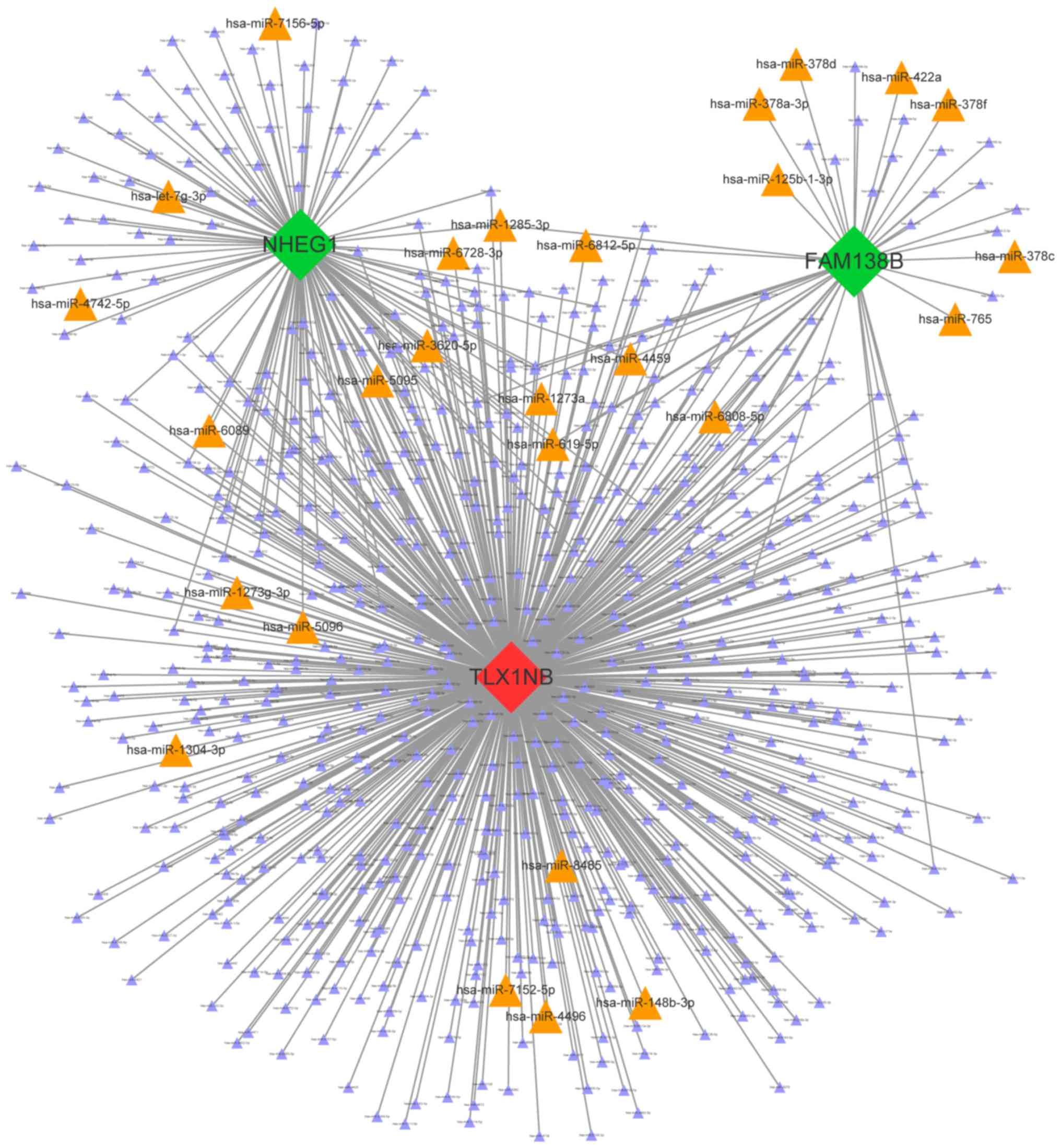
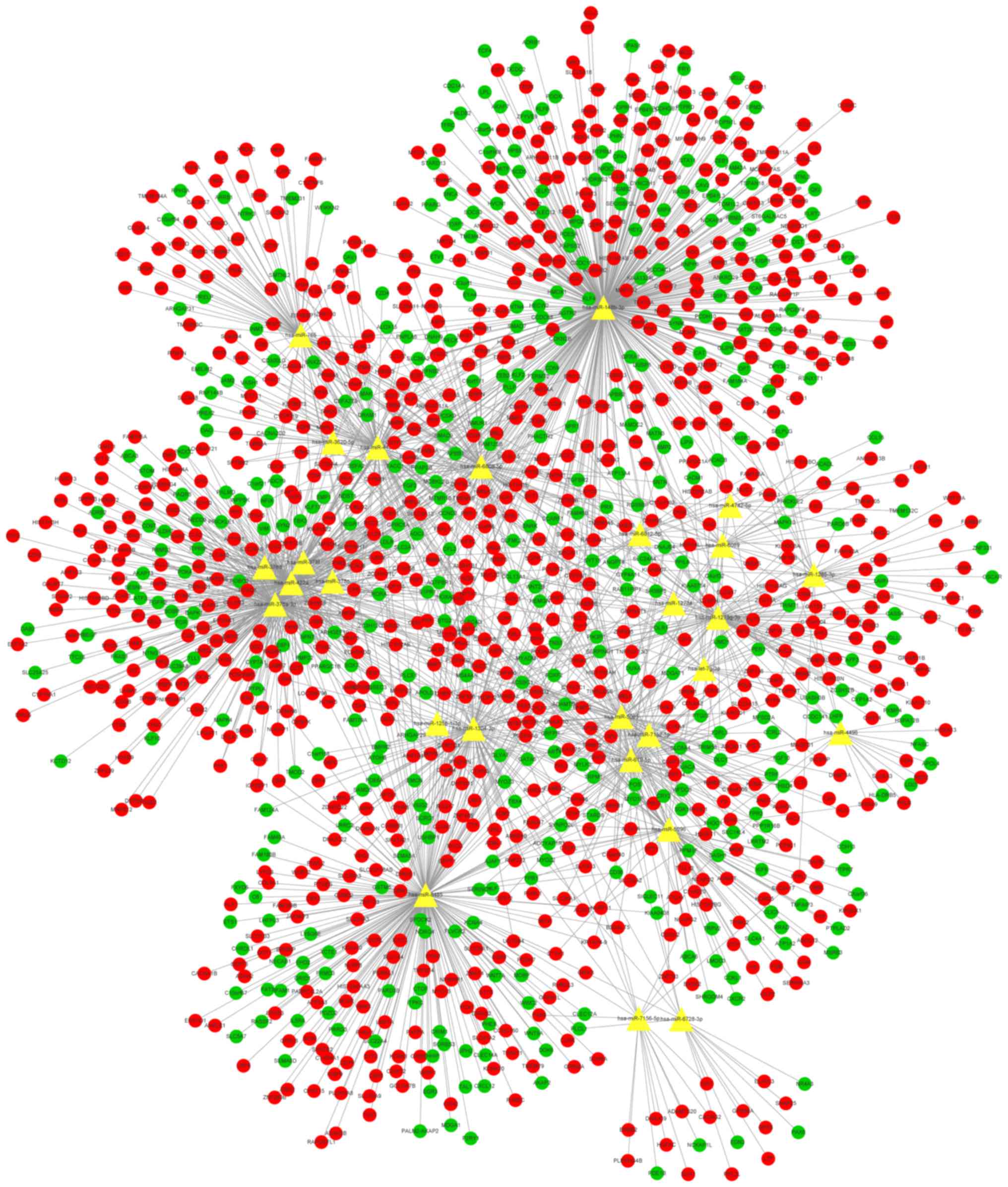
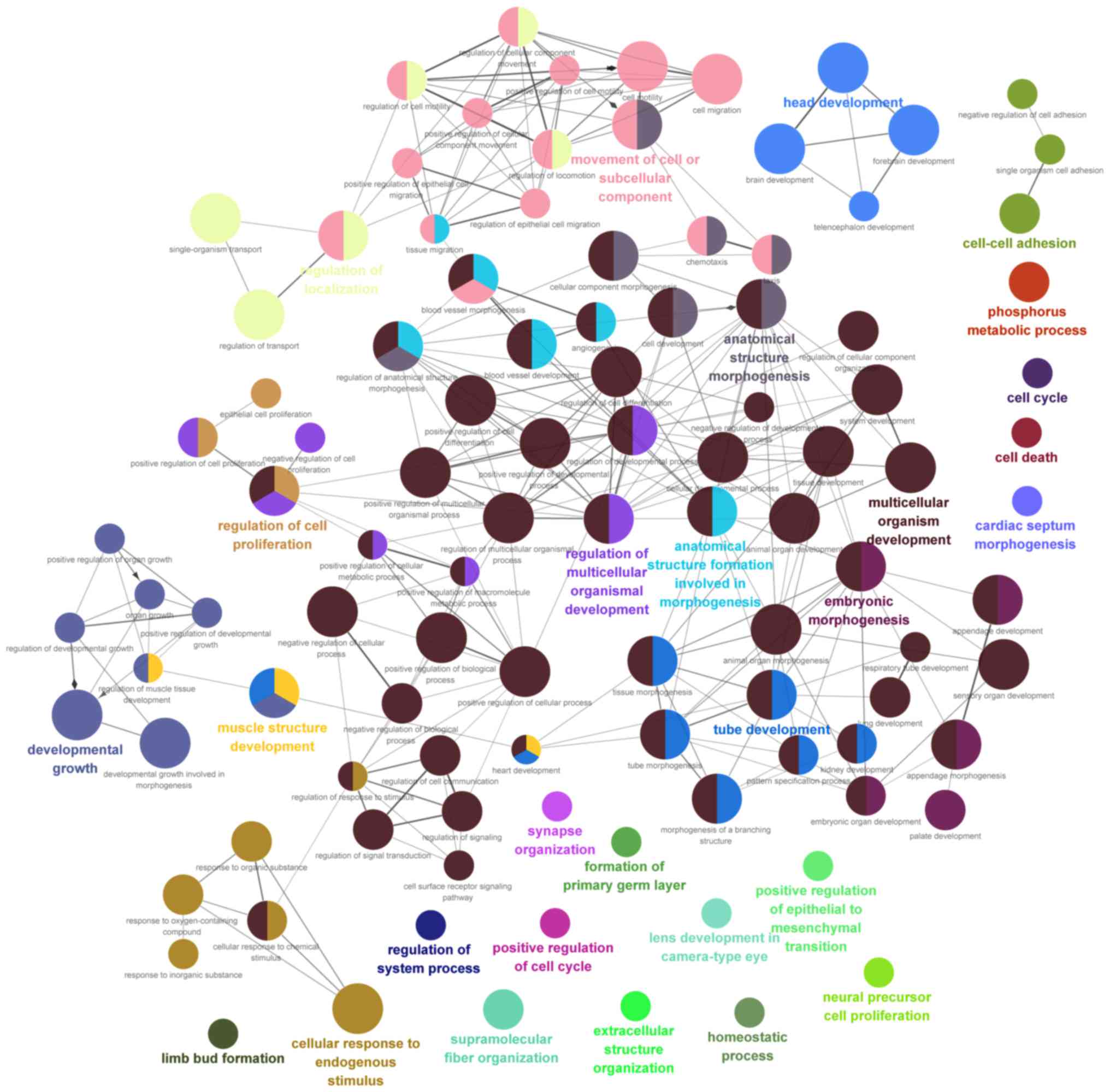
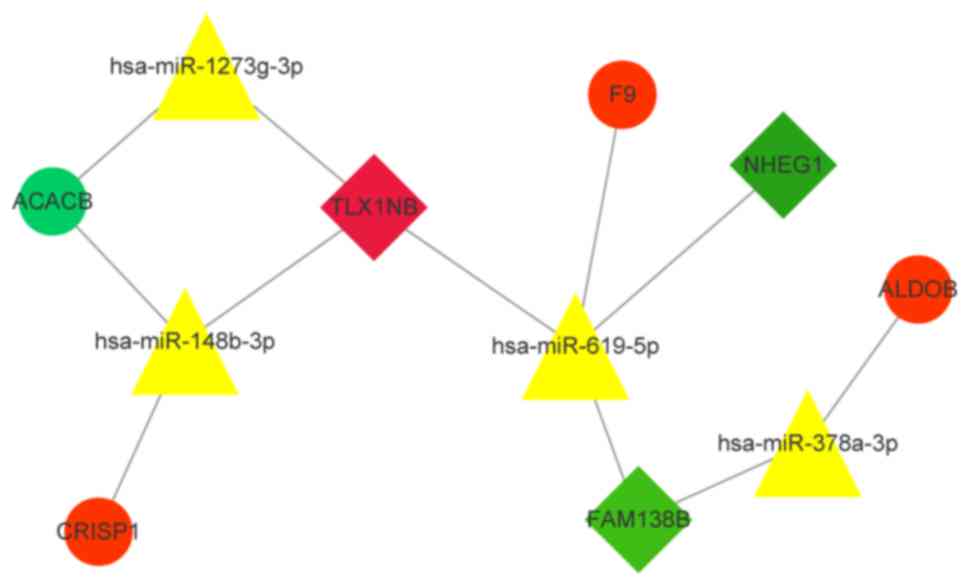
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lin JJ, Cardarella S, Lydon CA, Dahlberg
SE, Jackman DM, Jänne PA and Johnson BE: Five-Year survival in
EGFR-Mutant metastatic lung adenocarcinoma treated with EGFR-TKIs.
J Thorac Oncol. 11:556–565. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Qi L, Li Y, Qin Y, Shi G, Li T, Wang J,
Chen L, Gu Y, Zhao W and Guo Z: An individualised signature for
predicting response with concordant survival benefit for lung
adenocarcinoma patients receiving platinum-based chemotherapy. Br J
