Introduction
Oral cancer is a common malignant tumor observed in
the neck and the head area (1,2). Oral
squamous cell carcinoma (OSCC) is among the most frequent
pathological types of oral cancer (3,4). Despite
recent breakthroughs in surgical, chemoradiotherapeutic and
biological treatments, as well as molecular targeting therapy, the
long-term therapeutic impact remains poor (5,6).
Additionally, the clinical signs of ~2/3 patients with OSCC in the
early stages are not distinguishable as the disease is typically
diagnosed in the advanced stages (7).
The early diagnosis and treatment of OSCC serve substantial roles
in the improvement of the prognosis and survival rate of the
disease (8). Accordingly, the
detection of new molecular markers is suspected to be vital in the
improvement of the prognosis of OSCC.
Myristoylated alanine-rich C kinase substrate
(MARCKS) is regarded as a renowned specific substrate of protein
kinase C, extensively observed among different human tissues
(9). MARCKS is known to participate
in cell motility, phagocytosis and membrane trafficking, as well as
mitogenesis (10). Notably, previous
studies have shed light on the likely involvement of MARCKS in the
tumorigenesis and progression of different malignancies, including
brain cancer (11), thyroid cancer
(12), colon cancer (13), melanoma (14), glioblastoma cancer (15) and cholangiocarcinoma (16). However, relatively few studies are
available regarding the role of MARCKS in the proliferation of
OSCC.
In the present study, the levels of MARCKS protein
and mRNA were investigated in the OSCC tissue using western
blotting and reverse transcription-quantitative polymerase chain
reaction (RT-qPCR). The present study also evaluated the
association between the expression of MARCKS and the prognosis of
patients with OSCC. Additionally, the potential molecular mechanism
of MARCKS promoting the progression of OSCC was also further
analyzed.
Materials and methods
Tissue specimens
In the present study, OSCC tissues were immersed in
4% formalin for 4 h at room temperature and then were embedded in
paraffin. Subsequently, the tissue specimens were cut into sections
with a thickness of 4 µm and obtained from 128 patients with OSCC
(78 males and 50 females; Age range: 24–76 years old; Median age,
41 years). The tumor tissues and the corresponding normal
peri-tumor samples were obtained from patients who had undergone
surgical resection at the First and Second Affiliated Hospitals of
Anhui Medical University (Hefei, China) between June 2009 and
December 2012. The peri-tumor samples were used as negative
controls in the present study. Written informed consent was
obtained from each patient for the use of their tissue specimens in
the present study. Additionally, the present study was approved by
the Anhui Medical University Ethical Review Board, according to the
Declaration of Helsinki. None of the patients had undergone any
type of preoperative treatment prior to the curative surgery.
Independent classification of the pathological stage and tumor
grade (including poorly-, moderately- and well-differentiated) was
performed by two pathologists in accordance with the American Joint
Committee on Cancer Tumor-Node-Metastasis (TNM) staging system.
Immunohistochemistry
The MARCKS antibody (catalog no., ab52616; Abcam,
Cambridge, MA, USA) was used to detect MARCKS expression. All
sections were gradually deparaffinized and rehydrated with xylene
(twice for 10 min each time) and ethanol (100%, twice, for 5 min
each; 95%, 5 min; 90%, 5 min; 90%, 5 min; 85%, 5 min; 75%, 5 min)
at room temperature. Antigen retrieval was performed by heating the
sections in 10 mM sodium urinary citrate at 95°C for ~30 min.
Additionally, 0.3% hydrogen peroxide was applied to block
endogenous peroxidase activity for 10 min at room temperature.
According to the manufacturer's protocol, the MARCKS antibody was
diluted at 1:200 and was added dropwise to the sections, which were
subsequently incubated overnight at 4°C. Subsequently, the
avidin-biotin-peroxidase complex (Shanghai Guge Biotechnology Co.,
Ltd, Shanghai, China) was used for staining. PBS or normal mouse
serum was applied as a negative control. All sections were
subsequently counterstained with hematoxylin for 30 sec at room
temperature. Next, images of each of the sections were captured.
MARCKS expression was scored using a semi-quantitatively
immunostaining score system that was subsequently used in the
evaluation of the intensity of staining and the percentage of
stained cells. Additionally, the percentage score of stained cells
represents the proportion of positive cells: 5, >75% of cells
stained; 4, 50–75%; 3, 10–50%; 2, 5–10%; 1, <5%. The staining
intensities of stained cells were scored as follows: 0, no
staining; 1, weak positive staining; 2, moderate positive staining;
and 3, strong positive staining. Subsequently, the overall score
was calculated by multiplying the percentage score by the intensity
score (0–15). The expression level of MARCKS (high or low) was
determined by the total score as follows: high expression, ≥7; and
low expression, <7. Independent investigations of all these
scores were performed by two researchers and the average score was
statistically analyzed. Subsequently, the OLYMPUS light microscope
(BX41; ×200 magnification) was used to capture immunohistochemical
images.
Cell culture and transfection
Two human oral squamous cell carcinoma cell lines,
Cal27 and HN12, were cultured in Dulbecco's modified Eagle medium,
containing 10% fetal bovine serum (FBS), 1% penicillin and
streptomycin, in a dampened atmosphere of 5% CO2 at
37°C. RNA interference short interfering RNAs (siRNAs) were
obtained from RuiBo Biology Co., Ltd. (Guangzhou, China). Two
siRNAs (siRNA1, 5′-CTACACTTGGGCTCCTTTT-3′; and siRNA2,
5′-GGUGCCCAGUUCUCCAAGAUU-3′) were used to reduce the expression of
MARCKS. However, OSCC Cal27 and HN12 cells transfected with siRNA1
exhibited higher knockdown efficiency than those transfected with
siRNA2 (data not shown). The empty control cells were transfected
with non-targeting MARCKS siRNA (5′-CGCACCAGAACAAACACAUU-3′). In
order to deliver the targeted siRNA, Cal27 and HN12 cells were
incubated at 30–50% confluency, followed by the addition of 20 nM
siRNAs supplemented with 5 µl RNAiMAX (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA).
