Introduction
Oral squamous cell carcinoma (OSCC), one of the most
fatal types of cancer (1), is
identified with frequent lymph node and distant metastases
(2). Metastasis is a key prognostic
factor for OSCC and typically indicates a poor prognosis (3). However, the underlying molecular
mechanisms that regulate metastatic dissemination remain unclear,
despite a number of studies on the molecular mechanisms implicated
in OSCC development (4).
Forkhead box M1 (FOXM1), which belongs to the Fox
family of transcription factors, maintains a balance between
proliferation and apoptosis of cells (5). Mutation of FOXM1 prevents
differentiation, resulting in the malignant transformation of
undifferentiated cells (6).
Overexpression of FOXM1 is associated with the occurrence and
development of numerous cancer types (7). FOXM1 is an essential cell cycle
regulator in the G1/S and G2/M stage
transitions and mitosis (8), and thus
contributes to cell proliferation. Furthermore, FOXM1 is important
in tumor angiogenesis, epithelial-mesenchymal transition, invasion
and metastasis (9). Silencing of
FOXM1 inhibits the proliferation, invasion and migration of human
colorectal cancer (CRC) cells (10).
However, the precise functions and underlying molecular mechanisms
of FOXM1 in OSCC cells remain unclear.
In the present study, the expression level of FOXM1
was determined in OSCC cells, and the effect of FOXM1 gene
knockdown on the proliferation, migration and invasion of Tca8113
cells was investigated in vitro. The results of the present
study identified that the downregulation of FOXM1 suppressed the
activities of Tca8113 cells, and decreased the expression of
proteins associated with cell cycle and viability. These results
have suggested that FOXM1 is a therapeutic target for the treatment
of OSCC.
Materials and methods
Cell culture reagents
Human HaCaT, Tca8113 and SCC9 cells were purchased
from the American Type Culture Collection (Manassas, VA, USA). SCC9
cells were kept in a 1:1 mixture of Dulbecco's modified Eagle's
medium (DMEM) and Ham's F12 medium supplemented with 10% fetal
bovine serum (FBS), 1% penicillin/streptomycin solution (all from
Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA), and
400 ng/ml hydrocortisone (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany). HaCaT and Tca8113 cells were incubated at 37°C in an
atmosphere containing 5% CO2 with saturated humidity and
cultured as monolayers in RPMI-1640 medium (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS, 2 mM glutamine and 1%
penicillin/streptomycin. Trypsin-ethylenediaminetetra acetic acid
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to detach
cells.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from all cell lines was isolated using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and
reverse transcribed into cDNA using 5X PrimeScript RT Master mix
(Takara Bio, Inc., Otsu, Japan) at 37°C for 15 min and 85°C for 5
sec, according to the manufacturer's protocol. Gene expression
levels were determined using 2X SYBR Premix Ex Taq (Takara Bio,
Inc.) with a 7300 ABI RT-PCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.) for 40 cycles at 95°C for 30 sec, 95°C for
5 sec and 60°C for 31 sec. The relative mRNA levels were determined
using the 2−ΔΔcq method and normalized to the expression
of GAPDH (11). The sequence of the
primers were as follows: FOXM1, forward,
5′-ATACGTGGATTGAGGACCACT-3′ and reverse,
5′-TCCAATGTCAAGTAGCGGTTG-3′; GAPDH forward,
5′-ACTGCCACCCAGAAGACT-3′ and reverse,
5′-GCTCAGTGTAGCCCAGGAT-3′.
Design of FOXM1 RNA interference
(RNAi) sequence and construction of short hairpin
(sh)RNA-expressing lentiviral vectors
A total of three FOXM1-specific sequences were
selected using the online short interfering RNA tools (Invitrogen;
Thermo Fisher Scientific, Inc.; www.invitrogen.com/rnai) with the reference sequence
of FOXM1 (gene bank accession no. NM_021953). The target sequences
of FOXM1-1 (5′-TTGCAGGGTGGTCCGTGTAAA-3′), FOXM1-2
(5′-TTGCAGGGTGGTCCGTGTAAA-3′) and FOXM1-3
(5′-AGGACCACTTTCCCTACTTTA-3′) were homologous with those of
FOXM1-specific mRNA. The transfection of siRNA oligonucleotides was
performed with Lipofectamine2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocols.
Briefly, 16 µl of Lipofectamine2000 reagent was mixed with 400 µl
of Opti-MEM (Invitrogen; Thermo Fisher Scientific, Inc.) at room
temperature for 5 min and subsequently incubated with a mixture of
12 µl of 20 µM siRNA duplex and 400 µl of Opti-MEM for an
additional 20 min at room temperature. The complexes were then
applied to cultured cells at ~70% confluency on a 60-mm plate
containing 4 ml of RPMI-1640 medium. After 12 h incubation, the
medium was replaced with fresh RPMI-1640 medium supplemented with
10% fetal bovine serum and 1% penicillin/streptomycin. The chemical
synthesis of shRNA lentivirus vector construction was performed
according to a previous study (12).
The lentiviral shRNA was constructed harboring green fluorescent
protein. The invalid RNAi sequence (5′-GAGCTATGGCAGCTACCATCA-3′)
was used as a negative control. The appropriate insertion of the
specific shRNA was validated by sequencing.