Cancer. 115:1513–1519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou M, Wang X, Shi H, Cheng L, Wang Z,
Zhao H, Yang L and Sun J: Characterization of long non-coding
RNA-associated ceRNA network to reveal potential prognostic lncRNA
biomarkers in human ovarian cancer. Oncotarget. 7:12598–12611.
2016.PubMed/NCBI
|
|
6
|
Wang PL, Liu B, Xia Y, Pan CF, Ma T and
Chen YJ: Long non-coding RNA-Low expression in tumor inhibits the
invasion and metastasis of esophageal squamous cell carcinoma by
regulating p53 expression. Mol Med Rep. 13:3074–3082. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shang C, Guo Y, Zhang J and Huang B:
Silence of long noncoding RNA UCA1 inhibits malignant proliferation
and chemotherapy resistance to adriamycin in gastric cancer. Cancer
Chemother Pharmacol. 77:1061–1067. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Y, Zhang Y, Yang T, Zhao W, Wang N,
Li P, Zeng X and Zhang W: Long non-coding RNA MALAT1 for promoting
metastasis and proliferation by acting as a ceRNA of miR-144-3p in
osteosarcoma cells. Oncotarget. 8:59417–59434. 2017.PubMed/NCBI
|
|
9
|
Wang P, Zhi H, Zhang Y, Liu Y, Zhang J,
Gao Y, Guo M, Ning S and Li X: miRSponge: A manually curated
database for experimentally supported miRNA sponges and ceRNAs.
Database (Oxford). 2015(pii): bav0982015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: the Rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tan JY, Sirey T, Honti F, Graham B,
Piovesan A, Merkenschlager M, Webber C, Ponting CP and Marques AC:
Extensive microRNA-mediated crosstalk between lncRNAs and mRNAs in
mouse embryonic stem cells. Genome Res. 25:655–666. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Karreth FA and Pandolfi PP: ceRNA
cross-talk in cancer: When ce-bling rivalries go awry. Cancer
Discov. 3:1113–1121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Akcakaya P, Ekelund S, Kolosenko I,
Caramuta S, Ozata DM, Xie H, Lindforss U, Olivecrona H and Lui WO:
miR-185 and miR-133b deregulation is associated with overall
survival and metastasis in colorectal cancer. Int J Oncol.
