Introduction
Globally, gastric cancer (GC) is one of the most
common malignant tumors and remains the second-leading cause of
cancer-associated mortality (1). In
China, although the morbidity and mortality rates have declined in
recent years, GC continues to rank as the most common of all
malignant tumors (2). Evidence
indicates that gastric epithelial cells that were exposed to
inflammatory microenvironment in the long term could induce
neoplastic transformation (3,4). Stromal cells, one of the components of
inflammatory microenvironment, are notable players in the formation
of the cancer-associated inflammatory microenvironment (5). Research indicates that among these
stromal cells, mesenchymal stem cells (MSCs) and macrophages are
involved in the neoplastic transformation of epithelium in
inflammatory microenvironment (6,7).
MSCs, which are present in a number of tissues, are
adult stromal cells with self-renewal and multipotent
differentiation abilities (8), can be
recruited into inflamed tissue or tumors, forming a major component
of the inflammatory microenvironment (9,10), which
could have an oncogenic role in tumorigenesis. Cancer-associated
fibroblasts (CAFs) are a subpopulation of fibroblasts found in
tumor tissues, and have been demonstrated to be involved in tumor
growth and invasion (11). The
characteristics of CAFs have the following three features: i)
Expression of fibroblast markers, including vimentin, N-cadherin
and fibroblast-activating protein (FAP); ii) expression of
activation of marker, including α-smooth muscle actin (α-SMA); and
iii) increased cytokine expression, including inflammatory
cytokines (11,12). CAFs are mainly derived from MSCs
(11). In addition, MSCs have the
capacity to recruit monocytes/macrophages to tumors, promoting
their growth (13). Macrophages,
which are abundant during inflammation as immune regulatory cells,
could also have a central role in promoting the inflammatory
response and host defense. Thus, there has been increasing interest
in studying macrophages that mobilizes MSCs, leading to epithelial
lesions or cancer in the inflammatory microenvironment.
Epithelial-mesenchymal transition (EMT) is a
reversible biological process by which epithelial cells lose their
polarity, reduce intercellular adhesion and acquire characteristics
of mesenchymal cells (14). During
this process, expression of markers of polarized epithelial cells,
such as E-cadherin, are lost or lowered, whereas markers of
mesenchymal cells, such as vimentin and N-cadherin, are acquired or
increased. EMT serves a notable role in normal organ development
during embryonic development and wound healing (15,16).
However, dysfunctional EMT leads to disease states, including
fibrosis and carcinogenesis (17).
Evidence revealing that EMT is associated with stemness in cancer
cells indicated that epithelial cells that undergo EMT processes
acquire stem cell-like properties (18,19). These
stem cell-like cells in tumors are termed cancer stem cells (CSCs).
CSCs express specific markers for normal stem cells and are defined
by their self-renewal capacity. Specific markers for normal stem
cells are commonly used for identifying CSCs, including Nanog,
polycomb complex protein BMI-1 and SRY-box 2 (SOX2) (20–22). CSCs,
which are present in tumors as a unique subpopulation of cells,
possess the ability to initiate tumor growth and increase the
capacity for cellular migration and invasion. CSCs are therefore
tumorigenic, unlike other types of cancer cells.
Although interactions between macrophages and MSCs
have been reported (23,24), knowledge about the mechanism
underlying the macrophage-mediated regulation of MSCs phenotype and
certain functions in the inflammatory environment remains scant at
the present time. The present study was designed to establish a
co-culture system of MSCs isolated from human umbilical cord
(hucMSCs)-macrophages (THP-1 cells) to evaluate the biological
effects of macrophages on the phenotype and certain functions of
MSCs, and to investigate the effects of the induced MSCs on normal
gastric epithelial GES-1 cells in vitro. The results
demonstrated that MSCs were strongly induced by macrophages to
express CAF markers and improve the expression of the relevant
inflammatory cytokines. MSCs induced by macrophages
(macrophage-MSCs) promoted gastric epithelial cells to acquire
phenotypes of mesenchymal cells, increasing the expression of stem
cell markers and the ability to form cellular spheres, which are
associated with stem cell properties. The results of the present
study indicated that macrophages could induce MSCs to acquire
CAF-like phenotypes to remodel the inflammatory microenvironment,
which generated cells with stem cell properties via EMT-like
changes in gastric epithelial cells, inducing gastric lesions
and/or the potential for malignant transformation. The study of
macrophages/MSCs interaction in inflammatory environments could aid
the prevention of inflammation-associated gastric cancer and assist
the development of therapeutic applications for this disease.
Materials and methods
Cell culture
The gastric epithelial GES-1 cell line and the human
macrophage THP-1 cell line were purchased from the Institute of
Biochemistry and Cell Biology (Shanghai, China). We adopted the
adherent culture method of human umbilical cord tissues to acquire
MSCs from human umbilical cords (hucMSCs) (25). Briefly, fresh umbilical cords were
obtained from full-term infants delivered by cesarean section
following the obtainment of informed consent from their parents,
and were processed within 4 h of surgery. Umbilical cords were
rinsed several times in sterile PBS containing penicillin (100
U/ml) and streptomycin (100 µg/ml) to remove blood components. Cord
vessels (e.g., arteries and veins) in the umbilical cords were
surgically removed under sterile conditions. The washed cords were
then cut into pieces 1–3 mm2 in size and placed in
RPMI-1640 medium (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) containing 10% fetal bovine serum (FBS)
(Invitrogen; Thermo Fisher Scientific, Inc.), 1% penicillin and
streptomycin. The cord pieces were subsequently incubated at 37°C
in humid air with 5% (v/v) CO2. The medium was changed
every 3 days following the initial plating. After 5 days,
non-adherent cells were removed by washing and adherent cells were
cultured further. When well-developed colonies of fibroblast-like
cells reached 80% confluence, the cultures were trypsinized and
passaged into new flasks for further expansion. The isolated
hucMSCs were identified using the methods described by Qiao et
al (25). HucMSCs at passage 3
were selected for use in the present study. GES-1 and THP-1 cells
were cultured in RPMI-1640 medium containing 10% FBS at 37°C in
humidified air with 5% (v/v) CO2. Pregnant women who
were referred to the Maternity Ward of the First Affiliated
Hospital of Bengbu Medical College (Bengbu, China) to give birth in
March 2015 were studied, ages 25–35, median age 28 years. All
pregnant women with preeclampsia, sexually transmitted diseases,
hepatitis or infections were excluded from the present study. All
experimental protocols were approved by the Ethics Review Committee
of Bengbu Medical College (Bengbu, China).
Co-cultured macrophages/hucMSCs and
conditioned medium (CM) preparation
For the preparation of macrophage-hucMSCs CM,
hucMSCs were trypsinized and resuspended in RPMI-1640 supplemented
with 10% FBS, and then seeded into a culture flask
(3×105 cells/flask) for 12 h. THP-1 cells (ratio of
THP-1 cells to hucMSCs, 1:2) were added into the culture flask
containing hucMSCs with fresh medium and, following culturing for
48 h, the medium was discarded and THP-1 cells were washed off with
PBS. Next, fresh medium was added into the culture flask containing
macrophage-hucMSCs. The culture supernatant of macrophage-hucMSCs
was collected 48 h later, filtered with a 0.22-µm filter and stored
at −80°C until use. The culture supernatant from the
macrophage-hucMSCs was mixed with equal volume of fresh medium
containing 10% FBS for subsequent experiments. The
macrophage-hucMSCs were also collected for the following studies.
HucMSCs not induced with macrophages were used as the normal
control group.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from cells was extracted using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), and then cDNA
was synthesized using a Revert Aid™ First Strand cDNA
Synthesis kit (Fermentas; Thermo Fisher Scientific, Inc.) according
to the manufacturer's recommended protocol. qPCR was performed
using VeriQuest SYBR Green qRT-PCR Master Mix with Fluorescein
(Applied Biosystems; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. The thermocycling conditions were
described as follows: Initial denaturation at 95°C for 30 sec,
followed by 35 cycles of the three-step cycling program consisting
of 15 sec at 95°C, 30 sec at 60°C (N-cadherin, IL-6 and FAP) or
62°C (Vimentin, IL-8 and MCP-1) or 57°C (other genes) and 35 sec at
72°C, followed by a final extension step for 5 min at 72°C. Each
sample was analyzed in triplicate. All primers (Table I) used for RT-qPCR were designed by
Invitrogen; Thermo Fisher Scientific, Inc. β-actin was used as the
internal control. Relative quantification was performed using the
2−ΔΔCq method (26). All
experiments were repeated three times.
 | Table I.Primer sequences of target genes. |
Table I.