Cell extracts and western blot
analysis
Protein lysates were prepared on ice using a
radioimmunoprecipitation assay buffer [150 mM NaCl, 50 mM Tris-HCl
(pH 8.0), 1% NP40, 0.1% SDS and 0.5% sodium deoxycholate]
supplemented with 1 mg/ml aprotinin, 1 mM sodium orthovanadate and
0.1 mg/ml phenylmethylsulfonyl fluoride. Protein contents were
determined using a bicinchoninic acid protein assay system (Bio-Rad
Laboratories Inc., Hercules, CA, USA). Equal amounts of cell
extracts containing between 20 and 50 µg of total protein were
separated using 12% SDS-PAGE and transferred to 0.45-µm
nitrocellulose membranes (Osmonics, Westborough, MA, USA). The
membranes were blocked for 1 h in Blotto A (Beyotime Institute of
Biotechnology, Shanghai, China) at room temperature which consisted
of 5% non-fat milk powder in Tris-buffered saline and 0.05%
Tween-20 (TBS-T), containing 10 mM Tris-HCl (pH 8.0) and 150 mM
NaCl. Subsequently, membranes were incubated for 1 h at room
temperature in Blotto A containing a 1:1,000 dilution of the
following rabbit primary antibodies: Anti-FOXM1 (cat. no. 5436;
Cell Signaling Technology, Danvers, MA, USA), anti-GAPDH (cat. no.
ab8245; Abcam, Cambridge, MA, USA), anti-cyclin B1 (cat. no.
12231), anti-cyclin D1 (cat. no. 2926), anti-P27 (cat. no. 3686),
anti-P21 (cat. no. 2947) and anti-matrix metalloproteinase (MMP) 2
(cat. no. 40994) (all from Cell Signaling Technology Inc.).
Membranes were washed in TBS-T for 5 min and incubated for 1 h at
room temperature in Blotto A containing a 1:10,000 dilution of
peroxidase-conjugated anti-rabbit (cat. no. RPN4301) or anti-mouse
secondary antibody (cat. no. RPN4201) (GE Healthcare, Chicago, IL,
USA). Following washing in TBS-T, enhanced chemiluminescence was
performed according to the manufacturer's protocol.
MTT assay
The cytotoxic activity of LV-shFOXM1 was determined
on the basis of cytotoxicity to Tca8113 cells using an MTT assay.
Cells were seeded in 96-well plates (5×103 cells/well)
and treated with LV-shCON (control) or LV-shFOXM1. The following
day, 100 µl fresh RPMI-1640 medium was added to each well with 0.5
mg/ml MTT (Roche Applied Science, Manheim, Germany). After 4 h of
incubation at 37°C in a humidified atmosphere containing 5%
CO2, 150 µl solubilization solution (0.01 mol/l HCl in
100 g/l SDS) was added. Subsequently, cells were gently agitated
for 10 min at 37°C. The absorbance was determined at 450 nm on
microplate reader (model 550; Bio-Rad Laboratories, Inc.) as the
optical density of the plates. Cell viability was evaluated for
four consecutive days. Each MTT assay was performed in
triplicate.
Cell cycle analysis via flow
cytometry
Tca8113 cells, at the logarithmic phase, were seeded
in 6-well plates (6×105 cells/well) and incubated at
37°C in humid conditions containing 5% CO2 (v/v) for 24
h, followed by 24 h of treatment with LV-shFOXM1 or LV-shCON in
RPMI-1640. Subsequently, the cells were washed with ice-cold PBS
twice and fixed in ice-cold 70% (v/v) ethanol at −20°C overnight.
Following treatment with 10 mM Tris-HCl buffer (pH 7.5) with 1%
(w/v) RNase A (Sigma-Aldrich; Merck KGaA) for 15 min at 4°C the
cells were incubated with propidium iodide (Sigma-Aldrich; Merck
KGaA) for 15 min at 4°C. The cell cycle was analyzed using a flow
cytometer (BD Biosciences, Franklin Lakes, NJ, USA), and the data
were analyzed using ModFit LT 2.0 (Verity Software House, Topsham,
ME, USA).
Soft agar assays
The aim of this assay was to analyze the effect of
FOXM1-shRNA on the proliferation of Tca8113 cells. Briefly, cells
were subjected to soft agar assay in 6-well plates and 50,000
cells/well were added into each well, which consisted of a bottom
base layer (0.6% agar diluted in DMEM with 10% FBS) and top layer
(0.3% agarose diluted in DMEM with 10% FBS). The cells were
cultured at 37°C for 2 weeks. Colonies were scored following 2
weeks of cell incubation. Each protocol was repeated at least three
times.
Cell migration and invasion
assays
The assays were conducted using Transwell chambers
equipped with a pore size of 8 µm, according to the manufacturer's
protocol. After 24 h of infection, Tca8113 cells were resuspended
in serum-free DMEM (2×104 cells/well) and seeded into
the Transwell inserts either uncoated (migration assay) or coated
with growth factor-reduced Matrigel (BD Biosciences) (invasion
assay). The lower chambers were filled with 500 µl DMEM containing
10% FBS. After 24 h of incubation at 37°C, all cells on the upper
side of the insert filter were selected using a cotton swab, while
the invaded cells were 100% methanol-fixed for 20 min at room
temperature and stained with 0.1% crystal violet for 10 min at room
temperature. The manual cell counting was finished under an
inverted microscope (Olympus IX51; Olympus Corporation, Tokyo,
Japan) on five random fields (magnification, 40×; scale bar, 200
µm). Each protocol was performed in triplicate.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Significant differences between groups were examined using the
Student's t-test and the χ2 test. P<0.05 was
considered to indicate a statistically significant difference. All
statistical tests were conducted using SPSS software (version 17.0;
SPSS, Inc., Chicago, IL, USA).
