, and
regulation of a single target gene by several miRNAs
simultaneously. Therefore, miRNA mediated regulatory networks are
very complex. The target genes predicted by the DEmiRs in our study
were involved in multiple biological processes. We hypothesized
therefore that the aberrant expression of miRNAs may play an
important role in the development of MDD in ovarian cancer patients
by regulating various target genes and signaling pathways. The
target genes that were enriched in the risk pathways were
considered as the risk genes. One of the risk pathways, cAMP
signaling, controls a diverse range of cellular processes, and
studies have shown the frequent dysregulation of this pathway in
patients with MDD (14). The MAPK
signaling pathways regulate the expression of various genes through
phosphorylation cascades. A constitutively active MAPK signaling
pathway with mutations in the individual component(s) have been
identified in several malignancies, including ovarian cancer
(15). ERK1/2 signaling pathway, one
of five MAPK signaling pathways, plays a key role in the regulation
of cell growth and differentiation. Furthermore, Dwivedi et
al (16) found significantly
lower activity of ERK 1/2 in the prefrontal cortex and hippocampus
of suicidal patients with MDD compared to healthy individuals. The
dopaminergic synapse pathway is involved in the regulation the
dopamine transporter (DAT), and the deficiency of dopamine (DA) has
been correlated with MDD (17). The
density of DAT in the striatum of MDD patients is significantly
higher than that in normal individuals. The excessive levels of DAT
at the synaptic terminals increase the recovery of DA, and thus
reduce the level of DA in the synaptic gap resulting in symptoms of
MDD. The other risk signaling pathways also play important roles in
tumor progression and depression. Previous studies have shown that
reduced cGMP levels caused by inhibition of guanylate cyclase and
phosphodiesterase activity in the cGMP-PKG signaling pathway can
help alleviate symptoms of depression (18). In addition, inactivation of the
cGMP-PKG signal may promote tumor cell proliferation and
angiogenesis. Many studies have shown low expression levels of PKG
in various tumors including gastric cancer (19), lung cancer (20) and breast cancer (21). Over-activation of the PI3K/AKT
signaling pathway suppresses apoptosis and has been linked with
ovarian cancer development, invasion and metastasis (22). In addition, the PI3K/AKT pathway
exerts a protective action on the central nervous system.
Based on survival analysis of the risk genes, we
selected the following core genes involved in the survival of
ovarian cancer patients: AKT3, CAB39, CDKN1B, F2R, FGF1, JMJD1C,
MAGI2, MEF2A, TJP1, APAF1 and BDNF. As presented in Table I, All the core genes of the regulatory
network were involved in the top 15 significant biological process.
The damage to neural plasticity and neural cell regeneration may be
the pathophysiological basis for MDD in ovarian cancer.
Neurotrophic factors play a very important role in the development
and maintenance of peripheral and central nervous system. BDNF, or
brain-derived neurotrophic factor, is involved in nerve
regeneration (23). BDNF levels are
significantly decreased in the hippocampus and prefrontal cortex of
depression patients (17). Bachis
(24) correlated increased expression
of BDNF with hippocampal neurogenesis, and decreased expression
with anxiety disorder. FGF1 (Fibroblast growth factor 1) is a core
member of the FGFs family which play important roles in cell
proliferation, angiogenesis, morphogenesis and regeneration, and is
closely related to tumor development (25). FGF1 also has a nutritive function in
the regeneration of central nervous system and impaired neuronal
repair (26). AKT3, a member of the
AKT subfamily of serine/threonine protein kinases, is involved in
several biological processes including cell proliferation,
differentiation, apoptosis, and tumorigenesis (27,28). It
plays an important role in ovarian tumorigenesis via regulation of
VEGF secretion and angiogenesis (29). Moreover, one study (30) showed that AKT3 is the most abundant
AKT paralog in the brain during neurogenesis, and protects nerve
cells and promotes neurogenesis. Membrane-associated guanylate
kinase inverted 2 (MAGI2) is a scaffold protein with multiple
domains, and functions in raising and anchoring cell signaling
proteins. It is highly expressed in brain tissues, and is involved
in the formation and maintenance of synapses in the central nervous
system of vertebrates, and in the occurrence and development of
nervous system diseases. In addition, it has been reported as a
potential tumor suppressor (31–33). The
GO analysis indicated another function of MAGI2 as a negative
regulator of AKT signaling (Fig. 2).