Primer sequences of target genes.
| Genes | Primer sequences
(5′-3′) | Amplicon size
(bp) | Annealing
temperature (°C) |
|---|
| FAP | For:
5′-ATAGCAGTGGCTCCAGTCTC-3 |
|
|
|
| Rev:
5′-GATAAGCCGTGGTTCTGGTC-3 | 278 | 59 |
| α-SMA | For:
5′-CTGACTGAGCGTGGCTATTC-3′ |
|
|
|
| Rev:
5′-CCACCGATCCAGACAGAGTA-3′ | 452 | 58 |
| N-cadherin | For:
5′-AGCTCCATTCCGACTTAGACA-3′ |
|
|
|
| Rev:
5′-CAGCCTGAGCACGAAGAGTG-3′ | 165 | 60 |
| Vimentin | For:
5′-GACGCCATCAACACCGAGTT-3′ |
|
|
|
| Rev:
5′-CTTTGTCGTTGGTTAGCTGGT-3′ | 238 | 63 |
| β-actin | For:
5′-CACGAAACTACCTTCAACTCC-3′ |
|
|
|
| Rev:
5′-CATACTCCTGCTTGCTGATC-3′ | 265 | 56 |
| VEGF | For:
5′-GGGCAGAATCATCACGAAGT-3′ |
|
|
|
| Rev:
5′-TGGTGATGTTGGACTCCTCA-3′ | 211 | 58 |
| PBGF-B | For:
5′-CTGAACTCCATCGCCATCTT-3′ |
|
|
|
| Rev:
5′-GCAGGCTATGCTGAGAGGTC-3′ | 187 | 56 |
| TNF-α | For:
5′-CCGAGTGACAAGCCTGTAGC-3′ |
|
|
|
| Rev:
5′-AGGAGGTTGACCTTGGTCTG-3′ | 493 | 57 |
| GM-CSF | For:
5′-TTCTGCTTGTCATCCCCTTT-3′ |
|
|
|
| Rev:
5′-TGCCTGTATCAGGGTCAGTG-3′ | 206 | 58 |
| IL-8 | For:
5′-GCTCTGTGTGAAGGTGCAGTTT-3′ |
|
|
|
| Rev:
5′-TTCTGTGTTGGCGCAGTGT-3′ | 144 | 62 |
| IL-6 | For:
5′-TACATCCTCGACGGCATCTC-3′ |
|
|
|
| Rev:
5′-AGCTCTGGCTTGTTCCTCAC-3′ | 252 | 61 |
| MCP-1 | For:
5′-ACGGCCTTCCAAGGCAT-3′ |
|
|
|
| Rev:
5′-TTGTTACGCCGTCGCTGA-3′ | 103 | 63 |
Western blot analysis
Western blot analysis was performed as described
previously (27). Briefly, the cells,
collected as aforementioned, were immediately lysed with lysis
buffer (Cell Signaling Technology, Inc., Dancers, MA, USA)
supplemented with complete protease inhibitors (Shanghai Haoran
Biological Technology Co., Ltd., Shanghai, China) on ice. Lysed
cells were centrifuged at 4°C, 13,400 × g for 15 min, and then
total protein was collected. Protein concentrations were determined
to normalize different cells using the BCA protein assay kit (Merck
KGaA, Darmstadt, Germany). Aliquots containing identical amounts
(60 µg) of protein were fractionated using 12% SDS-PAGE and then
transferred onto PVDF membranes (EMD Millipore, Billerica, MA,
USA). The membranes were incubated with 5% skimmed milk to block
non-specific protein at room temperature for 1 h. Membranes were
incubated overnight at 4°C with primary antibodies at a dilution of
1:800 for rabbit polyclonal anti-N-cadherin (cat. no. BS2224;
Bioworld Technology, Inc., St. Louis Park, MN, USA), 1:1,000 for
rabbit polyclonal anti-E-cadherin (cat. no. BS1098; Bioworld
Technology, Inc.), 1:500 for rabbit polyclonal anti-vimentin (cat.
no. BS1855; Bioworld Technology, Inc.), 1:500 for rabbit polyclonal
anti-α-SMA (cat. no. BS8796; Bioworld Technology, Inc.), 1:800 for
rabbit polyclonal anti-B-cell lymphoma-2 (Bcl-2) (cat. no. BS70205;
Bioworld Technology, Inc.), 1:500 for rabbit polyclonal
anti-Bcl-2-associated X (Bax) (cat. no. BS1030; Bioworld
Technology, Inc.), 1:500 for rabbit polyclonal anti-SOX2 (cat. no.
BS6161; Bioworld Technology, Inc.), 1:1,000 for rabbit polyclonal
anti-Bmi-1 (cat. no. BS6015; Bioworld Technology, Inc.), 1:10,000
for rabbit polyclonal anti-β-actin (cat. no. AP0060; Bioworld
Technology, Inc.), 1:1,000 for mouse monoclonal anti-FAP (cat. no.
sc-71094; Santa Cruz Biotechnology, Inc.,) and 1:1,000 for rabbit
polyclonal anti-Nanog (cat. no. sc-33759; Santa Cruz Biotechnology,
Inc.). The membranes were then washed and incubated for 2 h at 37°C
in horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG
secondary antibodies (cat. no. L3012-2; Signalway Antibody Co.,
Ltd., Nangjing, China) or goat anti-mouse IgG secondary antibodies
(cat. no. L3032-2; Signalway Antibody Co., Ltd) at a dilution of
1:2,000. The signal was detected using HRP substrate (EMD
Millipore, Billerica, MA, USA) and analyzed using MD Image
Quant™ Software v1.5.2.0 (G-Box Chemi XT4; Syngene
Europe, Cambridge, UK).
Luminex assay/ELISA
Interleukin-8 (IL-8), IL-6, platelet-derived growth
factor (PDGF)-B, granulocyte macrophage colony-stimulating factor
(GM-CSF), monocyte chemoattractant protein (MCP)-1, vascular
endothelial growth factor (VEGF) and tumor necrosis factor (TNF) in
the CM of hucMSCs and macrophage-hucMSCs were evaluated using
MILLIPLEX® MAP Human Cytokine/Chemokine Magnetic Bead
Panel-Premixed 41 (EMD Millipore) according to the manufacturer's
protocol. ELISA (IL-6, cat. no. DKW12-1060-048; IL-8, cat. no.
DWK12-1080-048; GM-CSF, cat. no. DWK12-1730-048; and MCP-1, cat.
no. DWK12-1739-048; all reagents were purchased from Dakewe Biotech
Co., Ltd, Shenzhen, China) was subsequently used to measure the
levels of IL-6, IL-8, GM-CSF and MCP-1 in supernatant from hucMSCs
and macrophage-hucMSCs in accordance with the manufacturer's
instructions.
Immunofluorescence assay
Immunofluorescence assays were used to detect the
expression of E-cadherin in GES-1 cells cultured with the CM of
macrophage-hucMSCs (28). GES-1 cells
cultured with the CM of macrophage-hucMSCs in 24-well plates for 48
h were washed three times with PBS, fixed with 4% paraformaldehyde
at room temperature for 20 min, permeabilized with 0.1% Triton
X-100 for 5 min, blocked with 5% bovine serum albumin (Boster
Biological Technology, Pleasanton, CA, USA) at room temperature for
30 min, and then incubated with anti-E-cadherin antibodies at a
dilution of 1:100 (cat. no. BS1098; Bioworld Technology, Inc.) at
4°C overnight, followed by incubation with a fluorescein
isothiocyanate (FITC)-conjugated anti-rabbit secondary antibody at
dilution of 1:200 (F9887; Sigma-Aldrich; Merck KGaA) at 37°C for 1
h. The cells were then stained with DAPI (Beyotime Institute of
Biotechnology) for nuclear staining. Images were then acquired with
a Nikon TE300 Inverted Fluorescence Phase Contrast Microscope
(Nikon Corporation, Tokyo, Japan), magnification, ×200.