Results
FOXM1 mRNA is expressed at an
increased level in human OSCC
FOXM1 overexpression has been identified in human
head and neck squamous cell cancer, ovarian cancer and hepatoma
(13), but has rarely been
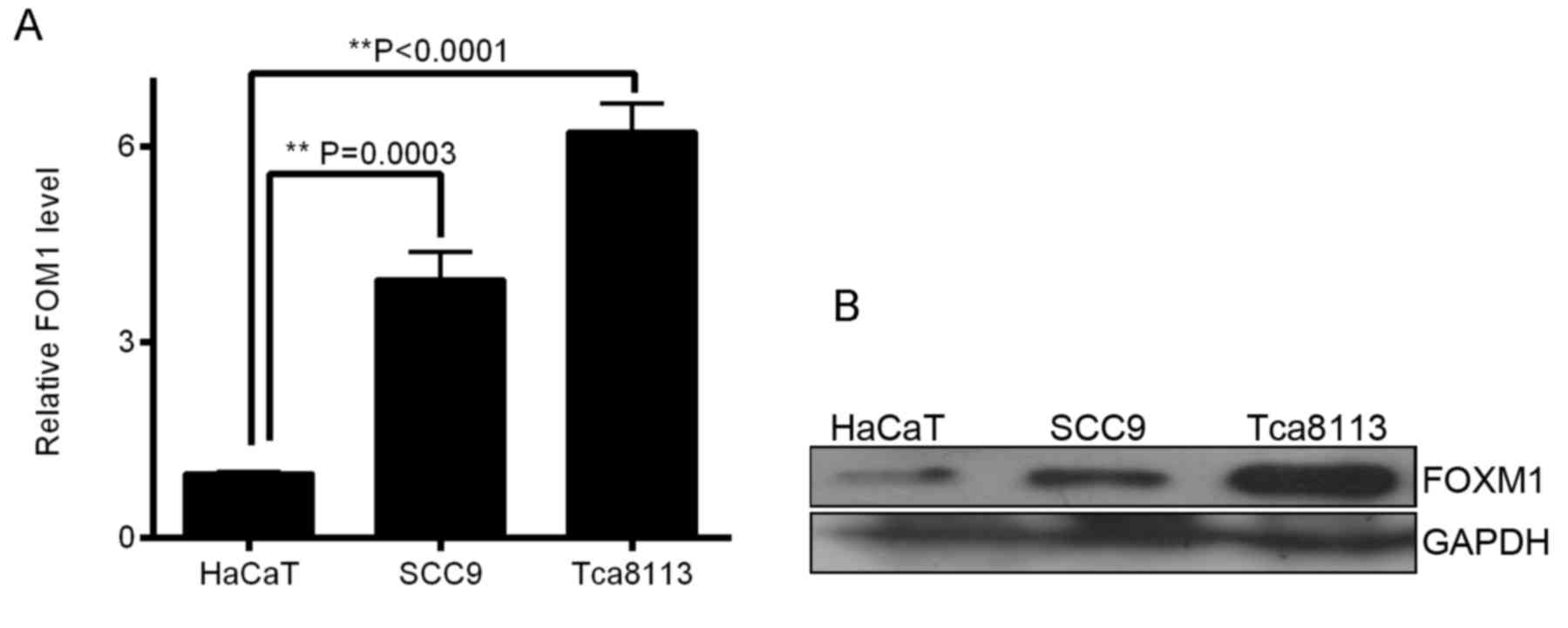
demonstrated in OSCC cells. In the present study, RT-qPCR revealed
a significant overexpression of FOXM1 mRNA in OSCC cells compared
with HaCaT cells (P<0.01; Fig.
1A). To validate these results, western blot analysis was
conducted to determine FOXM1 protein expression. The results
demonstrated that FOXM1 protein was markedly overexpressed in OSCC
cells compared with HaCaT cells (Fig.
1B). In the present study, RT-qPCR and western blot analysis
validated the expression of FOXM1 in OSCC cell lines compared with
human immortal keratinocyte cell line HaCaT (Fig. 1A and B), and indicated that Tca8113
was an appropriate cell line for RNAi targeting FOXM1 mRNA.
LV-shFOXM1-2 is the optimal
vector
Tca8113 cells were infected with four plasmids
separately for 48 h, including shFOXM1 (LV-shFOXM1-1, −2 and −3)
and LV-shCON. Subsequently, the expression of green fluorescent
protein in Tca8113 cells was observed under a fluorescent
microscope (Fig. 2A). RT-qPCR
revealed that LV-shFOXM1-2 significantly inhibited the expression
of FOXM1 mRNA compared with the shCON and shFOXM1 groups (Fig. 2B; P<0.0001). Western blot analysis
demonstrated that LV-shFOXM1-2 was the optimal vector and therefore
was selected for use in subsequent protocols (Fig. 2C), and termed LV-shFOXM1.