Due to its involvement in the nerve growth factor signaling
pathway, MAGI2 may play a role in the occurrence and development of
MDD. The interaction between the core genes and DEmiRs need to be
further experimentally validated.
 | Table I.Top 15 significant biological process
of risk genes. |
Table I.
Top 15 significant biological process
of risk genes.
| GOTERM_BP_DIRECT | Genes | P-value | Fold enrichment |
|---|
| Sensory perception
of sound | HOXA1, TJP1, USP53,
CDKN1B, MYO6, POU4F2, CACNA1D | 0.002 | 5.524 |
| Positive regulation
of transcription from RNA polymerase II promoter | CAMTA1, ZNF292,
MEF2A, HMGB2, MYO6, YY1, MET, CTCF, RORA, CSRNP3, NCOA6, LRP6,
POU4F2, FGF1, CLOCK KLF15, MEIS1, PPARGC1A, WBP2, HIF1A, | 0.002 | 2.140 |
| Apoptotic
process | MEF2A, PRKCE, TOX3,
PTEN, ZFP36L1, GHITM, CSRNP3, ATG5, BNIP2, MAP3K1, APAF1, FAS,
TNFAIP3, LTA | 0.003 | 2.591 |
| Neural tube
closure | COBL, KDM6A, LRP6,
APAF1, SDC4 | 0.006 | 6.815 |
| Signal
transduction | MRC1, FYB, MAGI2,
PPP2R5A, MET, CCL8, RASSF8, PRKCE, SDC4, HIF1A, STAC, PDE4B, RAP1A,
PPP2R5E, FAS, CSNK1G3, FGF1, CLOCK, PLAU, LTA, AKT3 | 0.007 | 1.898 |
| Positive regulation
of MAPK cascade | BNIP2, FAS, IL6R,
PRKCE, F2R | 0.007 | 6.478 |
| Transcription from
RNA polymerase II promoter | CAMTA1, MEF2A,
ZNF292, HIF1A, CSRNP3, POU4F2, CTCF, KLF15, MEIS1, CLOCK, NFX1 | 0.024 | 2.250 |
| Regulation of
transcription, DNA-templated | MEAF6, HMGB2,
MEF2A, ZKSCAN1, RORA, ZNF654, MEIS1, PPARGC1A, ZFP36L1, HOXA1,
ZFHX4, ZNF226, HIF1A, NR1D2, PNRC2, JMJD1C, TMPO, ZNF117, CLOCK,
ZNF257, ZNF493, ZNF267, KLF3 | 0.027 | 1.605 |
| Transcription,
DNA-templated | MEAF6, HMGB2,
MEF2A, CBX3, ZKSCAN1, RORA, ZNF654, HOXA1, ZNF226, NR1D2, TEAD4,
ASF1A, ZNF493, ZNF267, ZMYM2, YY1, HNF4G, TOX3, BRWD1, ZFHX4,
HIF1A, TRIM33, PNRC2, JMJD1C, ZNF117, CLOCK, ZNF257, KLF3 | 0.028 | 1.503 |
| Positive regulation
of cell migration | SEMA6D, RRAS2, F7,
FGF1, PLAU, F2R | 0.031 | 3.422 |
| Negative regulation
of neuron apoptotic process | BDNF, AMBRA1,
PPARGC1A, TOX3, F2R | 0.037 | 3.975 |
| Activation of
cysteine-type endopeptidase activity involved in apoptotic
process | CDKN1B, APAF1, FAS,
F2R | 0.044 | 5.058 |
| Negative regulation
of protein kinase B signaling | MAGI2, PTEN,
DLG1 | 0.048 | 8.509 |
| Nerve growth factor
signaling pathway | MAGI2, RAP1A | 0.073 | 26.238 |
| Intracellular
signal transduction | DGKA, FYB, STAC,
RGS6, SOCS6, CAB39, PRKCE, AKT3 | 0.089 | 2.083 |
In conclusion, we analyzed the miRNA expression
profiles of ovarian cancer patients with MDD, and eventually
constructed a miRNA-mRNA regulatory network through bioinformatics
analysis. The miRNA-mRNA regulatory network provides new insights
into the pathophysiological mechanisms of MDD in ovarian
cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by grants from
Chinese National Natural Science Foundation Grant (grant nos.