Migration assays
GES-1 cells (5×104 cells/200 µl/well)
suspended in RPMI-1640 medium (Invitrogen; Thermo Fisher
Scientific, Inc.) with 10% FBS were placed in the upper side of a
8.0-µm pore size Transwell insert (Corning Incorporated, Corning,
NY, USA) in 24-well culture plates, and then 600 µl medium with 10%
FBS, 600 µl CM from hucMSCs and 600 µl CM from the
macrophage-hucMSCs were placed in the lower well of separate
Transwell chambers. Following 12 h of incubation at 37°C, the GES-1
cells which migrated through the lower side of the inserts were
fixed with cold 4% paraformaldehyde (DingGuo Biotechnology Co.,
Ltd., Shanghai, China) at room temperature for 30 min and
subsequently stained with 0.1% crystal violet (Sigma-Aldrich; Merck
KGaA, Shanghai, China) for 15 min. Cells that migrated through the
inserts were counted on 10 different randomly chosen fields per
inset under a light microscope (Nikon Corporation), magnification,
×200. This experiment was repeated three times.
Apoptosis assay
GES-1 cells (1×105 cells) were collected
in the tubes following culturing with CM from hucMSCs or
macrophage-hucMSCs for 48 h. The treated GES-1 cells were washed
with PBS, and then Annexin V-FITC apoptosis assay kit (BioVision
Inc., San Francisco, CA, USA) was added into the tube, which was
gently mixed at room temperature for 10 min in the dark, according
to the manufacturer's protocol. Stained cells were analyzed using a
flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Data
analysis was performed with FlowJo software v10 (FlowJo LLC,
Ashland, OR, USA). All the experiments were conducted in
triplicate.
Cell colony formation assay
GES-1 cells (1,000 cells/well) were seeded in 6-well
plates with RPMI-1640 medium containing 10% FBS following
incubation with RPMI-1640 medium containing 10% FBS (control) or CM
from hucMSCs or CM from macrophage-hucMSCs for 48 h, and then all
groups were incubated at 37°C with 5% CO2 for 10 days.
The medium was changed every 3 days. The cell colonies were fixed
with methanol for 30 min, and then stained with 0.5% crystal violet
at room temperature for 15 min. The results are the mean values of
3 experiments in triplicate.
Sphere-forming assay
GES-1 cells were cultured with RPMI-1640 medium
containing 10% FBS (control), CM from hucMSCs or CM from
macrophage-hucMSCs for 48 h at 37°C. Subsequently, GES-1 cells from
all groups were harvested and plated at density of 2,000 cells/well
into 24-well culture plates coated with 10% poly-HEMA
(Sigma-Aldrich; Merck KGaA) solution in 100% ethanol and dried
overnight at 56°C with a serum-free RPMI-1640 Glutamax medium
(Invitrogen; Thermo Fisher Scientific, Inc.) containing 15 ng/ml
EGF (Bioworld Technology, Inc.), 10 ng/ml FGF (Bioworld Technology,
Inc.), 1:100 N-2 supplement 100X (Invitrogen; Thermo Fisher
Scientific, Inc.), 0.3% glucose, 5 mg/ml gentamicin (Sigma-Aldrich;
Merck KGaA), 50 IU/ml penicillin and 2.5 mg/ml amphotericin B
(Sigma-Aldrich; Merck KGaA), respectively, all groups were
incubated in humidified incubator at 37°C with 5% CO2
for 15 days. Every 3 days, 50% of the spent medium was removed and
replaced. The total number of spherical colonies (colonies with
diameters large than 50 µm were counted) obtained was quantitated
under an inverted microscope. This procedure was repeated at least
three times.
Statistical analysis
Data are presented as the mean ± standard deviation.
Statistical analysis between two groups was performed using
Student's t-test. For multiple group comparisons, an analysis of
variance was performed, followed by Bonferroni's post-hoc test.
SPSS 19.0 software (IBM Corp., Armonk, NY, USA) was used to analyze
the data. P<0.05 was considered to indicate a statistically
significant difference.
Results
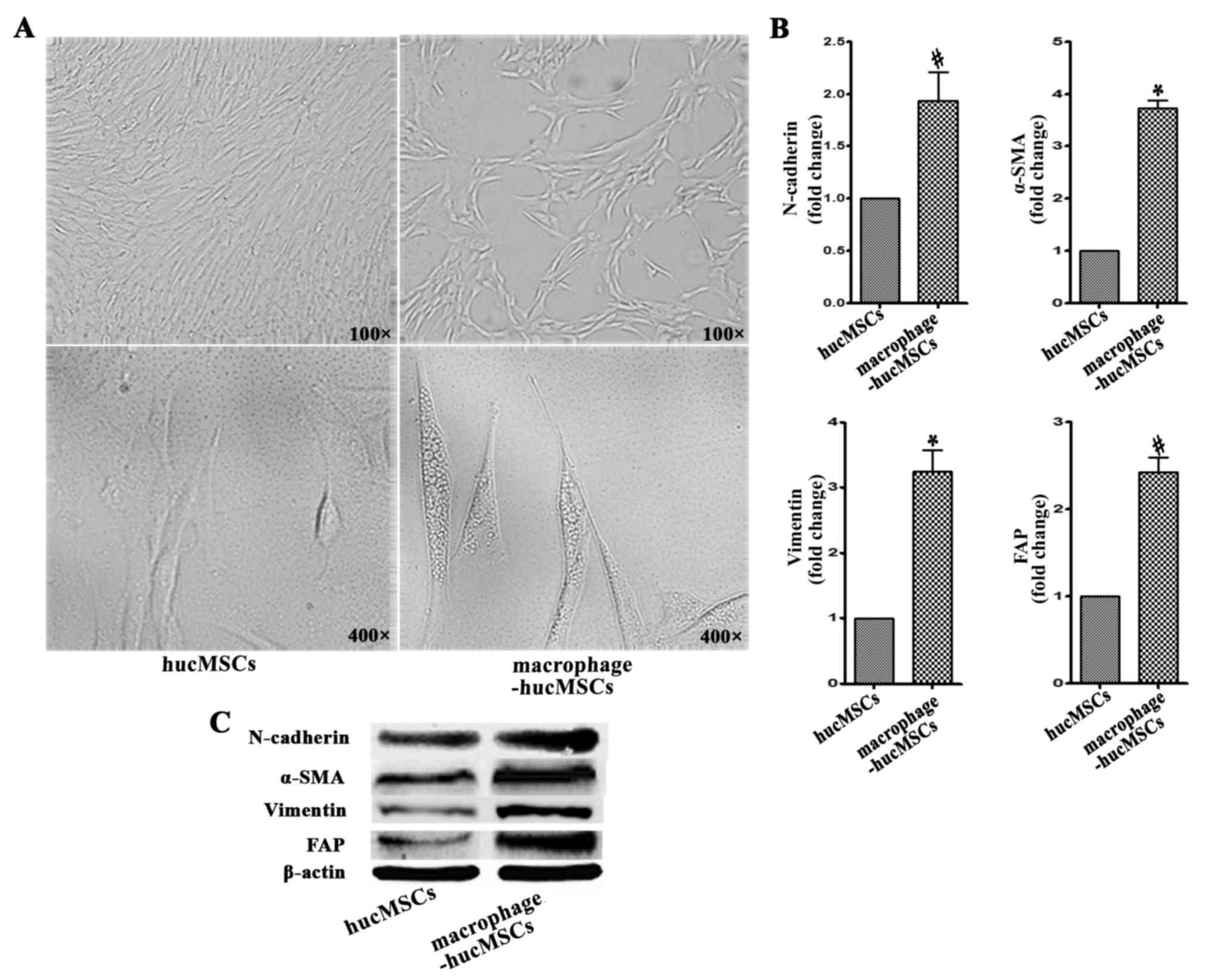
Macrophages induce the differentiation
of MSCs into CAFs
HucMSCs were co-cultured with macrophages (ratio of
hucMSCs to THP-1 cells, 2:1) for 48 h and subsequently observed
using an inverted microscope. The hucMSCs induced with macrophages
exhibited larger and more spindle-like morphology (Fig. 1A). The expression of FAP, α-SMA,
N-cadherin and vimentin, which are biomarkers for CAFs (12), was assessed by RT-qPCR in the induced
hucMSCs with macrophages. The results of RT-qPCR revealed that the
expression of FAP, α-SMA, N-cadherin and vimentin were upregulated
in the induced hucMSCs with macrophages (Fig. 1B). To confirm the protein expression
of FAP, a-SMA, N-cadherin and vimentin in the induced hucMSCs,
western blot analysis was performed, the results of which were
consistent with those of RT-qPCR (Fig.
1C).