RT-qPCR
Total RNA was extracted from the cell lines using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocol. The absorbance of RNA was measured at
260 nm using a NanoDrop spectrophotometer (ND-1000; Thermo Fisher
Scientific, Inc.), in order to determine the total RNA
concentration. Reverse transcription of 2 µg total RNA was
conducted using the Prime Script RT reagent kit, gDNA Eraser
(Takara Bio, Inc., Otsu, Japan). According to the manufacturer's
protocol, the ABI 7500 fast real-time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) and the SYBRGreen PCR
Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.)
were used with a first step at 95°C for 10 min, followed by 40
cycles with 95°C for 15 sec and 60°C for 1 min, with a fluorescent
reading at the end of this step to amplify the specific genes. The
primers were as follows: MARCKS forward, 5′-AGCCCGGTAGAGAAGGAGG-3′
and reverse, 5′-TTGGGCGAAGAAGTCGAGGA-3′; and GAPDH forward,
5′-CGTCCCGTAGACAAAATGGT-3′ and reverse, 5′-TTGATGGCAACAATCTCCAC-3′.
GAPDH was used as the interval control to calculate the relative
expression level of MARCKS using the comparative delta Cq
(2−ΔΔCq) method (17).
Furthermore, independent determination of each sample was performed
thrice, and the mean value of the expression levels was
calculated.
Transwell assay
Transwell cell migration assays were performed in
24-well plates with 8.0-µm permeable polycarbonate membranes.
Matrigel inserts were used (BD Biosciences, San Diego, CA, USA).
Cells were subsequently diluted with serum-free basic culture DMEM
(Gibco; Thermo Fisher Scientific, Inc.) at 5×105
cells/ml, followed by transferring 0.5–1 ml to each Transwell using
pipette guns. Additionally, culture medium with 10% FBS was
inserted into the lower chamber wells. Subsequently, the wells were
incubated at a temperature of 37°C in a moistened cell culture
incubator for 24 h. The non-invading cells on the upper side of the
membrane were removed by scrubbing. The invading cells were fixed
with 10% formalin for 10 min, followed by staining with 0.1%
crystal violet for 20 min at room temperature. The OLYMPUS light
microscope (BX41; ×200 magnification) was used to capture
images.
Cell colony formation assay
Cells in the logarithmic phase were diluted to
1–1.5×103 cells/ml by 10% FBS culture medium, followed
by drop by drop addition of 1 ml to each well of 6-well plates
supplemented with 1 ml culture medium. Subsequently, the cells were
incubated at a temperature of 37°C in a moistened cell culture
incubator for 2 weeks. The culture medium was replaced with new
medium. Following washing with PBS twice in 2 weeks, the cells were
fixed with absolute methanol for 15 mins, and staining with Giemsa
stock solution for 10 mins at room temperature.
Western blotting
Radioimmunoprecipitation assay (Beyotime Institute
of Biotechnology, Haimen, China) for the purpose of lysing the
accumulated cells and the protease inhibitor cocktail (Roche
Diagnostics, Basel, Switzerland) was added in order to prevent
denaturation. The cells were incubated on ice for 30 min. Next, the
samples were centrifuged at 12,000 × g at 4°C for ~30 min.
Subsequently, the total protein concentration was determined using
a bicinchoninic acid protein assay kit (Beyotime Institute of
Biotechnology). In order to break the protein structure, the
protein was heated at 100°C for 10 min. Subsequently, equal amounts
(40 µg) of protein were added to each well of 10% polyacrylamide
gels. Following electrophoresis, the protein was transferred to
nitrocellulose membranes, followed by blocking in 5% bovine serum
albumin or 5% skimmed milk diluted in Tris-buffered saline Tween
(TBST) for 2 h at room temperature. Next, the blocked membrane was
incubated with the following primary antibodies: MARCKS (dilution,
1:1,000; catalog no., 5607; Cell Signalling Technology, Inc.,
Danvers, MA, USA), phosphoinositide 3-kinase (PI3K; dilution,
1:1,000; catalog no., 4292; Cell Signalling Technology, Inc.),
protein kinase B (Akt; dilution, 1:1,000; catalog no., 9272; Cell
Signalling Technology, Inc.), phosphorylated PI3K (p-PI3K;
dilution, 1:1,000; catalog no., 13857; Cell Signalling Technology,
Inc.), p-Akt (dilution, 1:1,000; catalog no., 9611; Cell Signalling
Technology, Inc.) and β-actin (dilution, 1:1,000; catalog no.,
M20011; Abmart Co., Ltd., Shanghai, China) at 4°C overnight.
Subsequent to washing the membranes with TBST, the membranes were
incubated with a horseradish peroxidase-conjugated donkey anti-goat
secondary antibody (dilution, 1:5,000; catalog no., KC-RB-035;
Zhejiang Kangchen Biotech Co., Ltd., Wuhan, China) for ~60 min at
room temperature. Following washing of the membrane with TBST, the
membranes were visualized using an enhanced chemiluminescence kit
(Thermo Fisher Scientific, Inc.), followed by capturing the emitted
signals using KODAK X-OMAT BT Film (Kodak, Rochester, NY, USA). The
gray value of the protein was measured by employing ImageJ software
(National Institutes of Health, Bethesda, MD, USA).