39:311–318. 2011.PubMed/NCBI
|
|
14
|
Zhang J, Fan D, Jian Z, Chen GG and Lai
PB: Cancer specific long noncoding RNAs show differential
expression patterns and competing endogenous RNA potential in
hepatocellular carcinoma. PLoS One. 10:e01410422015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Y, Chen Z, Li MJ, Guo HY and Jing
NC: Long non-coding RNA metastasis-associated lung adenocarcinoma
transcript 1 regulates the expression of Gli2 by miR-202 to
strengthen gastric cancer progression. Biomed Pharmacother.
85:264–271. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu T, Zhang X, Yang YM, Du LT and Wang
CX: Increased expression of the long noncoding RNA CRNDE-h
indicates a poor prognosis in colorectal cancer, and is positively
correlated with IRX5 mRNA expression. Onco Targets Ther.
9:1437–1448. 2016.PubMed/NCBI
|
|
17
|
Han D, Gao X, Wang M, Qiao Y, Xu Y, Yang
J, Dong N, He J, Sun Q, Lv G, et al: Long noncoding RNA H19
indicates a poor prognosis of colorectal cancer and promotes tumor
growth by recruiting and binding to eIF4A3. Oncotarget.
7:22159–22173. 2016.PubMed/NCBI
|
|
18
|
Qiu JJ and Yan JB: Long non-coding RNA
LINC01296 is a potential prognostic biomarker in patients with
colorectal cancer. Tumour Biol. 36:7175–7183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH
and Guo RH: Decreased expression of long noncoding RNA MEG3 affects
cell proliferation and predicts a poor prognosis in patients with
colorectal cancer. Tumour Biol. 36:4851–4859. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Zhang M, Liang L, Li J and Chen YX:
Over-expression of lncRNA DANCR is associated with advanced tumor
progression and poor prognosis in patients with colorectal cancer.
Int J Clin Exp Pathol. 8:11480–11484. 2015.PubMed/NCBI
|
|
21
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gene Ontology Consortium, . The gene
ontology: Enhancements for 2011. Nucleic Acids Res. 40(Database
Issue): D559–D564. 2012.PubMed/NCBI
|
|
23
|
Kanehisa M: The KEGG database. Novartis
Found Symp. 247:91–103. 101–103. 119–128. 244–152. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Foulger RE, Osumi-Sutherland D, McIntosh
BK, Hulo C, Masson P, Poux S, Le Mercier P and Lomax J:
Representing virus-host interactions and other multi-organism
processes in the Gene Ontology. BMC Microbiol. 15:1462015.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang RX, Chen WJ, He RQ, Zeng JH, Liang L,
Li SK, Ma J, Luo DZ and Chen G: Identification of a RNA-Seq based
prognostic signature with five lncRNAs for lung squamous cell
carcinoma. Oncotarget. 8:50761–50773. 2017.PubMed/NCBI
|
|
26
|
Zhou M, Sun Y, Sun Y, Xu W, Zhang Z, Zhao
H, Zhong Z and Sun J: Comprehensive analysis of lncRNA expression
profiles reveals a novel lncRNA signature to discriminate
nonequivalent outcomes in patients with ovarian cancer. Oncotarget.