Macrophages upregulate the expression
of inflammatory cytokines in MSCs
The Luminex assay was conducted to determine the
levels of several cytokines in the supernatant from hucMSCs and
macrophage-hucMSCs. The results revealed that IL-8, PDGF-BB,
GM-CSF, IL-6, MCP-1, VEGF and TNF levels were all increased to a
certain degree in the CM from hucMSCs co-cultured with macrophages
in vitro, low levels of these cytokines were observed in
hucMSCs (Fig. 2A). To verify the
results of the Luminex assay, RT-qPCR was performed to detect the
mRNA levels of these cytokines and ELISA to examine the levels of
four of these cytokines (IL-8, GM-CSF, IL-6 and MCP-1; P<0.01;
Fig. 2A) in the induced hucMSCs with
macrophages. The results of RT-qPCR, as well as that of Luminex
analysis, revealed that the expression of these cytokine genes were
upregulated in the induced hucMSCs with macrophages (Fig. 2B). The results of ELISA detection of
four of these cytokines (IL-8, GM-CSF, IL-6 and MCP-1) were
identical to those of the Luminex assay, and in accordance with
those of RT-qPCR (Fig. 2C),
indicating that macrophages can enhance the expression of
inflammatory cytokines in MSCs.
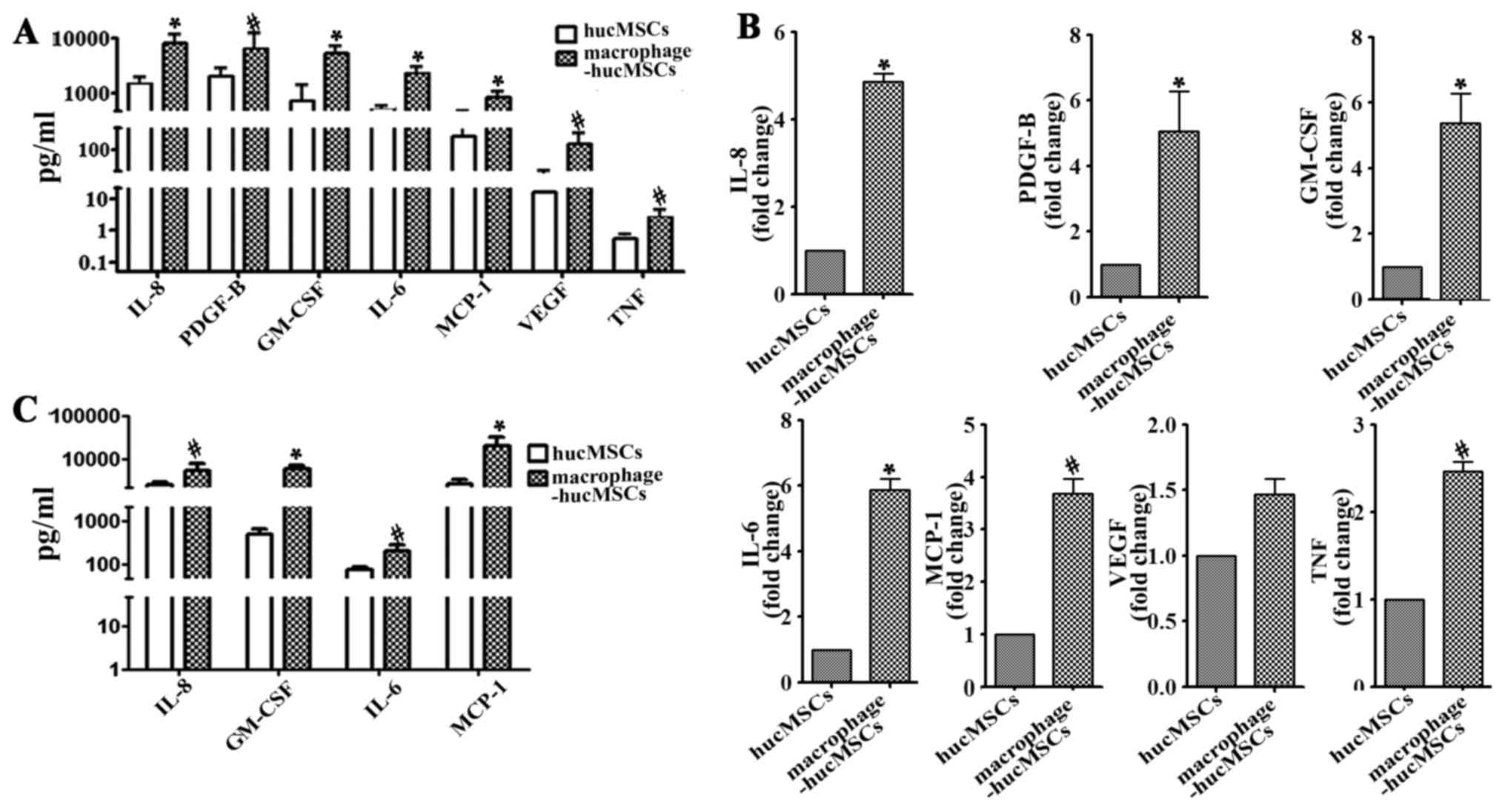 | Figure 2.High levels of IL-8, PDGF-B, GM-CSF,
IL-6, MCP-1, VEGF and TNF were presented in the supernatant from
macrophage-hucMSCs. (A) The expression of cytokines (IL-8,
P=0.0092; PDGF-B, P=0.0391; GM-CSF, P=0.0095; IL-6, P=0.0077;
MCP-1, P=0.0084; VEGF, P=0.0441; TNF, P=0.0244) in the supernatant
of the hucMSCs cultured with or without macrophages were measured
using a Luminex assay. (B) mRNA expressions of IL-8, PDGF-B,
GM-CSF, IL-6, MCP-1 and TNF significantly increased in the
macrophage-hucMSCs. (C) Supernatants from the two aforementioned
types of cells were assessed for IL-8, GM-CSF, IL-6 and MCP-1
expression by ELISA. Data are presented as the mean ± standard
deviation; *P<0.01 and #P<0.05, vs. hucMSCs (n=3).
IL-8, interleukin-8; PDGF-B, platelet-derived growth factor-B;
GM-CSF, granulocyte-macrophage colony-stimulating factor; MCP-1,
monocyte chemoattractant protein-1; VEGF, vascular endothelial
growth factor; TNF, tumor necrosis factor; macrophage-hucMSC, human
umbilical cord-derived mesenchymal stem cells pre-cultured with
macrophages for 48 h. |
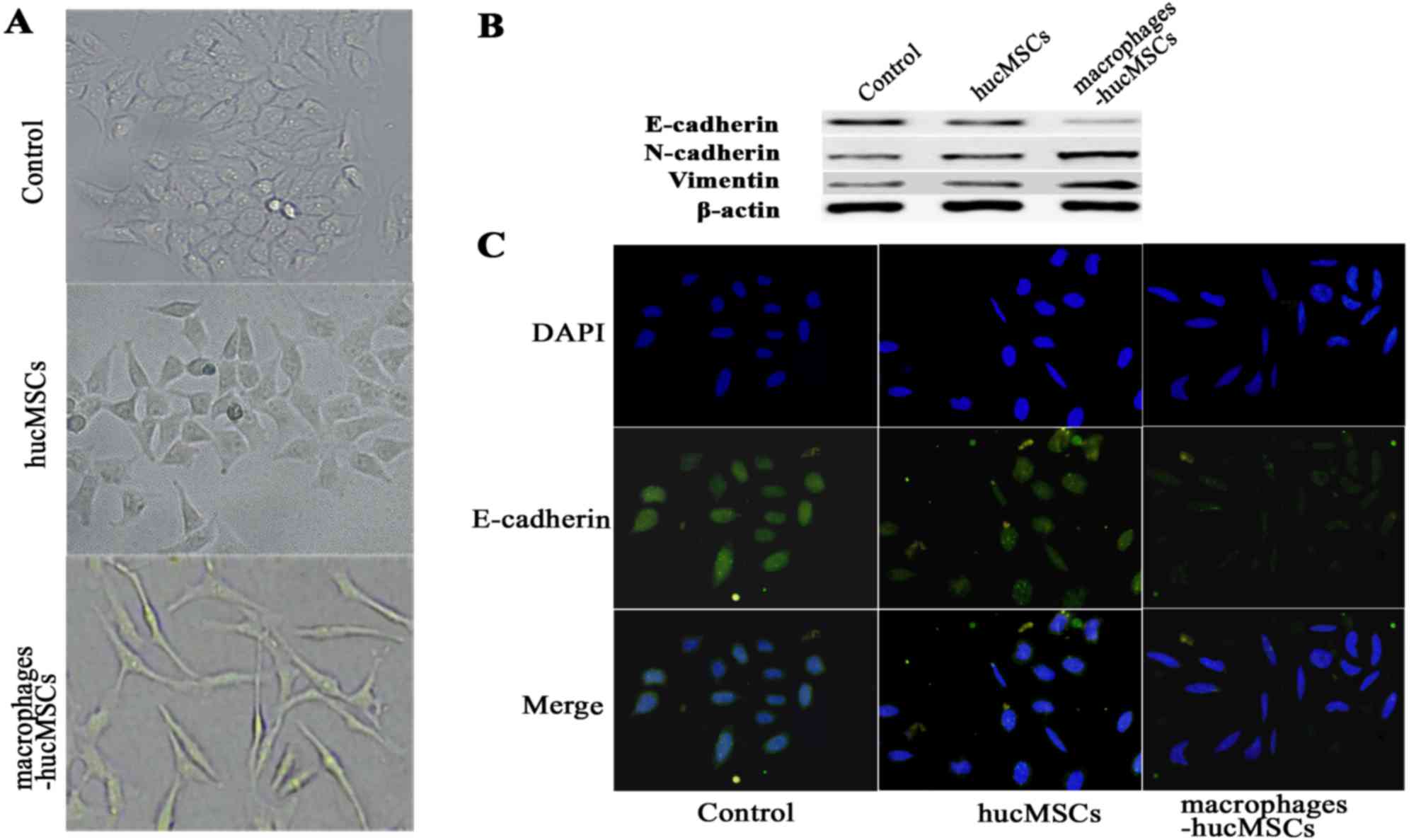
Macrophage-MSCs induce an EMT state in
GES-1 cells and stimulates gastric epithelial cell invasion
The results of the present study revealed that CM
from macrophage-hucMSCs induced GES-1 cell morphological changes,
characterized by a loss of polygonal shape, disruption of the
formation of cell clusters and the appearance of elongated cells,
indicating the presence of a mesenchymal phenotype (Fig. 3A). The quantification of cells
harboring a mesenchymal-like phenotype was ~85% in GES-1 cells.