81671541, 81273202 and 31400773), Clinical Medicine Science &
Technology Project of Jiangsu Province of China (grant no.
BL2013024), Jiangsu Provincial key research and development program
(grant no. BE2016721).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
CW conceived the idea for the study and wrote the
manuscript. YZ, YL, XY, MY, YM and ZP collected the data and
analyzed the data. SQ, SX, JY, PY, BW and QS conceived the idea for
the study and revised the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Massic MJ: Prevalence of depression in
patients with cancer. J Natl Cancer Inst Monogr. 57–71. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bailey RK, Geyen DJ, Scott-Gurnell K,
Hipolito MM, Bailey TA and Beal JM: Understanding and treating
depression among cancer patients. Int J Gynecol Cancer. 15:203–208.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bodurka-Bevers D, Basen-engquist K,
Carmack CL, Fitzgerald MA, Wolf JK, de Moor C and Gershenson DM:
Depression, anxiety, and quality of life in patients with
epithelial ovarian cancer. Gynecol Oncol. 78:302–308. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Price MA, Butow PN, Costa DS, King MT,
Aldridge LJ, Fardell JE, DeFazio A and Webb PM: Australian Ovarian
Cancer Study Group; Australian Ovarian Cancer Study Group Quality
of Life Study Investigators: Prevalence and predictors of anxiety
and depression in women with invasive ovarian cancer and their
caregivers. Med J Aust. 193 5 Suppl:S52–S57. 2010.PubMed/NCBI
|
|
5
|
Yang Y, Cui Y, Sang K, Dong Y, Ni Z, Ma S
and Hu H: Ketamine blocks bursting in the lateral habenula to
rapidly relieve depression. Nature. 554:317–322. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cui Y, Yang Y, Ni Z, Dong Y, Cai G,
Foncelle A, Ma S, Sang K, Tang S, Li Y, et al: Astroglial Kir4.1 in
the lateral habenula drives neuronal bursts in depression. Nature.
554:323–327. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lovejoy NC and Matties M: Pharmacokinetics
and pharmacodynamics of mood-altering drugs in patients with
cancer. Cancer Nurs. 19:407–418. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dwivedi Y, Rizavi HS, Roberts RC, Conley
RC, Tamminga CA and Pandey GN: Reduced activation and expression of
ERK1/2 MAP kinase in the post-mortem brain of depressed suicide
subjects. J Neurochem. 77:916–928. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nature Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
12
|
da Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gyorffy B, Lánczky A and Szállási Z:
Implementing an online tool for genome-wide validation of
survival-associated biomarkers in ovarian-cancer using microarray
data of 1287 patients. Endocr Relat Cancer. 19:197–208. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Plattner F, Hayashi K, Hernández A,
Benavides DR, Tassin TC, Tan C, Day J, Fina MW, Yuen EY, Yan Z, et
al: The role of ventral striatal cAMP signaling in stress-induced
behaviors. Nat Neurosci. 18:1094–1100. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wong KK: Recent developments in
anti-cancer agents targeting the Ras/Raf/MEK/ERK pathway. Recent
Pat Anticancer Drug Discov. 4:28–35. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dwivedi Y, Rizavi HS, Roberts RC, Conley
RC, Tamminga CA and Pandey GN: Reduced activation and expression of
ERK1/2 MAP kinase in the post-mortem brain of depressed suicide
subjects. J Neurochem. 77:916–928. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Taliaz D, Stall N, Dar DE and Zangen A:
Knockdown of brain-derived neurotrophic factor in specific brain
sites precipitates behaviors associated with depression and reduces
neurogenesis. Mol Psychiatry. 15:80–92. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ding L, Zhang C, Masood A, Li J, Sun J,
Nadeem A, Zhang HT, O'Donnell JM and Xu Y: Protective effects of
phosphodiesterase 2 inhibitor on depression- and anxiety-like
behaviors: involvement of antioxidant and anti-apoptotic mechanism.