7:32433–32448. 2016.PubMed/NCBI
|
|
27
|
Presson AP, Sobel EM, Papp JC, Suarez CJ,
Whistler T, Rajeevan MS, Vernon SD and Horvath S: Integrated
weighted gene co-expression network analysis with an application to
chronic fatigue syndrome. BMC Syst Biol. 2:952008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lossos IS, Czerwinski DK, Alizadeh AA,
Wechser MA, Tibshirani R, Botstein D and Levy R: Prediction of
survival in diffuse large-B-cell lymphoma based on the expression
of six genes. N Engl J Med. 350:1828–1837. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Alizadeh AA, Gentles AJ, Alencar AJ, Liu
CL, Kohrt HE, Houot R, Goldstein MJ, Zhao S, Natkunam Y, Advani RH,
et al: Prediction of survival in diffuse large B-cell lymphoma
based on the expression of 2 genes reflecting tumor and
microenvironment. Blood. 118:1350–1358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Meza R, Meernik C, Jeon J and Cote ML:
Lung cancer incidence trends by gender, race and histology in the
United States, 1973–2010. PLoS One. 10:e01213232015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kerr KM: Pulmonary adenocarcinomas:
Classification and reporting. Histopathology. 54:12–27. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aokage K, Yoshida J, Hishida T, Tsuboi M,
Saji H, Okada M, Suzuki K, Watanabe S and Asamura H: Limited
resection for early-stage non-small cell lung cancer as
function-preserving radical surgery: A review. Jpn J Clin Oncol.
47:7–11. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Provencio M, Isla D, Sanchez A and Cantos
B: Inoperable stage III non-small cell lung cancer: Current
treatment and role of vinorelbine. J Thorac Dis. 3:197–204.
2011.PubMed/NCBI
|
|
34
|
Ortea I, Rodriguez-Ariza A, Chicano-Galvez
E, Arenas Vacas MS and Jurado Gamez B: Discovery of potential
protein biomarkers of lung adenocarcinoma in bronchoalveolar lavage
fluid by SWATH MS data-independent acquisition and targeted data
extraction. J Proteomics. 138:106–114. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Navani N, Nankivell M, Lawrence DR, Lock
S, Makker H, Baldwin DR, Stephens RJ, Parmar MK, Spiro SG, Morris
S, et al: Lung cancer diagnosis and staging with endobronchial
ultrasound-guided transbronchial needle aspiration compared with
conventional approaches: An open-label, pragmatic, randomised
controlled trial. Lancet Respir Med. 3:282–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Y, Yang Y, Li L, Liu Y, Geng P, Li G
and Song H: LncRNA SNHG1 enhances cell proliferation, migration and
invasion in cervical cancer. Biochem Cell Biol. 96:38–43. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhao M, Sun D, Li X, Xu Y, Zhang H, Qin Y
and Xia M: Overexpression of long noncoding RNA PEG10 promotes
proliferation, invasion and metastasis of hypopharyngeal squamous
cell carcinoma. Oncol Lett. 14:2919–2925. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bao H, Guo CG, Qiu PC, Zhang XL, Dong Q
and Wang YK: Long non-coding RNA Igf2as controls hepatocellular
carcinoma progression through the ERK/MAPK signaling pathway. Oncol
Lett. 14:2831–2837. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lai Y, Xu P, Liu J, Li Q, Ren D, Zhang J
and Wang J: Decreased expression of the long non-coding RNA MLLT4
antisense RNA 1 is a potential biomarker and an indicator of a poor
prognosis for gastric cancer. Oncol Lett. 14:2629–2634. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Tian Z, Wen S, Zhang Y, Shi X, Zhu Y, Xu
Y, Lv H and Wang G: Identification of dysregulated long non-coding
RNAs/microRNAs/mRNAs in TNM I stage lung adenocarcinoma.
Oncotarget. 8:51703–51718. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen G, Peng L, Zhu Z, Du C, Shen Z, Zang
R, Su Y, Xia Y and Tang W: LncRNA AFAP1-AS functions as a competing
endogenous RNA to regulate RAP1B expression by sponging miR-181a in
the HSCR. Int J Med Sci. 14:1022–1030. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Tay Y, Rinn J and Pandolfi PP: The
multilayered complexity of ceRNA crosstalk and competition. Nature.
505:344–352. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li DS, Ainiwaer JL, Sheyhiding I, Zhang Z
and Zhang LW: Identification of key long non-coding RNAs as
competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma.
Eur Rev Med Pharmacol Sci. 20:2285–2295. 2016.PubMed/NCBI
|
|
44
|
Sui J, Li YH, Zhang YQ, Li CY, Shen X, Yao
WZ, Peng H, Hong WW, Yin LH, Pu YP and Liang GY: Integrated
analysis of long non-coding RNA-associated ceRNA network reveals
potential lncRNA biomarkers in human lung adenocarcinoma. Int J
Oncol. 49:2023–2036. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu B, Chen Y and Yang J: LncRNAs are
altered in lung squamous cell carcinoma and lung adenocarcinoma.