Western blot analysis of the expression of EMT markers in the GES-1
cells induced with CM from macrophage-hucMSCs was significantly
increased in the mesenchymal markers vimentin and N-cadherin, and
decreased in the epithelial marker E-cadherin (Fig. 3B). E-cadherin protein expression in
GES-1 cells treated with CM from macrophage-hucMSCs was also
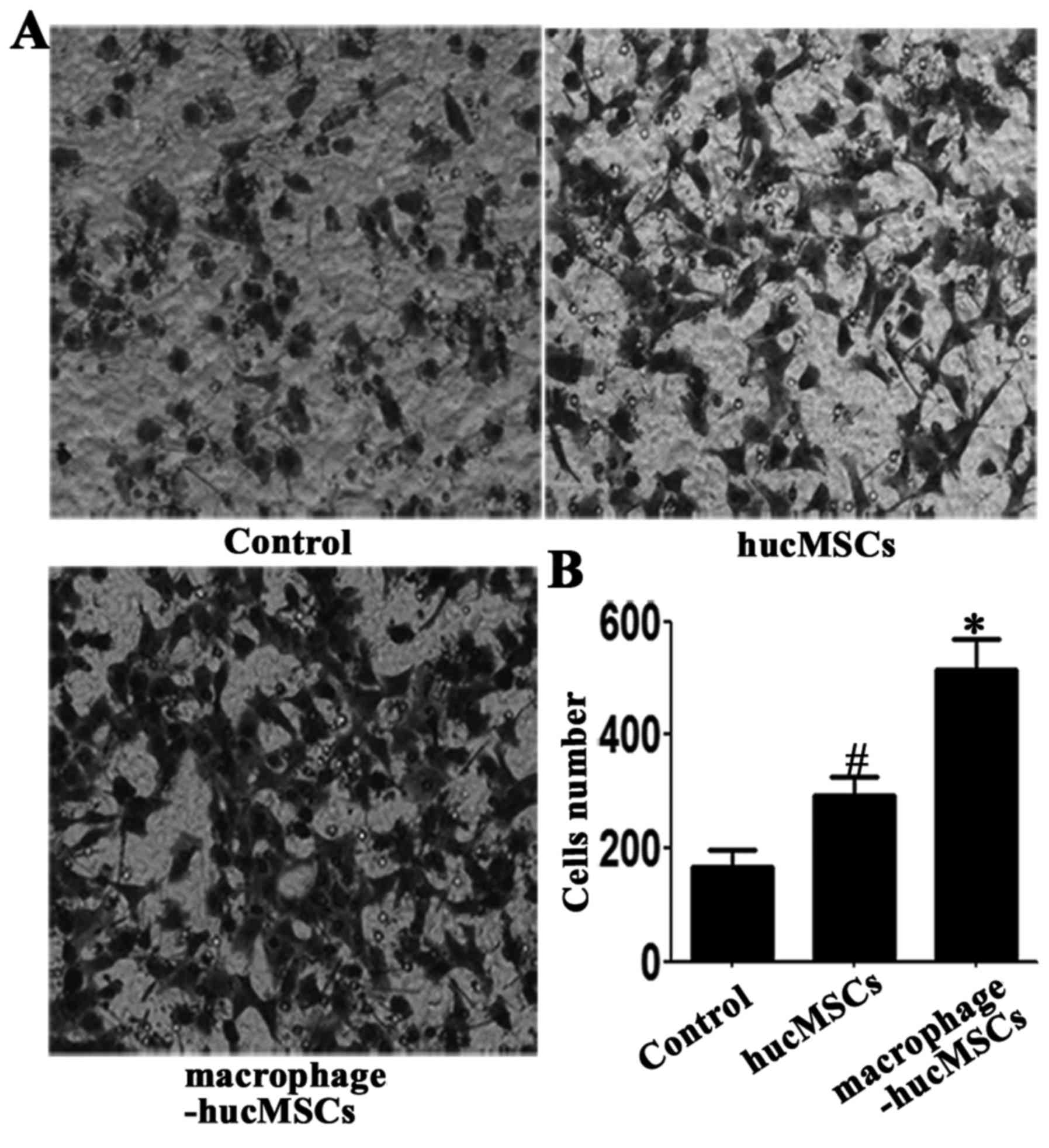
examined using an immunofluorescence assay (Fig. 3C). The migrated potential of the
treated GES-1 cells was assessed using Transwell assays. The
results revealed that the migration of GES-1 cells treated with CM
from macrophage-hucMSCs was greater than that of the control
counterparts (Fig. 4). GES-1 cells
treated with medium only served as controls.
Macrophage-MSCs reduce the apoptosis
of gastric epithelial cells
The apoptotic rate of the GES-1 cells treated with
CM from macrophage-hucMSCs was analyzed by annexin V-FITC/PI
staining. The results revealed that the apoptotic rate of the
induced GES-1 cells significantly decreased following exposure to
CM from macrophage-hucMSCs for 48 h (Fig.
5A and B). Western blot analysis also revealed the
downregulation of the pro-apoptotic protein Bax, along with the
upregulation of the anti-apoptotic protein Bcl-2 (Fig. 5C) in the GES-1 cells cultured with CM
from macrophage-hucMSCs, which indicated that macrophage-MSCs
inhibited the apoptosis of gastric epithelial cells.