Statistical analysis
SPSS statistical software (version 13.0; SPSS, Inc.,
Chicago, IL, USA) was used to analyze the immunohistochemical and
clinicopathological data. Data are presented as the mean ± standard
deviation of at least three independent experiments. Student's
t-tests were used to compare the levels of MARCKS mRNA in tumor and
corresponding peri-tumor samples. Additionally, χ2 was
used to compare immunohistochemistry results and
clinicopathological factors. The analysis of the survival curves
was performed using the Kaplan-Meier method. The long-rank test was
used for the determination of the discrepancy between different
groups. Furthermore, the Cox proportional hazards framework was
used to analyze the survival differences among various groups.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Abnormal expression of MARCKS in OSCC
tissues
In order to investigate the function of MARCKS in
the advancement of OSCC, the evaluation of the MARCKS mRNA and
protein expression levels was performed in the OSCC and the
corresponding normal tissues. The results of the present study
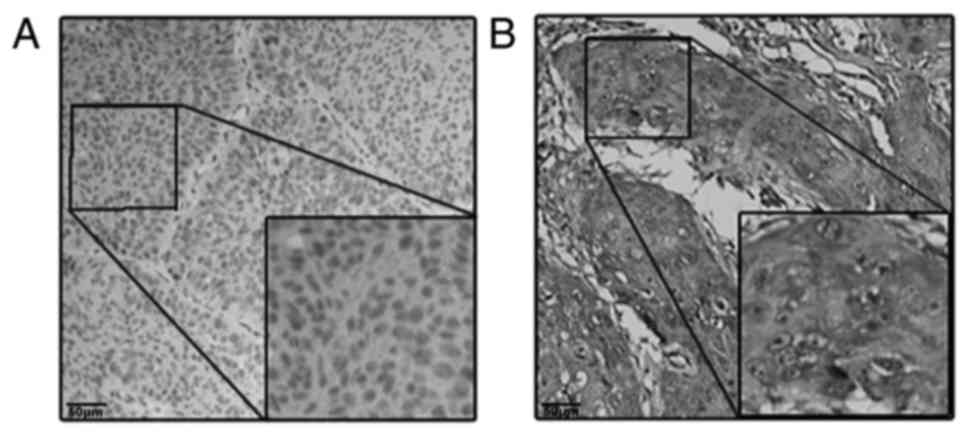
suggested that the higher protein expression level of MARCKS was
observed in the cytoplasm of tumor cells in 87 cases (67.9%) and in
the cytoplasm of matched peri-tumor normal cells in 21 cases
(16.4%), a difference that was statistically significant
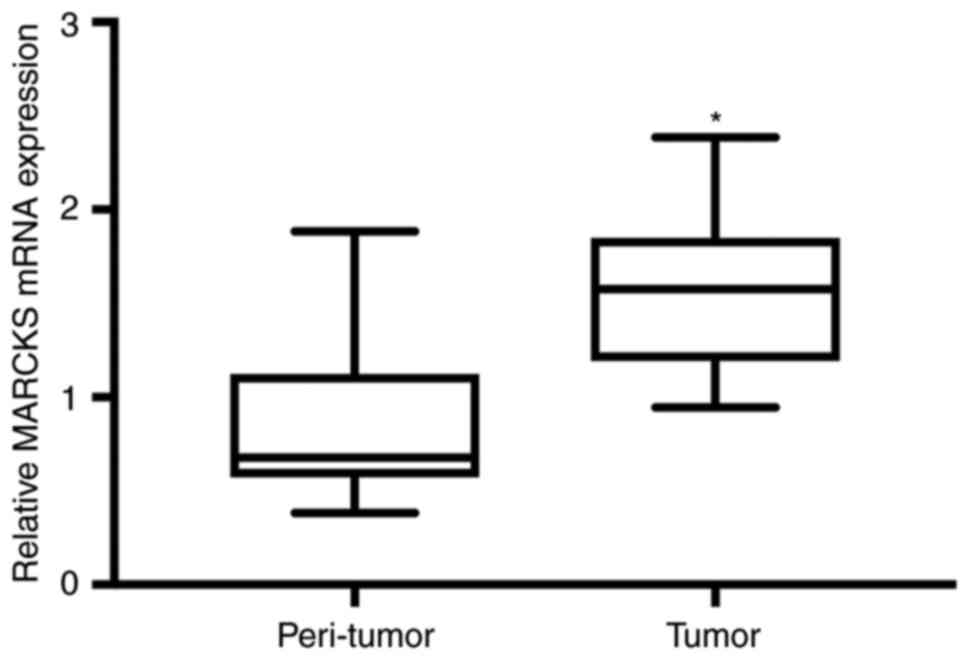
(P<0.001; Fig. 1). The same
results were observed in the assessment of MARCKS mRNA in 27
patients with OSCC using RT-qPCR (Fig.
2).
MARCKS is associated with lymphatic
metastasis and a poor prognosis of OSCC
Table I. shows that
χ2 test analysis revealed that the expression of MARCKS
was higher in the advanced-grade tumors (P<0.05), and that high
expression of MARCKS was correlated with lymphatic metastasis
(P<0.05). In summary, as suggested by the results of the present
study, MARCKS may have substantial impact on the metastasis or
invasion of OSCC.
 | Table I.MARCKS expression in relation to
clinical and pathological factors in 128 patients with OSCC. |
Table I.
MARCKS expression in relation to
clinical and pathological factors in 128 patients with OSCC.