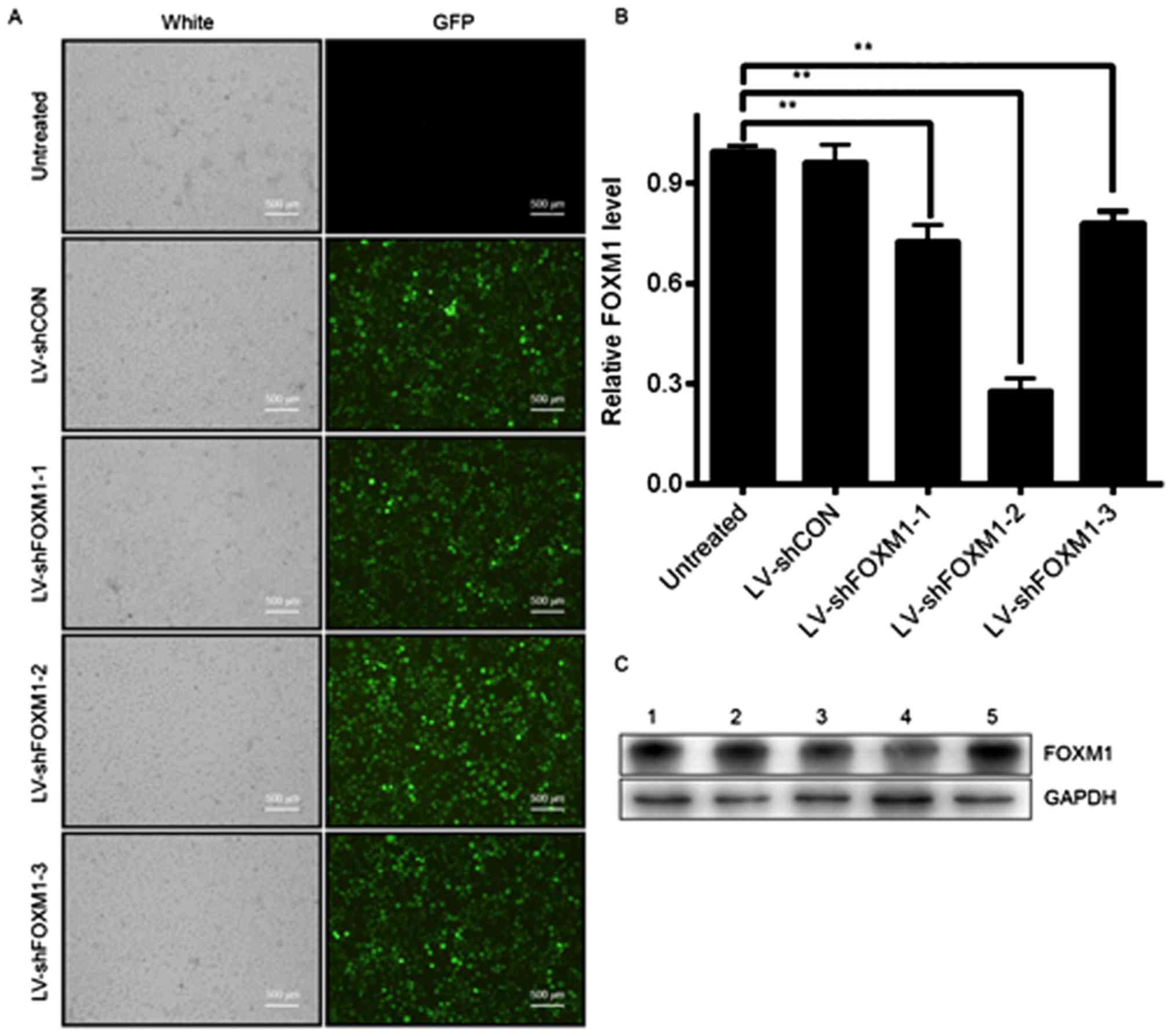 | Figure 2.Selection of the optimal LV-shFOXM1 in
Tca8113 cells. (A) GFP expression in Tca8113 cells after 48 h of
infection, determined under a fluorescence microscope. (B) mRNA
level of FOXM1 following lentiviral infection, determined using
reverse transcription-quantitative polymerase chain reaction. (C)
Protein level of FOXM1 determined using western blot analysis.
GADPH was used as the loading control. Lane 1, Tca8113 cells were
not infected; lane 2, Tca8113 cells were infected with LV-shCON;
lane 3, Tca8113 cells were infected with LV-shFOXM1-1; lane 4,
Tca8113 cells were infected with LV-shFOXM1-2; and lane 5, Tca8113
cells were infected with LV-shFOXM1-3. **P<0.01 vs. untreated.