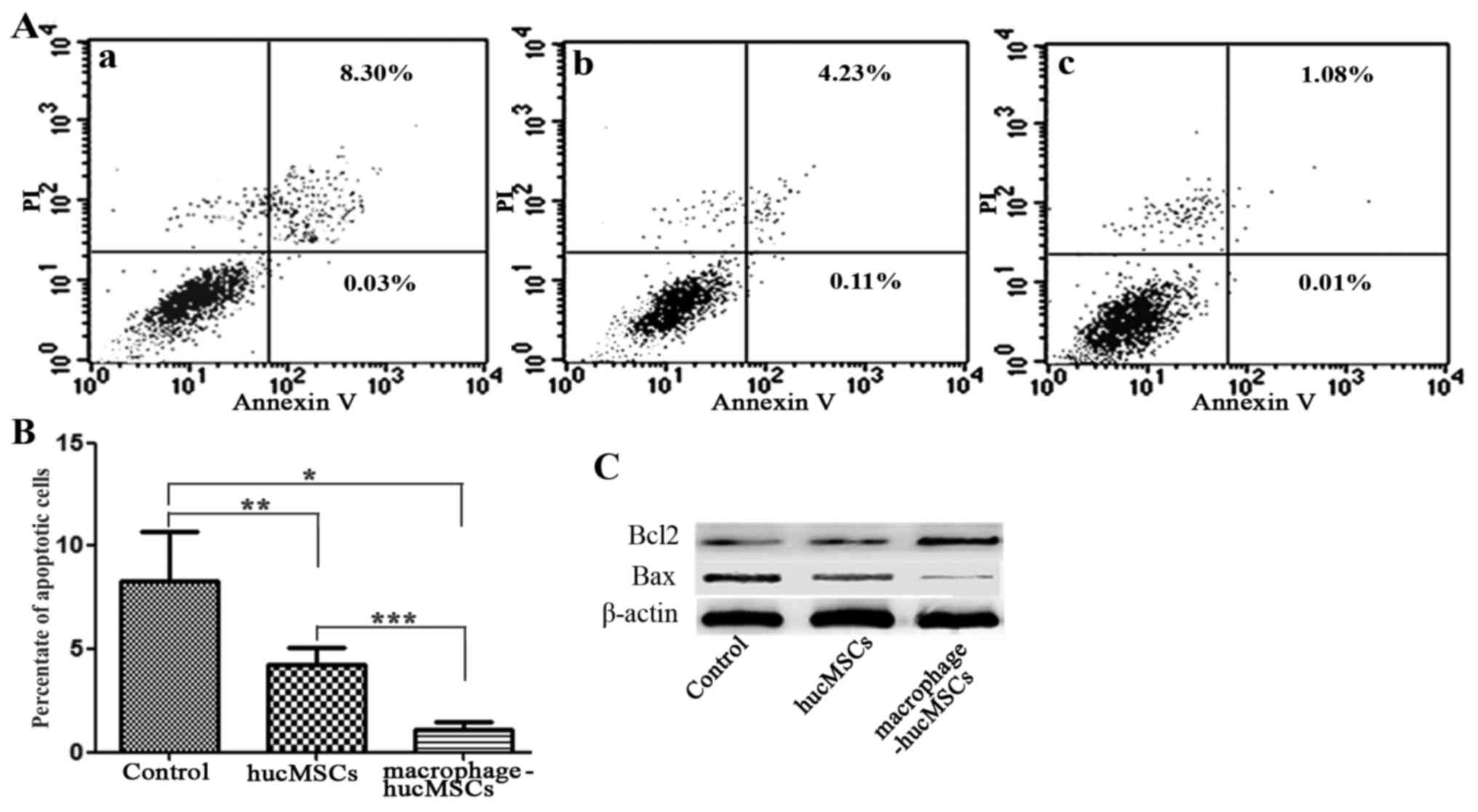 | Figure 5.Effects of macrophage-hucMSCs on the
apoptosis of gastric epithelial cells. (A) The apoptotic rate of
gastric epithelial GES-1 cells was assessed via flow cytometry 48 h
after treatment with (Aa) medium only (as the controls), (Ab) CM
from hucMSCs and (Ac) CM from macrophage-hucMSCs. The experiments
were performed in triplicate. (B) Histogram depicting the apoptotic
rate of the treated gastric epithelial cells with medium only, CM
from hucMSCs or CM from macrophage-hucMSCs (n=3). *P<0.01,
**P>0.05, ***P<0.05. (C) Bcl-2 and Bax protein expression of
GES-1 cells via western blot analysis 48 h after treatment with
medium only, CM from hucMSCs or CM from macrophage-hucMSCs. GES-1
cells treated with medium only served as control.
Macrophage-hucMSC, human umbilical cord-derived mesenchymal stem
cells pre-cultured with macrophages for 48 h; CM, conditioned
medium; Bcl-2, B-cell lymphoma-2; Bax, Bcl-2-associated X; PI,
propidium iodide. |
Macrophage-MSCs enhance the stem cell
properties of gastric epithelial cells
To investigate whether the gastric epithelial cells
induced with macrophage-MSCs had stem cell properties, a sphere
formation assay was performed to determine the presence of stem
cell-like cells in GES-1 cells treated with CM from
macrophages-hucMSCs. No sphere colonies were seen when GES-1 cells
were cultured with medium only (data not shown). By contrast, when
co-cultured with CM from macrophage-hucMSCs the total number of
GES-1 cell sphere colonies evidently increased (Fig. 6A and B). Western blot analysis also
revealed that the expression of stem cell-specific transcription
factors was elevated in the GES-1 cells treated with
macrophage-hucMSCs CM, compared with GES-1 cells and GES-1 cells
treated with CM from hucMSCs (Fig.
6C). To corroborate the effect of macrophages-MSCs increase on
promoting gastric epithelial stem cell properties, GES-1 cells,
GES-1 cells treated with CM from hucMSCs and GES-1 cells treated
with macrophage-hucMSCs were subjected to mono-cell
colony-formation assays in vitro. The results of these
assays revealed that GES-1 cells treated with CM from
macrophage-hucMSCs were able to produce larger-sized and a greater
number of mono-sphere colonies (Fig. 6D
and E). These data indicated that macrophages-MSCs enhance the
stem cell properties of gastric epithelial cells.
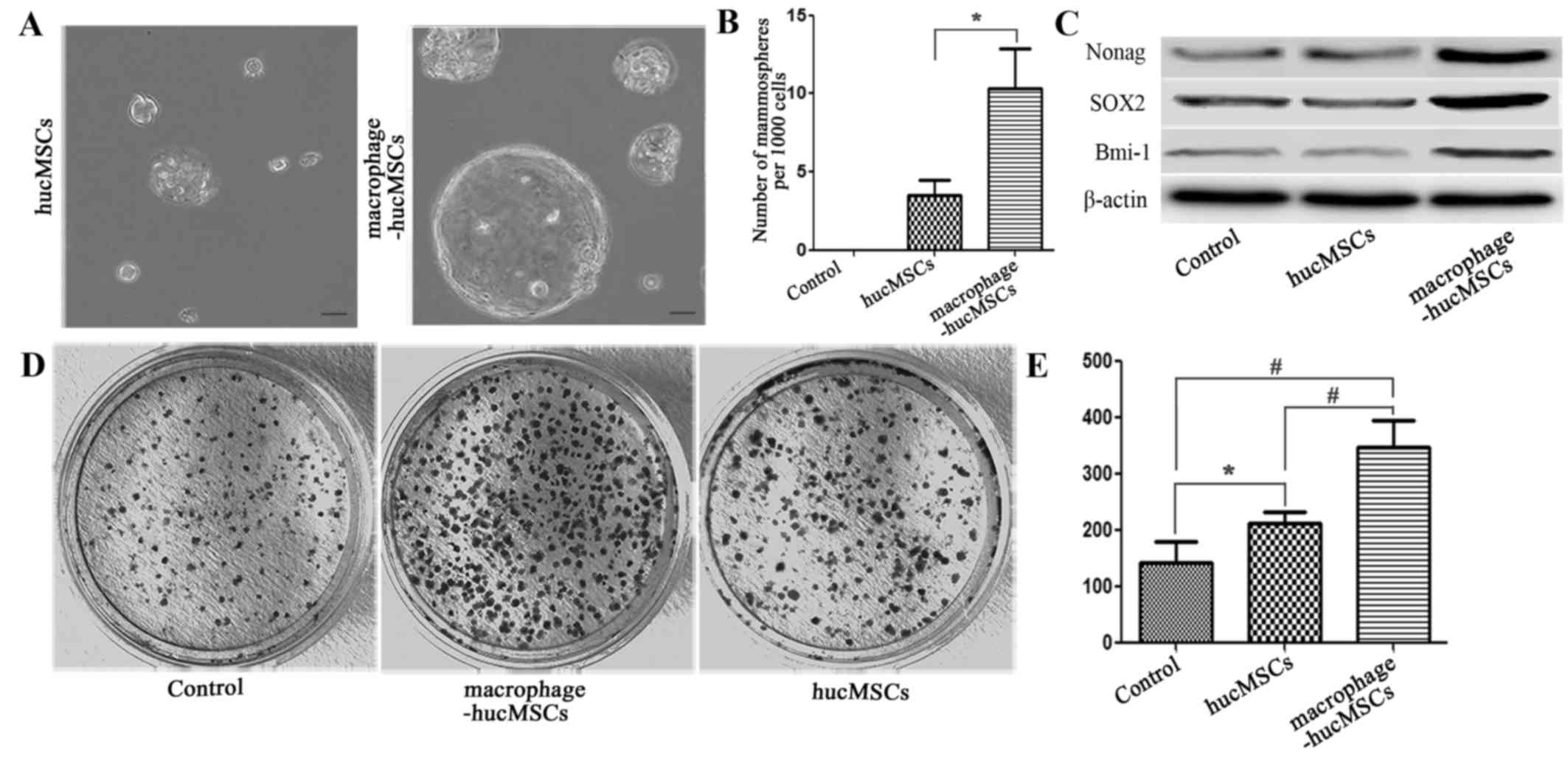 | Figure 6.Gastric epithelial cells incubated
with CM from macrophage-hucMSCs exhibit several properties of stem
cells. (A) GES-1 cells were incubated with CM from hucMSCs or
macrophage-hucMSCs for 48 h, and then seeded in non-adherent
culture conditions for spheroid formation. Depicted are
representative images of spheroid colonies after 15 days.