|
| MARCKS
expression |
|
|
|---|
|
|
|
|
|
|---|
| Variables | Low (n=41) | High (n=87) | χ2 | P-value |
|---|
| Sex |
|
| 1.371 | 0.242 |
| Male | 28 | 50 |
|
|
|
Female | 13 | 37 |
|
|
| Age (years) |
|
| 1.053 | 0.305 |
| ≤60 | 24 | 59 |
|
|
|
>60 | 17 | 28 |
|
|
| Size (cm) |
|
| 0.012 | 0.913 |
| ≤4 | 31 | 65 |
|
|
|
>4 | 10 | 22 |
|
|
| Smoking and
drinking |
|
| 0.001 | 0.969 |
| None | 19 | 40 |
|
|
| Yes | 22 | 47 |
|
|
| Differentiation |
|
| 0.230 | 0.631 |
| Well | 28 | 63 |
|
|
|
Moderate and poor | 13 | 24 |
|
|
| T
classification |
|
| 2.074 | 0.150 |
|
T1+T2 | 29 | 50 |
|
|
|
T3+T4 | 12 | 37 |
|
|
| N
classification |
|
| 7.420 | 0.006a |
| N0 | 32 | 46 |
|
|
|
N1-3 | 9 | 41 |
|
|
| TNM stage |
|
| 4.243 | 0.039a |
|
I–II | 23 | 32 |
|
|
|
III–IV | 18 | 55 |
|
|
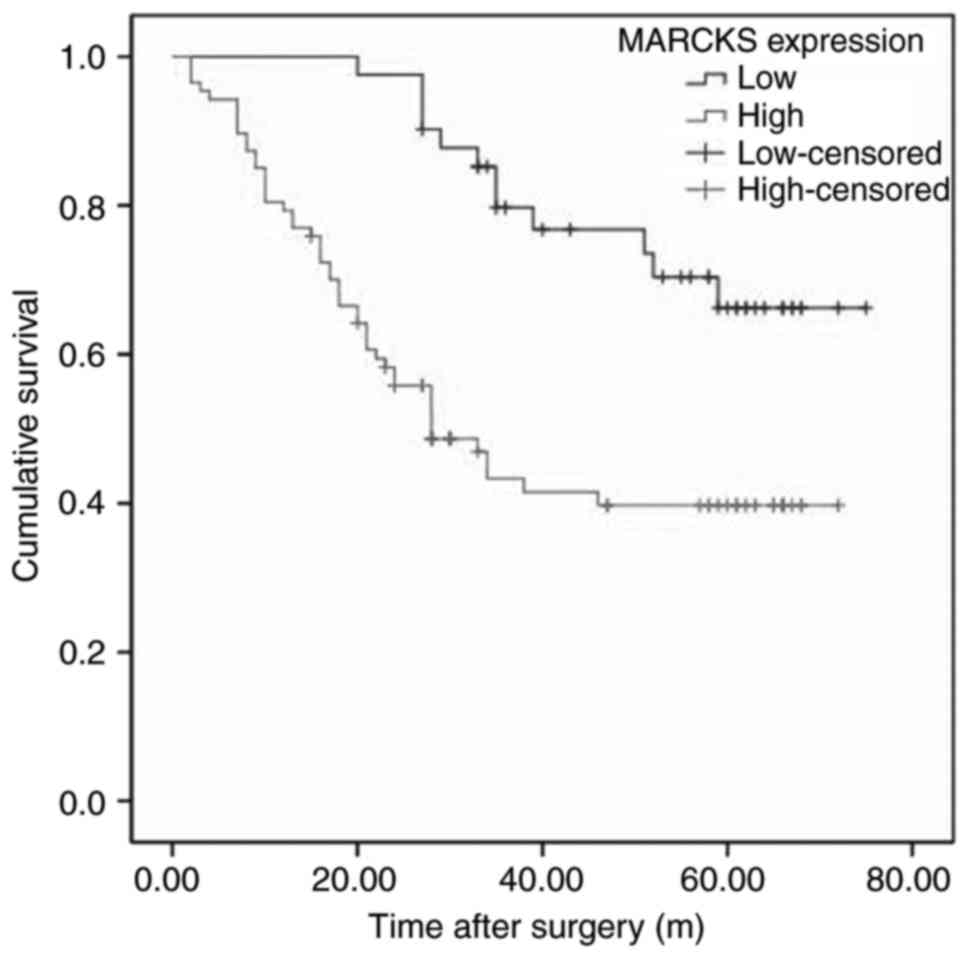
For further assessment of the use of MARCKS
expression in the prognosis of patients with OSCC, retrospective
analysis of the follow-up data of 128 cases was performed. Fig. 3 demonstrates that the OSCC patients
with higher MARCKS expression exhibited improved overall survival
(OS), as determined by Kaplan-Meier analysis (P<0.05). In
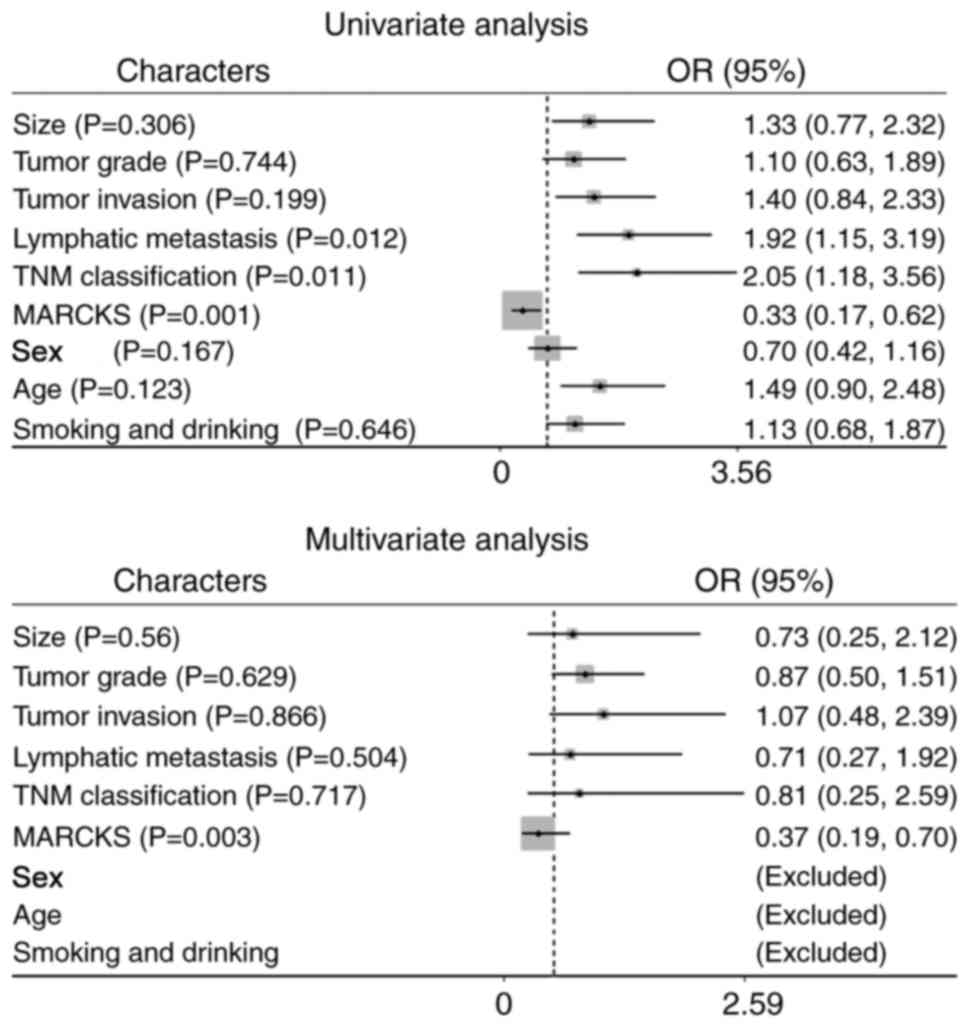
addition, univariate analysis demonstrated that the high MARCKS
expression level was an independent prognostic factor for a poor OS
(Fig. 4A, HR=0.33, 95% CI=0.17–0.62,
P=0.001), along with TNM stage and lymphatic metastasis (HR=2.05,
95% CI=1.18–3.56, P=0.011; HR=1.92, 95% CI=1.15–3.19, P=0.012,
respectively). Multivariate Cox regression analysis further
revealed the prognostic value of MARCKS (Fig. 4B, HR=0.37, 95% CI=0.19–0.70, P=0.003).