Oncotarget. 8:24275–24291. 2017.PubMed/NCBI
|
|
46
|
Zhang Y, Wen DY, Zhang R, Huang JC, Lin P,
Ren FH, Wang X, He Y, Yang H, Chen G and Luo DZ: A preliminary
investigation of PVT1 on the effect and mechanisms of
hepatocellular carcinoma: Evidence from clinical data, a
meta-analysis of 840 cases, and in vivo validation. Cell Physiol
Biochem. 47:2216–2232. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jiang R, Zhao C, Gao B, Xu J, Song W and
Shi P: Mixomics analysis of breast cancer: Long non-coding RNA
linc01561 acts as ceRNA involved in the progression of breast
cancer. Int J Biochem Cell Biol. 102:1–9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lu X, Huang C, He X, Liu X, Ji J, Zhang E,
Wang W and Guo R: A novel long Non-coding RNA, SOX21-AS1, indicates
a poor prognosis and promotes lung adenocarcinoma proliferation.
Cell Physiol Biochem. 42:1857–1869. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu X, Zang W, Cui S and Wang M:
Bioinformatics analysis of two microarray gene-expression data sets
to select lung adenocarcinoma marker genes. Eur Rev Med Pharmacol
Sci. 16:1582–1587. 2012.PubMed/NCBI
|
|
50
|
Heo JD, Oh JH, Lee K, Kim CY, Song CW,
Yoon S, Han JS and Yu IJ: Gene expression profiling in the lung
tissue of cynomolgus monkeys in response to repeated exposure to
welding fumes. Arch Toxicol. 84:191–203. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fukuyama R, Niculaita R, Ng KP, Obusez E,
Sanchez J, Kalady M, Aung PP, Casey G and Sizemore N: Mutated in
colorectal cancer, a putative tumor suppressor for serrated
colorectal cancer, selectively represses beta-catenin-dependent
transcription. Oncogene. 27:6044–6055. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Guerrero C, Rojas JM, Chedid M, Esteban
LM, Zimonjic DB, Popescu NC, Font de Mora J and Santos E:
Expression of alternative forms of Ras exchange factors GRF and
SOS1 in different human tissues and cell lines. Oncogene.
12:1097–1107. 1996.PubMed/NCBI
|
|
53
|
Langbein L, Heid HW, Moll I and Franke WW:
Molecular characterization of the body site-specific human
epidermal cytokeratin 9: cDNA cloning, amino acid sequence, and
tissue specificity of gene expression. Differentiation. 55:57–71.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lu JJ, Lu DZ, Chen YF, Dong YT, Zhang JR,
Li T, Tang ZH and Yang Z: Proteomic analysis of hepatocellular
carcinoma HepG2 cells treated with platycodin D. Chin J Nat Med.
13:673–679. 2015.PubMed/NCBI
|
|
55
|
Pan GQ, Ren HZ, Zhang SF, Wang XM and Wen
JF: Expression of semaphorin 5A and its receptor plexin B3
contributes to invasion and metastasis of gastric carcinoma. World
J Gastroenterol. 15:2800–2804. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Svensson RU, Parker SJ, Eichner LJ, Kolar
MJ, Wallace M, Brun SN, Lombardo PS, Van Nostrand JL, Hutchins A,
Vera L, et al: Inhibition of acetyl-CoA carboxylase suppresses
fatty acid synthesis and tumor growth of non-small-cell lung cancer
in preclinical models. Nat Med. 22:1108–1119. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li CY, Liang GY, Yao WZ, Sui J, Shen X,
Zhang YQ, Peng H, Hong WW, Ye YC, Zhang ZY, et al: Integrated
analysis of long non-coding RNA competing interactions reveals the
potential role in progression of human gastric cancer. Int J Oncol.
48:1965–1976. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Guo LL, Song CH, Wang P, Dai LP, Zhang JY
and Wang KJ: Competing endogenous RNA networks and gastric cancer.