LV-shFOXM1, lentivirus-short hairpin RNA Forkhead box M1; CON,
control; GFP, green fluorescent protein. |
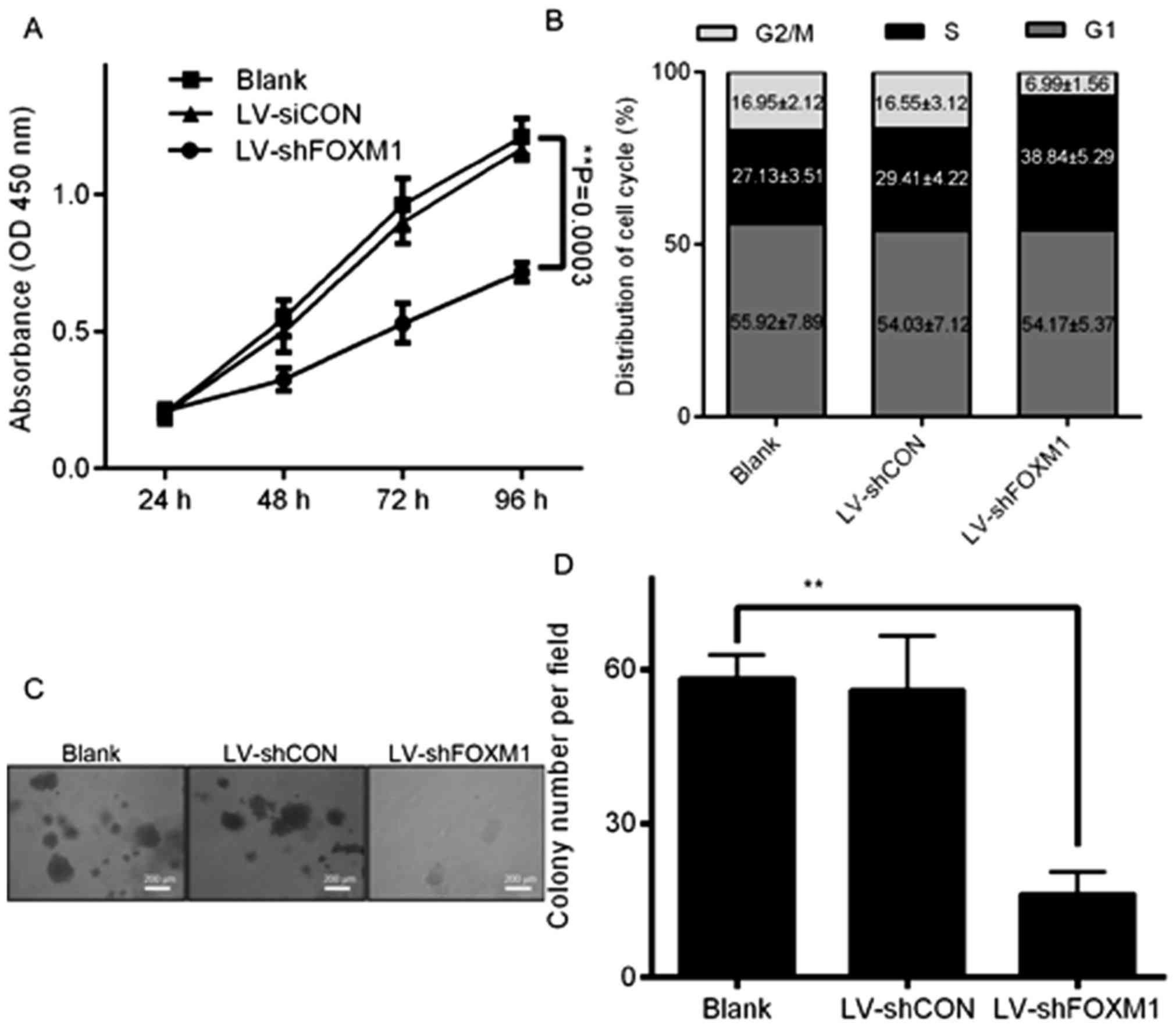
Downregulation of FOXM1, using
LV-shFOXM1, suppresses cell growth, colony formation and cell cycle
at the S-phase
FOXM1 is a key regulator of animal growth and cell
proliferation (13). The
downregulation of FOXM1 expression in Tca8113 cells significantly
inhibited cell proliferation, compared with the blank and LV-shCON
groups (P<0.05; Fig. 3A). The
association between the downregulation of FOXM1 expression and
alterations in cycle progression was analyzed in Tca8113 cells by
the flow cytometer. The proportion of S-phase cells following
LV-shFOXM1 infection was significantly decreased compared with the
blank and LV-shCON groups (P<0.05; Fig. 3B). Subsequently, whether FOXM1 gene
knockdown decreases the colony formation of Tca8113 cells was
investigated. After 24 h of infection with LV-shFOXM1, Tca8113
cells were seeded in 6-well plates and cultured for 14 days. The
results revealed that the number of colonies in LV-shFOXM1 Tca8113
cells were significantly decreased, compared with that of the
LV-shCON and untreated cells (Fig. 3C and
D). The results of the present study indicated that the
downregulation of FOXM1 expression may inhibit cell cycle
progression, growth and colony formation of Tca8113 cells.