In brief, the results of the present study demonstrated that the
expression pattern of MARCKS in OSCC can be applied as an
independent risk factor for the prognosis prediction of the
disease.
Inhibiting the expression of MARCKS
decreases the mobility and proliferation of OSCC cells
In order to investigate the fundamental molecular
mechanisms of the MARCKS-mediated progression of OSCC, siRNA was
used to decrease the MARCKS expression in the OSCC Cal27 and HN12
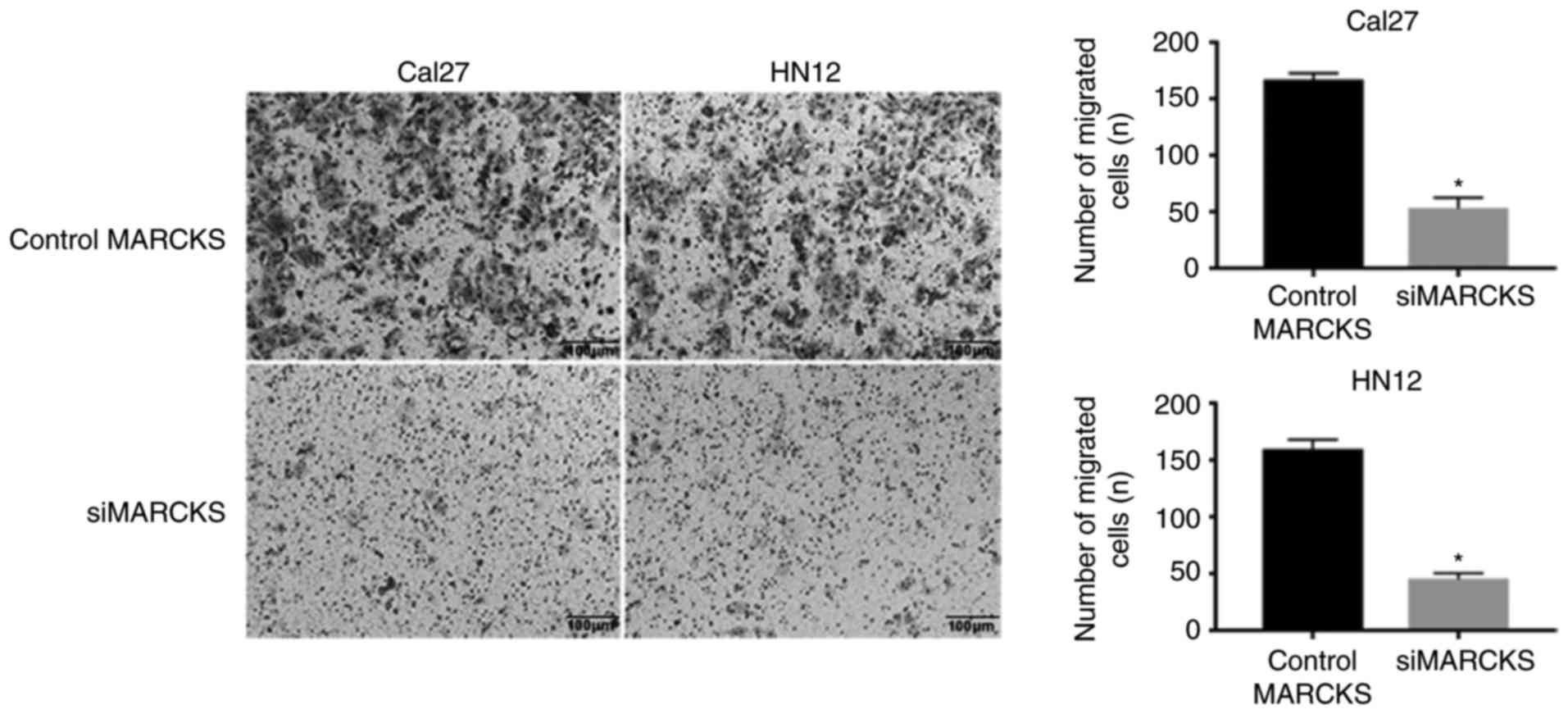
cell lines. Transwell migration and invasion assays suggested that
the downregulated expression level of MARCKS expressively inhibited
the tumor cell mobility compared with that of the control cells
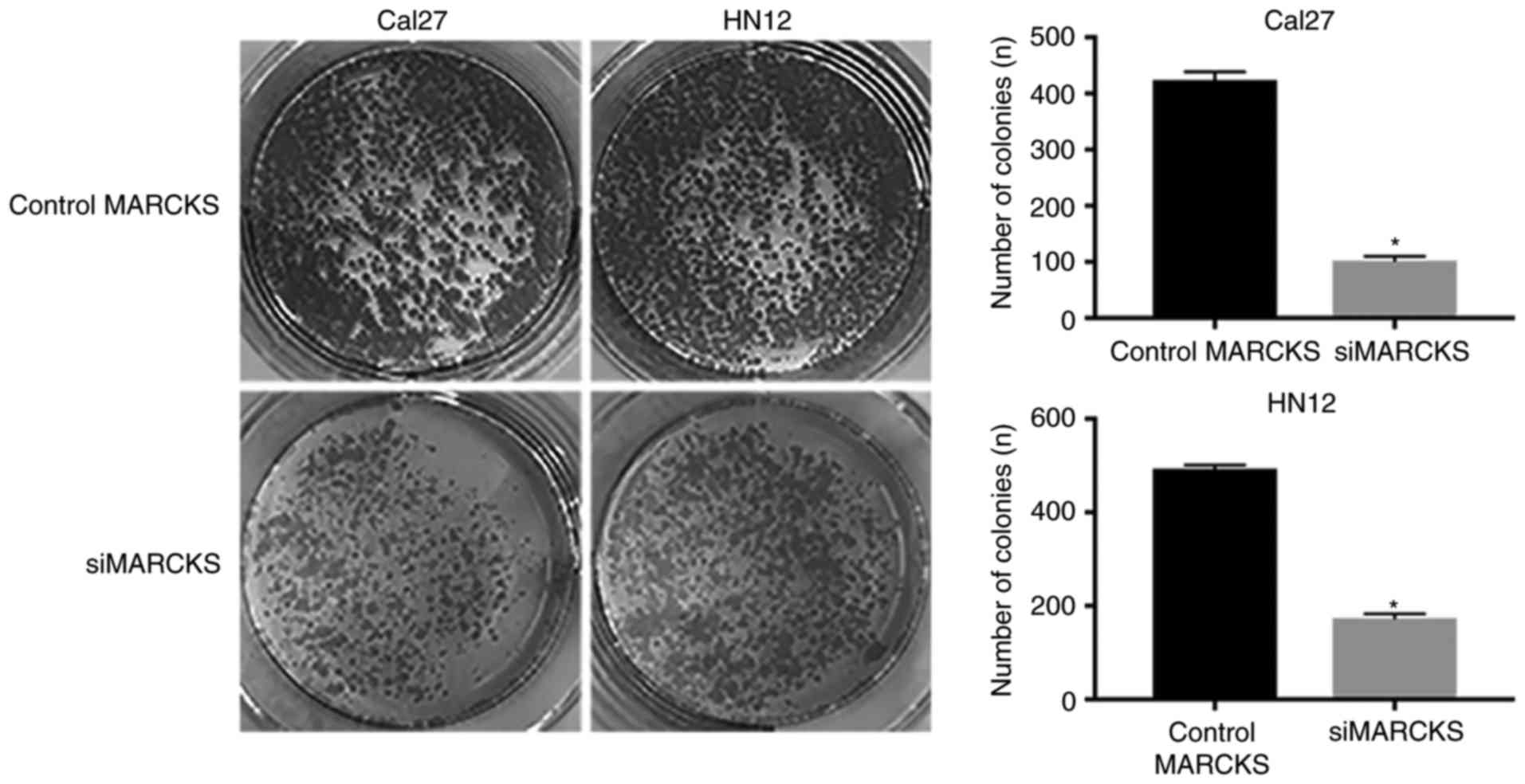
(Fig. 5). Furthermore, the colony
formation assay was employed to identify the impacts of MARCKS
expression on the OSCC cell proliferation. A notable difference was