World J Gastroenterol. 21:11680–11687. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hu Y, Tian H, Xu J and Fang JY: Roles of
competing endogenous RNAs in gastric cancer. Brief Funct Genomics.
15:266–273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Wu Q, Guo L, Jiang F, Li L, Li Z and Chen
F: Analysis of the miRNA-mRNA-lncRNA networks in ER+ and ER-breast
cancer cell lines. J Cell Mol Med. 19:2874–2887. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Song X, Cao G, Jing L, Lin S, Wang X,
Zhang J, Wang M, Liu W and Lv C: Analysing the relationship between
lncRNA and protein-coding gene and the role of lncRNA as ceRNA in
pulmonary fibrosis. J Cell Mol Med. 18:991–1003. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, De W, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Penhoet E, Rajkumar T and Rutter WJ:
Multiple forms of fructose diphosphate aldolase in mammalian
tissues. Proc Natl Acad Sci USA. 56:pp. 1275–1282. 1966; View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Asaka M, Kimura T, Meguro T, Kato M, Kudo
M, Miyazaki T and Alpert E: Alteration of aldolase isozymes in
serum and tissues of patients with cancer and other diseases. J
Clin Lab Anal. 8:144–148. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Tian YF, Hsieh PL, Lin CY, Sun DP, Sheu
MJ, Yang CC, Lin LC, He HL, Solórzano J, Li CF and Chang IW: High
expression of Aldolase B confers a poor prognosis for rectal cancer
patients receiving neoadjuvant chemoradiotherapy. J Cancer.
8:1197–1204. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wakil SJ and Abu-Elheiga LA: Fatty acid
metabolism: Target for metabolic syndrome. J Lipid Res. 50
Suppl:S138–S143. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Milgraum LZ, Witters LA, Pasternack GR and
Kuhajda FP: Enzymes of the fatty acid synthesis pathway are highly
expressed in in situ breast carcinoma. Clin Cancer Res.
3:2115–2120. 1997.PubMed/NCBI
|
|
68
|
Yahagi N, Shimano H, Hasegawa K, Ohashi K,
Matsuzaka T, Najima Y, Sekiya M, Tomita S, Okazaki H, Tamura Y, et
al: Co-ordinate activation of lipogenic enzymes in hepatocellular
carcinoma. Eur J Cancer. 41:1316–1322. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Swinnen JV, Brusselmans K and Verhoeven G:
Increased lipogenesis in cancer cells: New players, novel targets.
Curr Opin Clin Nutr Metab Care. 9:358–365. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Jones JE, Esler WP, Patel R, Lanba A, Vera
NB, Pfefferkorn JA and Vernochet C: Inhibition of Acetyl-CoA
carboxylase 1 (ACC1) and 2 (ACC2) reduces proliferation and De Novo
lipogenesis of EGFRvIII Human Glioblastoma cells. PLoS One.
12:e01695662017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Simioni P, Tormene D, Tognin G, Gavasso S,
Bulato C, Iacobelli NP, Finn JD, Spiezia L, Radu C and Arruda VR:
X-linked thrombophilia with a mutant factor IX (factor IX Padua). N
Engl J Med. 361:1671–1675. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Sarkar D and Fisher PB: AEG-1/MTDH/LYRIC:
Clinical significance. Adv Cancer Res. 120:39–74. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yoo BK, Emdad L, Su ZZ, Villanueva A,
Chiang DY, Mukhopadhyay ND, Mills AS, Waxman S, Fisher RA, Llovet
JM, et al: Astrocyte elevated gene-1 regulates hepatocellular
carcinoma development and progression. J Clin Invest. 119:465–477.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhu K, Dai Z, Pan Q, Wang Z, Yang GH, Yu
L, Ding ZB, Shi GM, Ke AW, Yang XR, et al: Metadherin promotes
hepatocellular carcinoma metastasis through induction of
epithelial-mesenchymal transition. Clin Cancer Res. 17:7294–7302.
2011. View Article : Google Scholar : PubMed/NCBI
|