Magnification, ×400; scale bar, 50 µm. (B) Quantification of the
soft agar colonies shown in (A). (C) Western blot analysis of
Nanog, SOX2, and BMI-1 in GES-1 cells (the controls) and GES-1
cells co-cultured with CM from hucMSCs and macrophage-hucMSCs after
48 h. The expression of all analyzed proteins is significantly
enhanced in the GES-1 cells cultured with CM from
macrophage-hucMSCs, compared with the control. (D) Representative
image of single-colony formations of GES-1 cells incubated with CM
from hucMSCs or CM from macrophage-hucMSCs. (E) The number of
colonies is depicted as mean ± standard deviation. *P<0.05,
#P<0.01; GES-1 cells treated with medium only served
as the control (magnification, ×100). Macrophage-hucMSC, human
umbilical cord-derived mesenchymal stem cells pre-cultured with
macrophages for 48 h; CM, conditioned medium; SOX2, SRY-box 2;
BMI-1, polycomb complex protein BMI-1. |
Discussion
Macrophages and MSCs, as two major components of the
cancer-associated inflammatory stroma, participate in the
remodeling of the inflammatory microenvironment, which may serve
notable roles in gastric epithelial cell lesions or their malignant
transformation (13,29). Previous research demonstrated that
Helicobacter pylori infection induced the differentiation of
MSCs into CAFs-like cells, accompanied by the release of
inflammatory cytokines (30).
Similarly, the results of the present study demonstrated that a
incubation with macrophages for a short time induced the typical
CAFs differentiation and enhance expression of inflammatory
cytokines, including IL-8, PDGF-B, GM-CSF, IL-6, VEGF and MCP-1
(31,32), in the MSCs. Together, the results of
the present study indicated that macrophages incubation may also
induce MSCs to acquire CAF-like features or pro-inflammation
phenotypes via the secretion of multiple cytokines in
vitro.
Epithelial-mesenchymal transition (EMT) is a process
by which epithelial cells are converted into mesenchymal cells by
losing their polarity, reducing cell-cell adhesion and gaining
improved migratory ability. The phenotypic changes of EMT in
epithelial cells include inhibition of the expression of epithelial
markers, such as E-cadherin, and the upregulation of the expression
of mesenchymal markers, such as vimentin and N-cadherin (33). EMT also promotes cellular cytoskeletal
rearrangement in a range of cancer cell lines and facilitates the
formation of epithelial lesions (15,33). The
phenotype of gastric epithelial cells cultured with CM from
macrophage-MSCs were analyzed, the results revealed that CM from
macrophage-MSCs not only induced morphological shifts in gastric
epithelial cells from an epithelial to a fibroblastic phenotype,
but also decreased the expression of E-cadherin and increased that
of vimentin and N-cadherin. In addition to a loss of epithelial
characteristics, EMT coincided with increased migration in the
treated gastric epithelial cells. These results indicated that
incubation with macrophage-MSCs conditioned medium results in
gastric lesions via induction of EMT. Previous studies demonstrated
that a single inflammatory factor could also induce EMT in gastric
epithelial cells, but the concentration of this factor, such as
IL-6, had to be fairly high (50 ng/ml) (34), compared with its level in CM from
macrophages-MSCs in the present study, in order to induce EMT in
GES-1 cells. Co-culture with normal hucMSCs but no macrophages
could lead to secretion of almost 1 ng/ml IL-6 in the supernatant;
however, the degree of induction of EMT in gastric epithelial cells
was weaker than that induced by macrophage-hucMSCs. In addition to
the increases in the concentrations of the aforementioned
inflammatory factors, other components in the CM from
macrophage-hucMSCs that were not quantified, such as exosomes, may
also be major contributors to the induction of EMT-like changes in
the CM, which should be the subject of further study.
Previous studies have demonstrated that EMT endows
cells with stem cell properties, and prevents apoptosis and
senescence (15,35). In the current study, assessment of the
expression of epithelial and mesenchymal markers revealed that CM
from macrophage-hucMSCs is responsible for signifiers of an EMT
phenotype, characterized by an increase in the expression of
mesenchymal markers and that of Nanog, BMI-1 and SOX2, three
stemness-associated genes, which are factors required for the
maintenance of self-renewal and pluripotency of embryonic stem
cells (20–22). The upregulation of these
stemness-associated factors indicated the acquirement of stem
cell-like properties in the gastric epithelial cells following
exposure to CM from macrophage-hucMSCs. Self-renewal of CSCs, one
of their fundamental attributes, has been demonstrated in
colony-formation and sphere-forming assays (36). Colony-formation and sphere-forming
assays revealed that cells had the proliferative activity to form
clones, such that the colony-formation rate reflects the proportion
of cells with stemness. The present study demonstrated that CM from
macrophage-hucMSCs significantly increased the colony-formation and
sphere-forming rate of gastric epithelial cells, and therefore the
proportion of cells with stemness properties within the treated
gastric epithelial cells. By contrast, the CM from hucMSCs without
macrophage co-culture did not appear to affect spheroid formation
in gastric epithelial cells. We hypothesized that the reasons for
this result may be similar to that of EMT in gastric epithelial
cells treated with CM from hucMSCs mentioned above. Apoptosis is a
pathophysiological process that scavenges useless or harmful cells
in the body under normal conditions, which regulates body
development and homeostasis of the internal environment. The
inhibition of apoptosis is a key tumorigenic mechanism, as is
abnormal cellular proliferation (37). The results of the present study
demonstrated that macrophage-hucMSCs downregulated the expression
of the pro-apoptotic gene Bax and enhanced the expression of the
apoptosis inhibitor Bcl-2 in gastric epithelial cells. The
macrophage-hucMSC-induced changes to the percentage of apoptotic
cells in the treated gastric epithelial cell were examined using
flow cytometry. These results further confirmed that incubation
with macrophage-MSCs CM significantly reduced the percentage of
apoptotic gastric epithelial cells. The results of the current
study indicated the ability of macrophage-hucMSCs to endow stemness
transformation upon gastric epithelial cells and facilitate their
potential tumorigenicity.
In summary, the results of the present study
indicated that macrophage-activated MSCs differentiated into
CAF-phenotype cells, resulting in gastric lesions and endowing
gastric epithelial cells with potential oncogenic properties via
EMT-like changes. The synergistic effects of these two types of
stromal components contribute to the generation of an activated
pro-inflammation phenotype via the secretion of multiple cytokines,
ultimately fostering the development of the gastric
cancer-associated inflammation. Although these results further
verified the multiple effects of macrophage-MSCs on gastric
epithelial cells, the precise molecular mechanisms involved in
these processes remain to be identified and require further
investigation.
Acknowledgements
The authors would like to thank Dr. Chaoqun Lian and
Dr. Hongtao Wang (Clinical Laboratory and Diagnostic Center,
Department of Clinical Laboratory Science, Bengbu Medical College,
Bengbu, China) for collecting materials, performing statistical
analysis and providing many suggestions.
Funding
The present study was supported by grants from
Bengbu Medical College's Natural Science Foundation (grant no.
BYKY1429ZD), the Natural Science Fund of Education Department of
Anhui province (grant no. KJ2017A226), Anhui Provincial Natural
Science Foundation (grant no. SBK201342044) and the Natural Science
Fund of Education Department of Anhui province (grant no.
KJ2016A466).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QZ and SC contributed the central idea, designed and
performed experiments, and wrote the initial draft of the paper.
WW, CW and FZ supervised the experimental procedures and analyzed
the data. YL made substantial contributions to analysis and
interpretation of data and given final approval of the version to
be published. FW made substantial contributions to design and
optimization of the experimental scheme and methods, revised the
paper critically for the central idea and frame structure regarding
relevant information from experimental data analysis. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
All pregnant women who participated in this study
signed informed consent forms. This study was reviewed and approved
by the Ethics Review Committee of Bengbu Medical College (Bengbu,
China).