observed in the tumor cell proliferation between the control group
and the MARCKS-knockdown group in the Cal27 and HN12 cells
(Fig. 6). Taken together, these data
indicated that MARCKS contributes toward the mobility and
proliferation of OSCC cells.
MARCKS may regulate the progression of
OSCC through the PI3K/Akt pathway
It has been demonstrated that MARCKS is involved in
regulating the PIP3 located upstream of the PI3K/Akt pathway.
Accordingly, we hypothesized that MARCKS regulates the progression
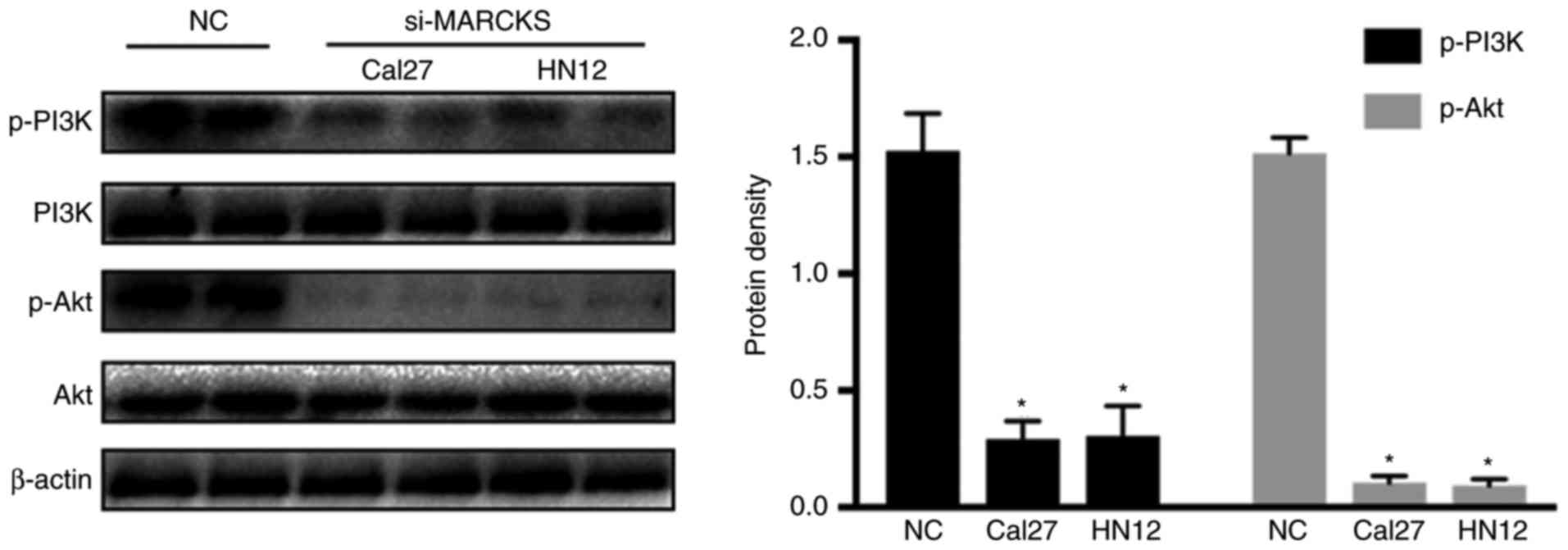
of OSCC through the PI3K/Akt pathway. Western blot analysis was
employed to identify the changes in p-PI3K and p-Akt in the OSCC
cell lines following interruption of MARCKS. Fig. 7 demonstrates that PI3K and Akt
phosphorylation were decreased while decreasing MARCKS expression
in the tumor cell lines. This result suggested that the PI3K/Akt
pathway was involved in the MARCKS-mediated progression of
OSCC.