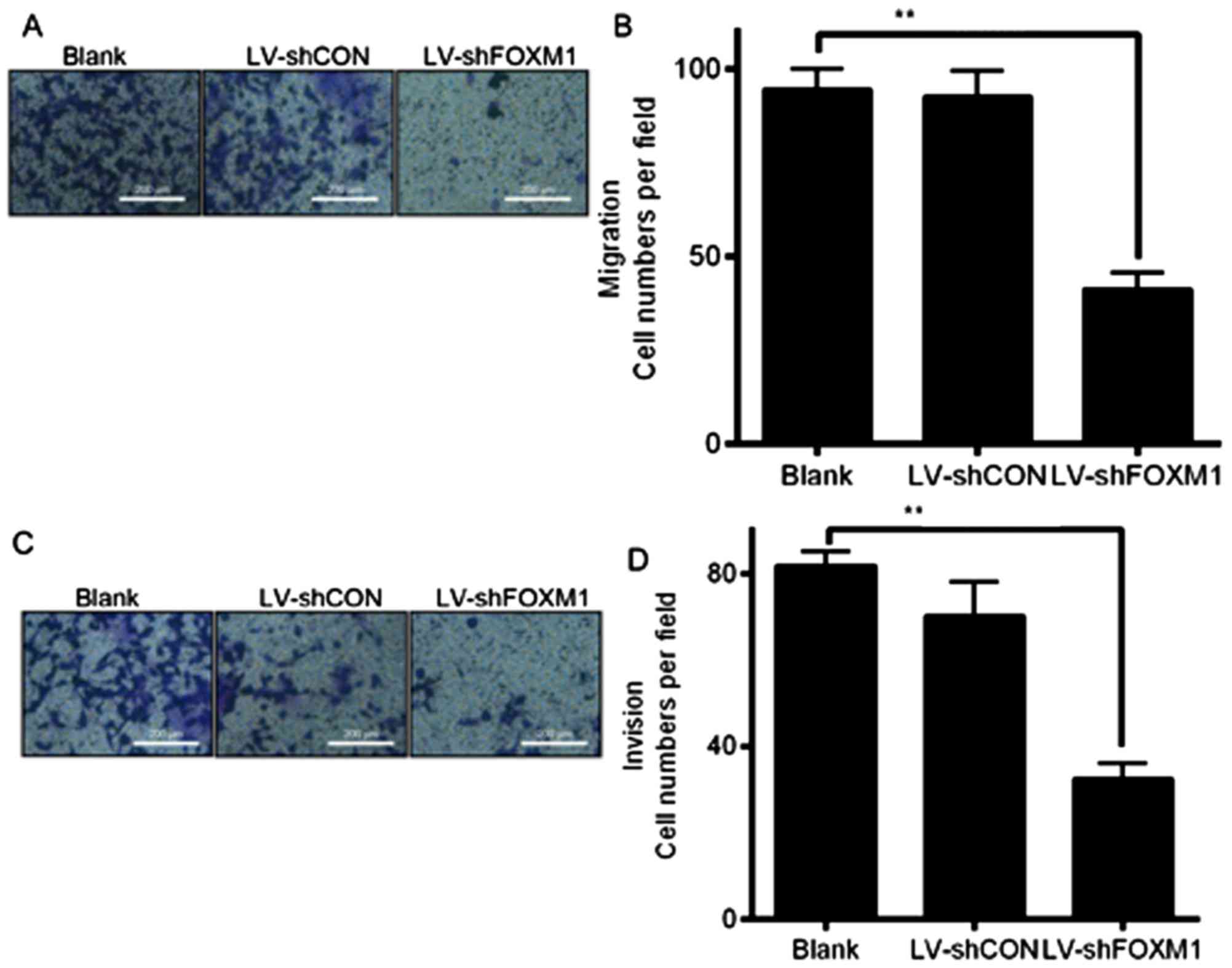
Downregulation of FOXM1 expression
inhibits cell migration and invasion
Cell migration and invasion assays were performed
using a Transwell system to determine whether the downregulation of
FOXM1 expression may affect the migratory and invasive abilities of
Tca8113 cells. The number of cells that migrated or invaded the
bottom of the well decreased significantly compard with the blank
and LV-shCON groups (P<0.05; Fig.
4). The results indicated that the downregulation of FOXM1,
following LV-shFOXM1 infection, inhibited the migratory and
invasive abilities of Tca8113 cells, compared with the blank and
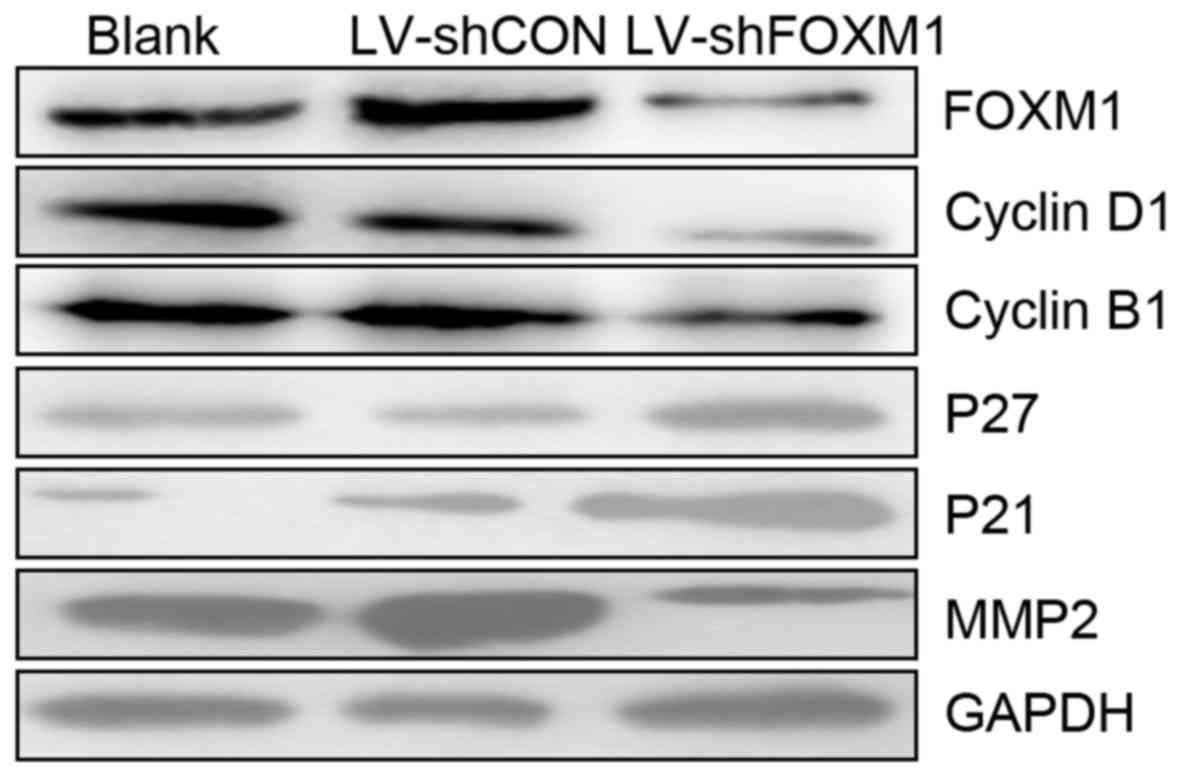
LV-shCON-infected cells. In addition, western blot analysis
revealed that the downregulation of the FOXM1 expression altered
expression of proteins associated with cell cycle. As presented in
Fig. 5, the expression levels of
cyclin B1, cyclin D1 and MMP-2 markedly decreased, but the
expression of p27 and p21 markedly increased (LV-shFOXM1 vs. blank
and LV-shCON groups).