Behav Brain Res. 268:150–158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wu M, Chen Y, Jiang L, Li Y, Lan T, Wang Y
and Qian H: Type II cGMP-dependent protein kinase inhibits
epidermal growth factor-induced phosphatidylinositol-3-kinase/Akt
signal transduction in gastric cancer cell. Oncol Lett.
6:1723–1728. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tao Y, Gu YJ, Cao ZH, Bian XJ, Lan T, Sang
JR, Jiang L, Wang Y, Qian H and Chen YC: Endogenous cGMP-dependent
protein kinase reverses EGF-induced MAPK/ERK signal transduction
through phosphorylation of VASP at Ser239. Oncol Lett. 4:1104–1108.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Karami-Tehrani F, Fallahian F and Atri M:
Expression of cGMP-dependent protein kinase, PKGIα, PKGIβ, and
PKGII in malignant and benign breast tumors. Tumor Biol.
33:1927–1932. 2012. View Article : Google Scholar
|
|
22
|
Li H, Zeng J and Shen K: PI3K/AKT/mTOR
signaling pathway as a therapeutic target for ovarian cancer. Arch
Gynecol Obstet. 290:1067–1078. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Polyakova M, Stuke K, Schuemberg K,
Mueller K, Schoenknecht P and Schroeter ML: BDNF as a biomarker for
successful treatment of mood disorders: A systematic &
quantitative meta-analysis. J Affect Disord. 174:432–440. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bachis A, Mallei A, Cruz MI, Wellstein A
and Mocchetti I: Chronic antidepressant treatments increase basic
fibroblast growth factor and fibroblast growth factor-binding
protein in neurons. Neropharmacology. 55:1114–1120. 2008.
View Article : Google Scholar
|
|
25
|
Jiao J, Zhao X, Liang Y, Tang D and Pan C:
FGF1-FGFR1 axis promotes tongue squamous cell carcinoma (TSCC)
metastasis through epithelial-mesenchymal transition (EMT). Biochem
Biophys Res Commun. 466:327–332. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Renaud F, Desset S, Oliver L,
Gimenez-Gallego G, Van Obberghen E, Courtois Y and Laurent M: The
neurotrophic activity of fibroblast growth factor 1 (FGF1) depends
on endogenous FGF1 expression and is independent of the
mitogen-activated kinase cascade pathway. J Biol Chem.
271:2801–2811. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cohen MM Jr: The AKT genes and their roles
in various disorders. Am J Med Genet A. 161A:2931–2937. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Madhunapantula SV and Robertson GP:
Targeting protein kinase-b3 (akt3) signaling in melanoma. Expert
Opin Ther Targets. 21:273–290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yeganeh PN, Richardson C,
Bahrani-Mostafavi Z, Tait DL and Mostafavi MT: Dysregulation of
AKT3 along with a small panel of mRNAs stratifies high-grade serous
ovarian cancer from both normal epithelia and benign tumor tissue.
Genes Cancer. 8:784–798. 2017.PubMed/NCBI
|
|
30
|
Poduri A, Evrony GD, Cai X, Elhosary PC,
Beroukhim R, Lehtinen MK, Hills LB, Heinzen EL, Hill A, Hill RS, et
al: Somatic activation of AKT3 causes hemispheric developmental
brain malformations. Neuron. 74:41–48. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hirao K, Hata Y, Ide N, Takeuchi M, Irie
M, Yao I, Deguchi M, Toyoda A, Sudhof TC and Takai Y: A novel
multiple PDZ domain-containing molecule interacting with
N-methyl-D-aspartate receptors and neuronal cell adhesion proteins.
J Biol Chem. 273:21105–21110. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hirao K, Hata Y, Yao I, Deguchi M, Kawabe
H, Mizoguchi A and Takai Y: Three isoforms of synaptic scaffolding
molecule and their characterization. Multimerization between the
isoforms and their interaction with N-methyl-D-aspartate receptors
and SAP90/PSD-95-associated protein. J Bol Chem. 275:2966–2972.
2000. View Article : Google Scholar
|
|
33
|
Hu Y, Li Z, Guo L, Wang L, Zhang L, Cai X,
Zhao H and Zha X: MAGI-2 Inhibits cell migration and proliferation
via PTEN in human hepatocarcinoma cells. Arch Biochem Biophys.
467:1–9. 2007. View Article : Google Scholar : PubMed/NCBI
|