Discussion
MARCKS is a protein kinase C substrate that is
implicated in the adhesion, secretion and motility of cells using
the regulation of the actin cytoskeletal structure (18). MARCKS protein was primarily identified
in the rat brain synaptosomes by Ueno et al (19). A previous study reported that MARCKS
serves a pivotal role in different malignant tumors (20). Previous studies revealed that abnormal
MARCKS expression is associated with the diagnosis and prognosis of
patients with cancer (21,22). Manai et al (21) reported MARCKS-overexpression in
inflammatory breast cancer, and that this was associated with a
poor metastasis-free survival. Hanada et al (22) also concluded that MARCKS could serve
as a potential biomarker for the human primary lung squamous cell
carcinoma using high-throughput screening methodology. Furthermore,
drug resistance of tumors could be regulated by MARCKS expression.
Chen et al (23) suggested
that knockdown of MARCKS decreased the IC50 of regorafenib in the
renal cancer cells. Taken together, these results suggested that
MARCKS participates in the malignant biological properties and may
serve as a promising therapeutic target.
The PI3K/Akt signalling pathway was involved in cell
proliferation, survival, motility and metabolism, tumorigenesis,
and tumor progression (24–26). Phosphatidylinositol-4,5-bisphosphate
(PIP2), together with phosphatidylinositol-4 and 5-bisphosphate
(PIP3), functions as a pivotal link in the PI3K/Akt signalling
pathway. A recent study demonstrated that MARCKS is persistently
hyperphosphorylated by either a PKC-dependent and/or a
PKC-independent manner, resulting in its cytosolic retention, as
well as the accessibility of PIP2 to PI3K (27). This further progress increases Akt
activation, giving rise to tumor cell proliferation and acquired
resistance. The present study developed the preliminary
investigation of the effects of MARCKS on OSCC progression.
The present study revealed high MARCKS expression in
OSCC tumor specimens for the purpose of being positively associated
with the advanced tumor stage and lymph node metastasis.
Additionally, an association was observed between high MARCKS
expression and a decreased overall survival rate in patients with
OSCC. Univariate and multivariate regression analyses supported the
value of MARCKS as an independent risk prognostic factor of OSCC.
Furthermore, the present study investigated the impact of MARCKS
expression on the metastasis and proliferation of OSCC cells
through the use of siRNA interruption methodology. A substantially
decreased growth rate in the Cal27-si-MARCKS and HN12-si-MARCKS
cells was determined by comparing it with that of the respective
control cells. In addition, the present study determined apparent
variations in tumor cell migration and infiltrations between the
MARCKS inhibiting and control cells. These results supported the
hypothesis that upregulated MARCKS enhances the malignant
phenotype. With regards to the role of MARCKS in cancer, the
available mechanistic studies have drawn attention to the Akt
pathway. In this way, the present study also determined the
associated between the protein expression level of MARCKS, and the
phosphorylation of PI3K and Akt using western blot analysis.
The results of the present study suggested that
MARCKS may be an independent prognosis factor for patients with
OSCC. MARCKS aids in improving tumor cell invasion and metastasis
via the PI3K/Akt pathway. However, more comprehensive
investigations are required for unravelling the mechanism through
which MARCKS regulates the tumorigenesis and progression of
OSCC.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Anhui
Technological Research for Public Welfare Project Funding (grant
no. 1704f0804023).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
CJ performed the majority of the study and was a
major contributor in writing the manuscript. CJ, RX, HW, YK and JZ
performed the experiment. MS, DD, WJ and FX collected and analyzed
the clinical data. CJ and JH made substantial contributions to the
design of the work, drafting the manuscript and revising it
critically for important intellectual content. All authors gave
final approval of the version to be published.
Ethics approval and consent to
participate
All human samples were collected with patients'
written informed consent and approval from the Institute Research
Medical Ethics Committee of Anhui Medical University (Hefei,
China).
Patient consent for publication
Written informed consent was obtained from all
subjects for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kademani D: Oral cancer. Mayo Clin Proc.