Patient consent for publication
Written informed consent was obtained from all
participants for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Qu Y, Dang S and Hou P: Gene methylation
in gastric cancer. Clin Chim Acta. 424:53–65. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jing JJ, Liu HY, Hao JK, Wang LN, Wang YP,
Sun LH and Yuan Y: Gastric cancer incidence and mortality in
Zhuanghe, China, between 2005 and 2010. World J Gastroenterol.
18:1262–1269. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Atsumi T, Singh R, Sabharwal L, Bando H,
Meng J, Arima Y, Yamada M, Harada M, Jiang JJ, Kamimura D, et al:
Inflammation amplifier, a new paradigm in cancer biology. Cancer
Res. 74:8–14. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rokavec M, Wu W and Luo JL: IL6-mediated
suppression of miR-200c directs constitutive activation of
inflammatory signaling circuit driving transformation and
tumorigenesis. Mol Cell. 45:777–789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mao Y, Keller ET, Garfield DH, Shen K and
Wang J: Stromal cells in tumor microenvironment and breast cancer.
Cancer Metastasis Rev. 32:303–315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fiaschi T, Marini A, Giannoni E, Taddei
ML, Gandellini P, De Donatis A, Lanciotti M, Serni S, Cirri P and
Chiarugi P: Reciprocal metabolic reprogramming through lactate
shuttle coordinately influences tumor-stroma interplay. Cancer Res.
72:5130–5140. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fiaschi T, Giannoni E, Taddei ML, Cirri P,
Marini A, Pintus G, Nativi C, Richichi B, Scozzafava A, Carta F, et
al: Carbonic anhydrase IX from cancer-associated fibroblasts drives
epithelial-mesenchymal transition in prostate carcinoma cells. Cell
Cycle. 12:1791–1801. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cao H, Xu W, Qian H, Zhu W, Yan Y, Zhou H,
Zhang X and Xu X, Li J, Chen Z and Xu X: Mesenchymal stem cell-like
cells derived from human gastric cancer tissues. Cancer Lett.
274:61–71. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Glaire MA, EI-Omar EM, Wang TC and
Worthley DL: The mesenchymal in malignancy: A partner in the
initiation, progression and dissemination of cancer. Pharmacol
Ther. 136:131–141. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brennen WN, Denmeade SR and Isaacs JT:
Mesenchymal stem cells as a vector for the inflammatory prostate
microenviroment. Ednocr Relat Cancer. 20:R269–290. 2013. View Article : Google Scholar
|
|
11
|
De Veirman K, Rao L, De Bruyne E, Menu E,
Van Valckenborgh E, Van Riet I, Frassanito MA, Di Marzo L, Vacca A
and Vanderkerken K: Cancer associated fibroblasts and tumor growth:
Focus on multiple myeloma. Cancers (Basel). 6:1363–1381. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Madar S, Goldstein I and Rotter V: Cancer
associated fibroblasts'-more than meets the eye. Trends Mol Med.
19:447–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ren G, Zhao X, Wang Y, Zhang X, Chen X, Xu
C, Yuan ZR, Roberts AI, Zhang L, Zheng B, et al: CCR2-dependent
recruitment of macrophages by tumor-educated mesenchymal stromal
cells promotes tumor development and is mimicked by TNFα. Cell Stem
Cell. 11:812–824. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lee JM, Dedhar S, Kalluri R and Thompson
EW: The epithelial-mesenchymal transition: New insights in
signaling, development, and disease. J Cell Biol. 172:973–981.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thiery JP, Acloque H, Huang PY and Nieto
MA: Epithelial- mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Acloque H, Adams MS, Fishwick K,
Bronner-Fraser M and Nieto MA: Epithelial-mesenchymal transitions:
The importance of changing cell state in development and disease. J
Clin Invest. 119:1438–1449. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu KJ and Yang MH: Epithelial-mesenchymal
transition and cancer stemness: The Twist1-Bmil connection. Biosci
Rep. 31:449–455. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bessède E, Staedel C, Amador Acuña LA,
Nguyen PH, Chambonnier L, Hatakeyama M, Belleannée G, Mégraud F and
Varon C: Helicobacter pylori generates cells with cancer stem cell
properties via epithelial-mesenchymal transition-like changes.
Oncogene. 33:4123–4131. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial- mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jeter CR, Badeaux M, Choy G, Chandra D,
Patrawala L, Liu C, Calhoun-Davis T, Zaehres H, Daley GQ and Tang
DG: Functional evidence that the self-renewal gene NANOG regulates
human tumor development. Stem Cells. 27:993–1005. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu Y, Futtner C, Rock JR, Xu X, Whitworth
W, Hogan BL and Onaitis MW: Evidence that SOX2 overexpression is
oncogenic in the lung. PLoS One. 5:e110222010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qiao B, Chen Z, Hu F, Tao Q and Lam AK:
BMI-1 activation is crucial in hTERT-induced epithelial-mesenchymal
transition of oral epithelial cells. Exp Mol Pathol. 95:57–61.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jia XH, Du Y, Mao D, Wang ZL, He ZQ, Qiu
JD, Ma XB, Shang WT, Ding D and Tian J: Zoledronic acid prevents
the tumor-promoting effects of mesenchymal stem cells via MCP-1
dependent recruitment of macrophages. Ocotarget. 6:26018–26028.
2015.
|
|
24
|
Kovach TK, Dighe AS, Lobo PI and Cui Q:
Interactions between MSCs and immune cells: Implications for bone
healing. J Immunol Res. 2015:7525102015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qiao C, Xu W, Zhu W, Hu J, Qian H, Yin Q,
Jiang R, Yan Y, Mao F, Yang H, et al: Human mesenchymal stem cells
isolated from the umbilical cord. Cell Biol Int. 32:8–15. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang T, Zhang X, Wang M, Zhang J, Huang F,
Cai J, Zhang Q, Mao F, Zhu W, Qian H and Xu W: Activation of
mesenchymal stem cells by macrophages prompts human gastric cancer
growth through NF-κB pathway. PLoS One. 9:e975692014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang F, Wang M, Yang T, Cai J, Zhang Q,
Sun Z, Wu X, Zhng X, Zhu W, Qian H and Xu W: Gastric cancer-derived
MSC-secreted PDGF-DD promotes gastric cancer progression. J Cancer
Res Clin Oncol. 140:1835–1848. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang M, Cai J, Huang F, Zhu M, Zhang Q,
Yang T, Zhang X, Qian H and Xu W: Pre-treatment of human umbilical
cord-derived mesenchymal stem cells with interleukin-6 abolishes
their growth-promoting effect on gastric cancer cells. Int J Mol
Med. 35:367–375. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Q, Wang M, Huang F, Yang T, Cai J,
Zhang X, Zhu W, Qian H and Xu W: H. pylori infection-induced MSC
differentiation into CAFs promotes epithelial-mesenchymal
transition in gastric epithelial cells. Int J Mol Med.
32:1465–1473. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Räsänen K and Vaheri A: Activation of
fibroblasts in cancer stroma. Exp Cell Res. 316:2713–2722. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Augsten M, Hägglöf C, Olsson E, Stolz C,
Tsagozis P, Levchenko T, Frederick MJ, Borg A, Micke P, Egevad L
and Ostman A: CXCL14 is an autocrine growth factor for fibroblasts
and acts as a multi-modal stimulator of prostate tumor growth. Proc
Natl Acad Sci USA. 106:3414–3419. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transitions: At the crossroads of
development and tumor metastasis. Dev Cell. 14:818–829. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang Q, Shi H, Huang F, Wang M, Qian H,
Zhu W and Xu WR: Effects of IL-6 on epithelial-mesenchymal
transition of HGC-27 cells in vitro. Chin J Clin Lab Sci.
31:617–620. 2013.(In Chinese).
|
|
35
|
Morel AP, Lièvre M, Thomas C, Hinkal G,
Ansieau S and Puisieux A: Generation of breast cancer stem cells
through epithelial-mesenchymal transition. PLoS One. 3:e28882008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Martins-Neves SR, Lopes ÁO, do Carmo A,
Paiva AA, Simões PC, Abrunhosa AJ and Gomes CM: Therapeutic
implications of an enriched cancer stem-like cell population in a
human osteosarcoma cell line. BMC Cancer. 12:1392012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Si PH and Hu QC: Progress of RNA interfere
in gene therapy for oral tumor. Int J Stomatol. 134:281–283.
2007.
|