Discussion
The overexpression of FOXM1 expression or activity
has been associated with the occurrence and development of numerous
types of cancer (14–17). Activation of FOXM1, as an independent
poor prognostic factor, is associated with the proliferation and
metastasis of human colon cancer cells, and decreased overall
survival and metastasis-free survival rates in patients with CRC
(18). However, the function of FOXM1
in OSCC remains unclear. In the present study, the mRNA and protein
expression level of FOXM1 in OSCC cells was determined using
RT-qPCR and western blot analysis, respectively, and was identified
to be expressed at increased levels in OSCC cells compared with the
human immortal keratinocyte cell line, HaCaT. The results suggested
that FOXM1 may be a novel factor in OSCC development.
RNAi-mediated gene silencing is used as the therapy
for a variety of diseases (19–21).
Lentivirus vectors are considered to be a vehicle for efficient
gene delivery in research and gene therapy, due to their ability to
transform dividing and non-dividing cells, provide stable transgene
expression and exhibit decreased toxicity (22). In the present study, a recombinant
shRNA-expressing lentiviral vector was constructed, and its effects
on the growth and invasion of OSCC cells were observed.
Lentivirus-mediated RNAi, which specifically downregulated FOXM1,
was identified to be a possible effective treatment for OSCC, as
OSCC cells exhibited decreased growth and invasion following the
silencing of FOXM1.
FOXM1 has been identified to be a key regulator of
the G1/S and G2/M stage transitions during
the cell cycle and is required for proliferative expansion during
tumor development (23). Cell
proliferation, mediated by cell cycle machinery, depends on the
equilibrium between cyclin-dependent kinase (CDK) regulators, in
particular CDK inhibitors (CDKI) and positive factors (cyclins)
(24). In the present study, cyclins
B1 and D1, and CDKIs (p21 and p27) were selected. The
downregulation of FOXM1 expression, using LV-shFOXM1, markedly
decreased the protein expression levels of cyclins B1 and D1, and
increased that of p27 and p21. The results of the present study
indicated that FOXM1 affected the cycle of Tca8113 cells by
mediating the expression levels of a number of cyclins and CDKIs. A
previous study suggested that silencing the expression of FOXM1 may
serve a role in the suppression of proliferation (25).
Cell migratory and invasive abilities are key to
cancer invasion and metastasis, which involves the breakdown of
extracellular matrix and basement membranes, enabling tumor cells
to migrate. Proteolytic enzymes and MMPs have been identified to be
involved in cancer invasion and metastasis (26,27). The
results of the present study identified that downregulation of
FOXM1 expression by LV-shFOXM1 significantly decreased the
migratory and invasive abilities of Tca8113 cells. In addition, the
expression of MMP2 was inhibited by the downregulation of FOXM1
expression. Therefore, the loss of invasive abilities of OSCC
cells, following silencing of FOXM1, may be induced partially by
the inhibition of MMP-2.
The results of the present study demonstrated that
the downregulation of FOXM1 expression by FOXM1-specific shRNA
decreased cell proliferation and the invasive abilities of Tca8113
cells, resulted in the downregulation of cyclins B1, D1 and MMP-2,
and upregulated p27 and p21 expression. The results of the present
study identified FOXM1 as a functionally important component in the
occurrence and development of OSCC, and may be a novel target for
the treatment of OSCC.
Acknowledgements
The authors would like to thank Dr. Shihai Liu,
Medical Animal Lab, the Affiliated Hospital of Qingdao University
(Qingdao, China) for assisting with the statistical analysis.
Funding
The present study was supported by Qingdao
postdoctoral application research funded project (grant no.
2014011218; Qingdao, China).
Availability of data and materials
All data generated or analyzed during the present
study are included in this published article.