82:878–887. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ong TK, Murphy C, Smith AB, Kanatas AN and
Mitchell DA: Survival after surgery for oral cancer: A 30-year
experience. Br J Oral Maxillofac Surg. 55:911–916. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Montero PH and Patel SG: Cancer of the
oral cavity. Surg Oncol Clin N Am. 24:491–508. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saeed NR and Gold JA: Oral squamous cell
carcinoma. BMJ. 308:1372–1373. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang ZQ, Liu K, Huo ZJ, Li XC, Wang M, Liu
P, Pang B and Wang SJ: A cell-targeted chemotherapeutic
nanomedicine strategy for oral squamous cell carcinoma therapy. J
Nanobiotechnology. 13:632015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Xi S and Grandis JR: Gene therapy for the
treatment of oral squamous cell carcinoma. J Dent Res. 82:11–16.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lo WY, Tsai MH, Tsai Y, Hua CH, Tsai FJ,
Huang SY, Tsai CH and Lai CC: Identification of over-expressed
proteins in oral squamous cell carcinoma (OSCC) patients by
clinical proteomic analysis. Clin Chim Acta. 376:101–107. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zygogianni AG, Kyrgias G, Karakitsos P,
Psyrri A, Kouvaris J, Kelekis N and Kouloulias V: Oral squamous
cell cancer: Early detection and the role of alcohol and smoking.
Head Neck Oncol. 3:22011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Taniguchi H, Manenti S, Suzuki M and
Titani K: Myristoylated alanine-rich C kinase substrate (MARCKS), a
major protein kinase C substrate, is an in vivo substrate of
proline-directed protein kinase(s). A mass spectroscopic analysis
of the post-translational modifications. J Biol Chem.
269:18299–18302. 1994.PubMed/NCBI
|
|
10
|
Brooks G: The role of 80K/MARCKS, a
specific substrate of protein kinase C, in cell growth and tumour
progression. Pigment Cell Res. 7:451–457. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jarboe JS, Anderson JC, Duarte CW, Mehta
T, Nowsheen S, Hicks PH, Whitley AC, Rohrbach TD, McCubrey RO, Chiu
S, et al: MARCKS regulates growth and radiation sensitivity and is
a novel prognostic factor for glioma. Clin Cancer Res.
18:3030–3041. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ulbrich C, Pietsch J, Grosse J, Wehland M,
Schulz H, Saar K, Hübner N, Hauslage J, Hemmersbach R, Braun M, et
al: Differential gene regulation under altered gravity conditions
in follicular thyroid cancer cells: Relationship between the
extracellular matrix and the cytoskeleton. Cell Physiol Biochem.
28:185–198. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rombouts K, Carloni V, Mello T, Omenetti
S, Galastri S, Madiai S, Galli A and Pinzani M: Myristoylated
alanine-rich protein kinase C substrate (MARCKS) expression
modulates the metastatic phenotype in human and murine colon
carcinoma in vitro and in vivo. Cancer Lett. 333:244–252. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen X and Rotenberg SA: PhosphoMARCKS
drives motility of mouse melanoma cells. Cell Signal. 22:1097–1103.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Micallef J, Taccone M, Mukherjee J, Croul
S, Busby J, Moran MF and Guha A: Epidermal growth factor receptor
variant III-induced glioma invasion is mediated through
myristoylated alanine-rich protein kinase C substrate
overexpression. Cancer Res. 69:7548–7556. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Techasen A, Loilome W, Namwat N, Takahashi
E, Sugihara E, Puapairoj A, Miwa M, Saya H and Yongvanit P:
Myristoylated alanine-rich C kinase substrate phosphorylation
promotes cholangiocarcinoma cell migration and metastasis via the
protein kinase C-dependent pathway. Cancer Sci. 101:658–665. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Glaser M, Wanaski S, Buser CA, Boguslavsky
V, Rashidzada W, Morris A, Rebecchi M, Scarlata SF, Runnels LW,
Prestwich GD, et al: Myristoylated alanine-rich C kinase substrate
(MARCKS) produces reversible inhibition of phospholipase C by
sequestering phosphatidylinositol 4,5-bisphosphate in lateral
domains. J Biol Chem. 271:26187–22693. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ueno E and Rosenberg P: Beta-Bungarotoxin
blocks phorbol ester-stimulated phosphorylation of MARCKS, GAP-43
and synapsin I in rat brain synaptosomes. Toxicon. 33:747–762.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Woo JH, Wilsbach K, Nordin A, Lorincz A
and Gabrielson E: Increased expression of MARCKS in cancer cells
represents a potential target for treatment. Cancer Res.
64:2004.PubMed/NCBI
|
|
21
|
Manai M, Thomassin-Piana J, Gamoudi A,
Finetti P, Lopez M, Eghozzi R, Ayadi S, Lamine OB, Manai M, Rahal
K, et al: MARCKS protein overexpression in inflammatory breast
cancer. Oncotarget. 8:6246–6257. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hanada S, Kakehashi A, Nishiyama N, Wei M,
Yamano S, Chung K, Komatsu H, Inoue H, Suehiro S and Wanibuchi H:
Myristoylated alanine-rich C-kinase substrate as a prognostic
biomarker in human primary lung squamous cell carcinoma. Cancer
Biomark. 13:289–298. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen CH, Fong LWR, Yu E, Wu R, Trott JF
and Weiss RH: Upregulation of MARCKS in kidney cancer and its
potential as a therapeutic target. Oncogene. 36:3588–3598. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hers I, Vincent EE and Tavaré JM: Akt
signalling in health and disease. Cell Signal. 10:1515–1527. 2011.
View Article : Google Scholar
|
|
25
|
Fruman DA and Rommel C: PI3K and cancer:
Lessons, challenges and opportunities. Nat Rev Drug Discov.
13:140–156. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
26
|
Carnero A, Blanco-Aparicio C, Renner O,
Link W and Leal JF: The PTEN/PI3K/AKT signalling pathway in cancer,
therapeutic implications. Curr Cancer Drug Targets. 8:187–198.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ziemba BP, Burke JE, Masson G, Williams RL
and Falke JJ: Regulation of PI3K by PKC and MARCKS: Single-molecule
analysis of a reconstituted signaling pathway. Biophys J.
110:1811–1825. 2016. View Article : Google Scholar : PubMed/NCBI
|