Authors' contributions
JQ performed signaling pathway analysis, RT-qPCR and
wrote the manuscript. QL, JZ and LL performed the migration and
invasion experiments. AZ and HP carried out the soft agar analysis.
XY, as the corresponding author, designed the protocol and revised
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Neville BW and Day TA: Oral cancer and
precancerous lesions. CA Cancer J Clin. 52:195–215. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fusco A and Fedele M: Roles of HMGA
proteins in cancer. Nat Rev Cancer. 7:899–910. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Quan J, Johnson NW, Zhou G, Parsons PG,
Boyle GM and Gao J: Potential molecular targets for inhibiting bone
invasion by oral squamous cell carcinoma: A review of mechanisms.
Cancer Metastasis Rev. 31:209–219. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bella L, Zona S, Nestal de Moraes G and
Lam EW: FOXM1: A key oncofoetal transcription factor in health and
disease. Semin Cancer Biol. 29:32–39. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Z, Ahmad A, Li Y, Banerjee S, Kong D
and Sarkar FH: Forkhead box M1 transcription factor: A novel target
for cancer therapy. Cancer Treat Rev. 36:151–156. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Halasi M and Gartel AL: FOX(M1) news-it is
cancer. Mol Cancer Ther. 12:245–254. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Leung TW, Lin SS, Tsang AC, Tong CS, Ching
JC, Leung WY, Gimlich R, Wong GG and Yao KM: Over-expression of
FoxM1 stimulates cyclin B1 expression. FEBS Lett. 507:59–66. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shi M, Cui J and Xie K: Signaling of
miRNAs-FOXM1 in cancer and potential targeted therapy. Curr Drug
Targets. 14:1192–1202. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang HG, Xu XW, Shi XP, Han BW, Li ZH,
Ren WH, Chen PJ, Lou YF, Li B and Luo XY: Overexpression of
forkhead box protein M1 (FOXM1) plays a critical role in colorectal
cancer. Clin Transl Oncol. 18:527–532. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dull T, Zufferey R, Kelly M, Mandel RJ,
Nguyen M, Trono D and Naldini L: A third-generation lentivirus
vector with a conditional packaging system. J Virol. 72:8463–8471.
1998.PubMed/NCBI
|
|
13
|
Korver W, Schilham MW, Moerer P, van den
Hoff MJ, Dam K, Lamers WH, Medema RH and Clevers H: Uncoupling of S
phase and mitosis in cardiomyocytes and hepatocytes lacking the
winged-helix transcription factor Trident. Curr Biol. 8:1327–1330.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jin H, Li XJ, Park MH and Kim SM:
FOXM1-mediated downregulation of uPA and MMP9 by
3,3′-diindolylmethane inhibits migration and invasion of human
colorectal cancer cells. Oncol Rep. 33:3171–3177. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Quan M, Wang P, Cui J, Gao Y and Xie K:
The roles of FOXM1 in pancreatic stem cells and carcinogenesis. Mol
Cancer. 12:1592013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu H: Forkhead box transcription factors
in embryonic heart development and congenital heart disease. Life
Sci. 144:194–201. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gormally MV, Dexheimer TS, Marsico G,
Sanders DA, Lowe C, Matak-Vinković D, Michael S, Jadhav A, Rai G,
Maloney DJ, et al: Suppression of the FOXM1 transcriptional
programme via novel small molecule inhibition. Nat Commun.
5:51652014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chu XY, Zhu ZM, Chen LB, Wang JH, Su QS,
Yang JR, Lin Y, Xue LJ, Liu XB and Mo XB: FOXM1 expression
correlates with tumor invasion and a poor prognosis of colorectal
cancer. Acta Histochem. 114:755–762. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rao DD, Wang Z, Senzer N and Nemunaitis J:
RNA interference and personalized cancer therapy. Discov Med.
15:101–110. 2013.PubMed/NCBI
|
|
20
|
Swanton C, Nicke B and Downward J: RNA
interference, DNA methylation, and gene silencing: A bright future
for cancer therapy? Lancet Oncol. 5:653–654. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Z, Rao DD, Senzer N and Nemunaitis J:
RNA interference and cancer therapy. Pharm Res. 28:2983–2995. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Root DE, Hacohen N, Hahn WC, Lander ES and
Sabatini DM: Genome-scale loss-of-function screening with a
lentiviral RNAi library. Nat Methods. 3:715–719. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kalin TV, Ustiyan V and Kalinichenko VV:
Multiple faces of FoxM1 transcription factor: Lessons from
transgenic mouse models. Cell Cycle. 10:396–405. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wakino S, Kintscher U, Kim S, Yin F, Hsueh
WA and Law RE: Peroxisome proliferator-activated receptor gamma
ligands inhibit retinoblastoma phosphorylation and G1->S
transition in vascular smooth muscle cells. J Biol Chem.
275:22435–22441. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu M, Dai B, Kang SH, Ban K, Huang FJ,
Lang FF, Aldape KD, Xie TX, Pelloski CE, Xie K, et al: FoxM1B is
overexpressed in human glioblastomas and critically regulates the
tumorigenicity of glioma cells. Cancer Res. 66:3593–3602. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rubin MA: Insights into the mechanism of
organ-specific cancer metastasis. Cancer Discov. 4:1262–1264. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kessenbrock K, Wang CY and Werb Z: Matrix
metalloproteinases in stem cell regulation and cancer. Matrix Biol.
44–46. 184–190. 2015.
|